Analysis of Tumor Specimens at the Time of Acquired Resistance to EGFR TKI therapy in 155 patients with EGFR mutant Lung Cancers (original) (raw)
. Author manuscript; available in PMC: 2014 Apr 15.
Abstract
Purpose
All patients with EGFR mutant lung cancers eventually develop acquired resistance to EGFR tyrosine kinase inhibitors (TKIs). Smaller series have identified various mechanisms of resistance, but systematic evaluation of a large number of patients to definitively establish the frequency of various mechanisms has not been performed.
Experimental Design
Patients with lung adenocarcinomas and acquired resistance to erlotinib or gefitinib enrolled onto a prospective biopsy protocol and underwent a re-biopsy after the development of acquired resistance. Histology was reviewed. Samples underwent genotyping for mutations in EGFR, AKT1, BRAF, ERBB2, KRAS, MEK1, NRAS and PIK3CA, and FISH for MET and HER2.
Results
Adequate tumor samples for molecular analysis were obtained in 155 patients. Ninety-eight had second-site EGFR T790M mutations (63%, 95% CI 55-70%) and four had small cell transformation (3%, 95% CI 0-6%). MET amplification was seen in 4/75 (5%, 95% CI 1-13%). HER2 amplification was seen in 3/24 (13%, 95% CI 3-32%). We did not detect any acquired mutations in PIK3CA, AKT1, BRAF, ERBB2, KRAS, MEK1, or NRAS. (0/88, 0%, 95% CI 0-4%). Overlap among mechanisms of acquired resistance was seen in 4%.
Conclusions
This is the largest series reporting mechanisms of acquired resistance to EGFR TKI therapy. We identified EGFR T790M as the most common mechanism of acquired resistance, while MET amplification, HER2 amplification, and small cell histologic transformation occur less frequently. More comprehensive methods to characterize molecular alterations in this setting are needed to improve our understanding of acquired resistance to EGFR TKIs.
Keywords: EGFR mutant lung cancer, lung adenocarcinoma, targeted therapy, acquired resistance, tyrosine kinase inhibitor therapy
Introduction
In patients with lung cancer whose tumors harbor activating EGFR mutations, treatment with EGFR TKIs induces initial tumor shrinkage, with progression of cancer after a median of 8-16months1-4. Various mechanisms of resistance to erlotinib and gefitinib have been identified, and understanding these mechanisms is critical to developing treatment strategies in the acquired resistance setting. The most frequently reported mechanism of acquired resistance is the EGFR T790M point mutation within exon 205,6. Small cell histologic transformation has also been associated with the development of acquired resistance7,8. MET amplification and HER2 amplification are also seen and illustrate the upregulation of parallel signaling pathways9-11. Rare secondary BRAF mutations12 have also been implicated. The frequency and overlap of these mechanisms of resistance is not well-characterized, as all reports have been in relatively small series of patients8.
Rebiopsy of growing tumors at clinical progression has become increasingly important as the results may better predict prognosis13,14 or direct a change in therapy7. Understanding why acquired resistance occurs is essential as new therapies focus on alternative means of EGFR inhibition and inhibition of parallel signaling pathways to prevent or circumvent resistance. To characterize the frequency of the various mechanisms of acquired resistance in a single population, we report the updated results of a prospective clinical trial initiated in 2004 to determine mechanisms of acquired resistance in patients with _EGFR_-mutant lung cancers who had an initial response to erlotinib or gefitinib13.
Methods
Patients
Patients had lung adenocarcinoma with a documented EGFR mutation, received treatment with single agent erlotinib or gefitinib, had either prolonged stable disease (>3 months) or a partial response to therapy, and developed radiographic progression while on EGFR TKI. The primary objective of the study was to characterize mechanisms of acquired resistance to EGFR TKI therapy. Biopsies were obtained in the least invasive manner possible, and typically consisted of either a fine needle aspiration or core biopsy done with image guidance, or rarely excisional biopsies. Fluid from malignant effusions was collected to create cell blocks. If a surgical procedure was clinically indicated, tissue from the procedure was obtained for analysis. Protocol #04-103 was approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center, and all patients signed informed consent. Partial data from a subset of patients on this protocol have been reported previously11,13,15.
Tissue Analysis
All samples underwent histologic review. If required, additional diagnostic immunohistochemical stains were performed at the discretion of the pathologist. For molecular analysis, genomic DNA was extracted from the tumor samples which included fresh, frozen, and formalin fixed paraffin embedded (FFPE) tissue specimens. Cytologic samples were used to create cell blocks from which DNA was extracted. The initially identified sensitizing EGFR mutation was confirmed using previously described methods16-18.
Standard sequencing and/or fragment analysis were used to identify EGFR T790M using techniques previously described15,18. In July 2009, we began locked nucleic acid based PCR sequencing to improve the sensitivity of EGFR T790M detection19. Beginning in January 2009, a mass spectrometry-based mutation profiling assay (Sequenom), was used on all samples. This assay identifies 92 specific point mutations in 8 genes: EGFR, BRAF, PIK3CA, AKT1, ERBB2, MEK1, NRAS and KRAS. If additional material was available on samples prior to 2009, locked nucleic acid -based sequencing and Sequenom analysis were performed. For a subset of patients with available tissue, direct sequencing of EGFR exons 18-21 was also available. Available unstained FFPE tumor tissue was analyzed by a dual-color FISH assay using a MET/CEP7 probe cocktail15,20. A MET/CEP ratio was established based on a count of at least 200 cells. Samples with a MET:CEP7 ratio greater than 2 were considered to have MET amplification (low amplification≤3, high amplification>3). Assessment of HER2 gene copy number was also performed on available unstained FFPE tumor tissue using the Vysis PathVysion HER2 DNA Probe Kit (Abbott Laboratories, Abbott Park, IL) and scored according to previously published criteria21,22. Tumors were classified as amplified if the HER2/CEP17 ratio per cell was >2 or homogeneously staining regions with >15 copies in >10% of cells were present. At least 40 cells were analyzed for each case.
Statistical Methods
Medical records were reviewed to obtain clinical information. Overall survival from the time of advanced cancer diagnosis and post-progression survival following the development of acquired resistance were calculated using Kaplan-Meier methodology, with patients censored if they were alive at the time of last follow-up, February 2012. Univariate comparisons between groups were performed using the log-rank test.
To evaluate whether patients enrolled on the protocol were representative of patients with advanced NSCLCs, we assembled a reference cohort of all patients diagnosed with advanced NSCLC at our institution during a similar time period (2004-2009), whose tumors harbored EGFR mutations, and were not enrolled in this acquired resistance protocol. The two groups were compared with respect to clinical characteristics using the chi-square test.
Results
Enrollment and rebiopsy
From August, 2004 to January, 2012, 175 patients were enrolled and 162 underwent rebiopsy at the time of acquired resistance (see Table 1). Seven patients had biopsy samples with either insufficient tumor content for molecular analysis (n=5) or no evidence of the previously present EGFR sensitizing mutation upon rebiopsy (n=2). One lung core biopsy had insufficient tumor cells. One patient’s core bone biopsy was inadequate for DNA analyses due to decalcification. Two patients had core biopsies of the lung with tumor evident where the sensitizing mutation could not be identified. Two samples, a brain resection and a pneumonectomy specimen, had no viable tumor cells due to tumor necrosis. Finally, one patient had a core biopsy of the adrenal gland that failed to sample the tumor, whereas the corresponding FNA revealed small cell transformation, but there was insufficient material for molecular testing. A total of 155 patients had biopsy samples that were sufficient for molecular analysis, including fine needle aspirations, core biopsies, surgical samples and cytology from malignant effusions. There was only one serious adverse event, a pneumothorax requiring pigtail catheter placement.
Table 1.
Procedures performed
| Procedures Performed | N=162 |
|---|---|
| Adrenalectomy | 3 |
| Brain resection | 10 |
| Lung resection | 4 |
| Lymph node excision | 5 |
| Autopsy | 2 |
| Biopsies | |
| Lung/Chest Wall (Core/FNA) | 82 (64/18) |
| Bone (Core/FNA) | 9 (7/2) |
| Liver (Core/FNA) | 13 (10/3) |
| Lymph node (Core/FNA) * | 9 (2/7) |
| Other (Core/FNA) ** | 9 (4/5) |
| Fluid | |
| Pleural | 14 |
| Peritoneal | 1 |
| CSF | 1 |
Baseline clinical and molecular characteristics
Pre-treatment clinical and molecular characteristics of the 155 patients are described in Table 2. Compared with a reference group of patients with EGFR mutant lung cancers diagnosed in the same time period (data not shown), patients enrolled on the protocol were significantly younger, were more likely to be never smokers, had significantly fewer pack years, and were more likely to have EGFR exon 19 deletions.
Table 2.
Clinical and Molecular Characteristics
| Patient Characteristics | |
|---|---|
| N=155 | |
| Age | |
| Median | 57 |
| Range | 33-81 |
| Sex-n (%) | |
| Female | 100 (66) |
| Male | 55 (35) |
| Race-n (%) | |
| White | 113 (73) |
| Asian | 30 (19) |
| Black | 11 (7) |
| Other | 1 (1) |
| Smoking history-n (%) | |
| Never smoker | 107 (69) |
| Former smoker | 48 (31) |
| Current smoker | 0 |
| Median pack yr | 10 |
| Range | 0-96 |
| Pretreatment EGFR mutation- n (%) | |
| Exon 19 deletion | 103 (66) |
| Exon 21 L858R | 46 (30) |
| Exon 21 L861Q | 1 |
| Other/double mutations | |
| Exon 19 deletion + T790M | 1 |
| Exon 19 deletion + PIK3CA | 1 |
| Exon 21 L861Q and Exon 20 R776H | 1 |
| Exon 21 L833V and Exon 21 H835L | 1 |
| Exon 21 L858R and Exon 20 R776H | 1 |
Pre-biopsy course
The 155 patients began EGFR TKI therapy from February 1999 to January 2011. One hundred ten patients (71%) were started on EGFR TKI as first line therapy. Twenty four patients (15%) received EGFR TKI as second line therapy, and 9 patients (6%) as third or fourth line therapy. Twelve patients (8%) received EGFR TKI as adjuvant or maintenance therapy after surgery and/or chemotherapy. The majority of patients (78%, 121/155) received single agent EGFR TKI therapy, while 22% (34/155) were initially treated concurrently with EGFR TKI and cytotoxic chemotherapy. The patients on this protocol had documented clinical progression on EGFR TKI between March, 2004 and December, 2011. The median time from start of EGFR TKI to clinical progression was 13 months with a range of 2 to 73 months. The time to progression on EGFR TKI was similar for patients with EGFR exon 19 deletions versus EGFR L858R mutations, 15 and 17 months respectively (p=0.99). One patient with EGFR exon 19 deletion and EGFR T790M at baseline had a time to progression on EGFR TKI of 2 months. Another patient with EGFR exon 19 deletion and a PIK3CA mutation had a time to progression of 5 months. No other patients had either EGFR T790M or PIK3CA mutations in their pre-TKI specimens. There was no difference in time to progression on EGFR TKI for patients with secondary EGFR T790M mutations and patients with other mechanisms of resistance, 16 and 17 months respectively (p=0.37). Ninety one percent (141/155) of patients continued their EGFR TKI after evidence of clinical progression.
Findings at the time of acquired resistance
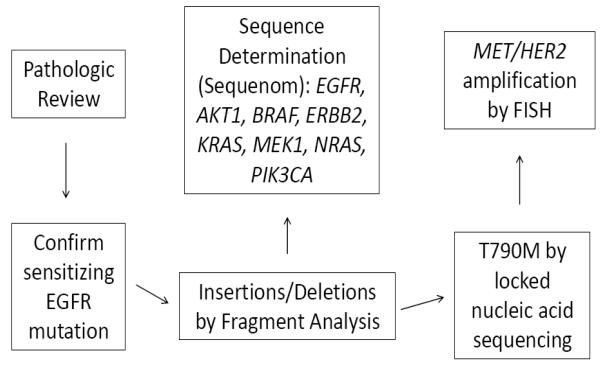
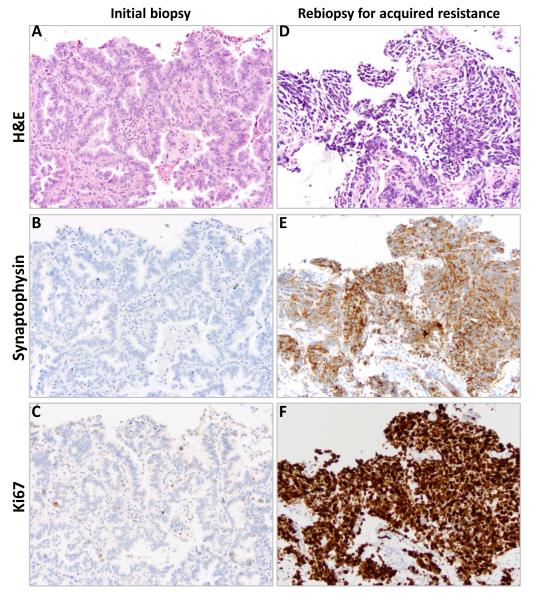
All 155 samples were tested for known mechanisms of acquired resistance. Not all analyses were conducted on all specimens due to the limited amount of tissue available. Figure 1 illustrates the prioritization of analyses conducted. All specimens underwent pathologic review by a thoracic pathologist. Four of 155 had evidence of small cell histologic transformation (3%, 95% CI 0 to 6%). Samples with small cell histology had further immunostains performed including synaptophysin, chromogranin, and CD56 to confirm the diagnosis (Figure 2). There was no evidence of small cell lung cancer in any of the pretreatment biopsy specimens. Morphologic changes consistent with epithelial-to-mesenchymal transition were not seen, although no immunohistochemical stains for vimentin or E-cadherin were performed.
Figure 1.
Assay prioritization.
Figure 2.
A patient’s tumor tissue at initial lung biopsy (A-C) and biopsy at acquired resistance (D-F) with small cell histologic transformation. Hematoxylin and eosin stained material demonstrating histologic changes from initial biopsy (A) to biopsy at acquired resistance (D) with immunohistochemical staining for CD56 and Ki67 on initial biopsy (B, C) and biopsy at acquired resistance (E, F).
We identified a second-site EGFR T790M mutation in 98 of 155 samples (62%, 95% CI 55-70%). One patient had an acquired T854A mutation [observed incidence <1% (5% CI <1 to 4%]. In 88 patients, no acquired mutations were identified in PIK3CA, AKT1, BRAF, ERBB2, MEK1, NRAS and KRAS (0%, 95% CI 0 to 4%).
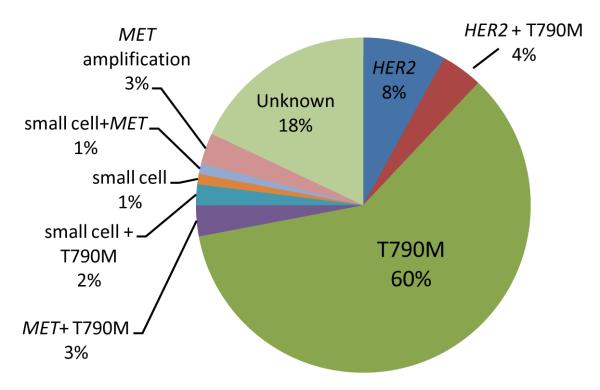
Sufficient tissue was available for MET FISH analysis for 75 patients. Four of 75 patients tested had MET amplification, using the standard MET/CEP7 ratio of greater than 2 (5%, 95% CI 1 to 13%). 24 patients had sufficient tissue available for HER2 FISH analysis. 2 of 24 patients had evidence of HER2 amplification using the standard HER2/CEP17 ratio of greater than 2 (13%, 95% CI 3-32%). Baseline samples for the 4 _MET_-amplified and 2 _HER_-amplified cases were not available for FISH analysis. The frequency of the different mechanisms of resistance was similar when comparing patients with baseline EGFR L858R mutations versus EGFR exon 19 deletions. The relative percentages of the observations are illustrated in Figure 3.
Figure 3.
The relative frequencies of the various mechanisms of acquired resistance. Composite pie chart with percentages compiled from tests with varying denominators.
Overlap of Mechanisms of Resistance
4% of samples had more than one mechanism of resistance identified. Tumors from two patients had both small cell histologic transformation and EGFR T790M. One patient had small cell histologic transformation and MET amplification. In these patients showing small cell histologic transformation, there was no residual adenocarcinoma component in the biopsies studied, indicating that the aforementioned acquired genetic alterations were present in the small cell carcinoma. Two patients had EGFR T790M and MET amplification by FISH; both molecular findings were identified in the same tumor samples. One patient had two acquired resistance samples; one sample was EGFR T790M positive and the other had evidence of HER2 amplification. The autopsy specimen was EGFR T790M positive, but negative for HER2 amplification, and the adrenalectomy sample had evidence of HER2 amplification, but was EGFR T790M negative. The time to progression on EGFR TKI for patients with two mechanisms of resistance ranged from 7 to 24 months.
Outcomes
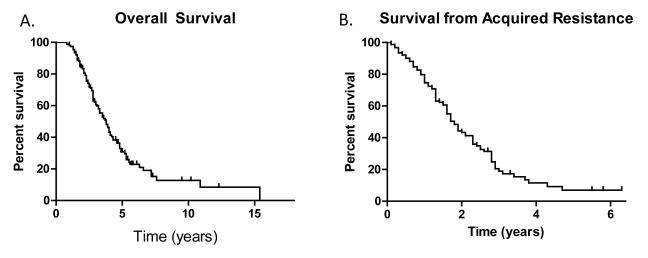
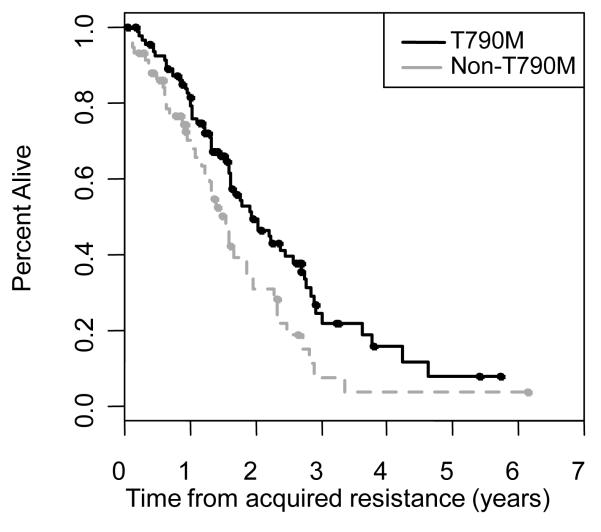
Sixty-four percent (99/155) of patients have died as of February 2012. The median overall survival from diagnosis of stage IV disease was 3.8 years (95% CI 3.1-5.1 years) (Figure 4). The median post-progression survival was 1.7 years (95% CI 1.6-2.0 years) (Figure 4). In patients with EGFR T790M, the median post-progression survival was 1.9 years (95% CI 1.6-2.6 years) and was 1.6 years (95% CI 1.2-1.8 years) for those without EGFR T790M (p=0.015) (Figure 5).
Figure 4.
A) Overall survival (from the development of Stage IV disease), and B) post progression survival (from the time of acquired resistance)
Figure 5.
Post progression survival for EGFR T790M+ patients and non-T790M+ patients
Discussion
To provide a more precise understanding of mechanisms of acquired resistance to EGFR TKIs and their overlap in a single cohort, we report this prospective study of 155 patients. While the most commonly observed mechanism of resistance is EGFR T790M, our data demonstrate that tumors biopsied at the time of clinical acquired resistance to EGFR TKIs display a spectrum of mechanisms, with rare overlap among mechanisms.
EGFR T790M is the most common mechanism of acquired resistance, seen in nearly two-thirds of cases. MET amplification is uncommon and is less frequent than initially proposed10. Acquired mutations in EGFR other than T790M, including T854A, D761Y and L747S, are infrequent23-25. Except for the one patient with a baseline PIK3CA mutation, no additional patients in this large series had a mutation in PIK3CA, AKT1, BRAF, HER2, MEK1, NRAS and KRAS, which is consistent with another series that found no mutations in MEK1, NRAS or KRAS, but did identify a secondary BRAF mutation in 2 of 195 acquired resistance samples12. Point mutations in these genes including PIK3CA appear to be infrequent, having been occasionally reported in other series8,12 . HER2 amplification may be a more common finding at the time of resistance, seen in 13% of cases in our series. Pretreatment samples were not available for HER2 FISH testing to confirm that amplification was not present prior to treatment. However, HER2 testing performed on 99 untreated lung adenocarcinomas has been reported, with only 1 tumor (1%) displaying HER2 amplification pretreatment11.
Small cell histologic transformation was also less frequent than previously reported8. Interestingly, 3 of 4 cases of small cell histologic transformation occurred in addition to another mechanism of resistance. It is unclear if small cell transformation is merely associated with the development of resistance or is itself a causal mechanism. Small cell histologic transformation may be a phenomenon unique to TKI therapy as it has not been reported in patients who are treated and progress on cytotoxic chemotherapy, but re-biopsy data on such patients are limited. The patients with small cell histologic transformation all had a relatively aggressive clinical course as typically seen with small cell lung cancer. Two of the four patients received platinum based doublet therapy and had partial responses after completion of six cycles of therapy. One patient received adjuvant cisplatin and etoposide after metastasectomy and had a 13 month disease free interval prior to developing recurrent disease. One patient had a poor performance status and was treated with single agent etoposide without response. Although the role of small cell histologic transformation in acquired resistance is presently unclear, histologic transformation is important to identify because it significantly alters our treatment recommendations.
MET amplification was seen concurrently with another mechanism of resistance in 3 of 4 cases. Two patients had both EGFR T790M and MET amplification and one patient had MET amplification and evidence of small cell transformation. This raises the question as to whether MET amplification alone is sufficient to induce resistance to EGFR TKI therapy. Previous reports suggest a reciprocal relationship between EGFR T790M and MET amplification, with overlap of the two mechanisms becoming less frequent with increasing MET amplification (≥4-fold)26. This reciprocal relationship was not seen in our cohort, but is limited by the small number of patients with MET amplification. In two samples with EGFR T790 and MET, one had high level MET amplification (>10-fold) and one had moderate amplification (4-fold), and the sample with MET amplification only was determined by qPCR, and not quantified by FISH. Finally, one patient had different mechanisms of resistance, EGFR T790M and HER2 amplification, identified in two distinct resistance samples with the same EGFR sensitizing mutation. This suggests that either genetic alteration may be sufficient to impart clinical resistance, but different mechanisms of resistance may exist in distinct clones of a patient’s cancer. Intratumor heterogeneity is a significant limitation of basing treatment decisions on molecular analysis of a single tumor biopsy in the metastatic setting27.
While many aspects of our findings are similar to other series, there are several differences. The median time to progression on EGFR TKI of 13 months in our cohort is similar to previously published EGFR mutant cohorts.1-3 Despite this and other similarities to a similar cohort of patients with advanced EGFR mutant lung cancers, these patients were somewhat younger with less tobacco exposure and were more likely to have EGFR exon 19 deletion. These factors have been associated with improved survival in previous studies28-30 which may explain the patients’ ability to enroll in a prospective trial at the time of development of acquired resistance.
The majority of patients (91%) continued EGFR TKI therapy after clinical progression, sometimes in combination with chemotherapy. This treatment approach is consistent with preclinical data that continued EGFR inhibition is indicated due to the presence of a heterogenous population of tumor cells, with varied sensitivity to EGFR inhibition31 as well as clinical data demonstrating rapid clinical progression (flare) after discontinuation of EGFR TKI32,33. Recent retrospective analyses support the clinical observation of improved outcomes with continued EGFR TKI therapy after radiologic progression34-36. Prospective evaluation of continued EGFR TKI after clinical progression will be important and will inform future clinical practice.
Since this protocol opened, our understanding of the mechanisms of resistance has grown significantly, leading to some limitations in our analysis. Sufficient tissue from re-biopsies was not obtained for all the molecular tests currently performed. Histologic analysis and testing for EGFR T790M was performed on every sample, but FISH for MET and HER2 and sequenom for other acquired oncogene mutations were performed on only a portion of the samples. This limits the accuracy of the relative frequency of the various mechanisms we present, and potentially underestimates the prevalence of overlap among the different mechanisms of resistance. Our analysis did not encompass alterations in autocrine or paracrine growth pathways that may also confer resistance37,38. Epithelial to mesenchymal transition has been described as a potential mechanism of resistance8, although we did not note any evidence of spindle-like mesenchymal morphology on histologic review of our samples. However, we did not perform immunohistochemistry for e-cadherin or vimentin which limits the conclusions that can be made. We confirmed the presence of the EGFR sensitizing mutation in the pre-treatment tumor sample of each patient included in this analysis, but we were unable to assess for HER2 or MET amplification, or perform comprehensive genotyping on the majority of pre-treatment specimens. Consequently, we cannot say with certainty that the genetic changes identified were acquired after treatment with an EGFR TKI.
This prospective study confirms that standardized, step-wise molecular testing at the time of acquired resistance is feasible and safe. We plan to perform comprehensive next generation based sequencing based mutation profiling as well as protein and gene expression analysis on specimens where no mechanism of acquired resistance was identified. Other proposed mechanisms of acquired resistance to EGFR TKI therapy including upregulated AXL and HGF expression and MAPK1 amplification will need to be tested and validated in our larger data set37-40. While the intent of these studies is to discover additional mechanisms of resistance to EGFR inhibition, the ultimate purpose is to better inform treatment decisions in the acquired resistance setting. Our growing knowledge of oncogenes and the molecular basis of resistance to kinase inhibition will allow for personalized treatment strategies in both the initial and resistance settings.
Statement of translational relevance.
Responses to EGFR tyrosine kinase inhibitors (EGFR TKIs) in EGFR mutant lung cancer are limited by acquired resistance. The goal of this prospective study was to characterize the mechanisms of resistance by performing repeat tumor biopsies at clinical disease progression. Previously published reports included small numbers of patients, and do not adequately establish the frequency of the various mechanisms of resistance. This is the largest reported cohort of patients with EGFR mutant lung cancer and acquired resistance to EGFR TKI therapies that have had comprehensive mutational analysis on acquired resistance biopsy samples. Our findings indicate that second site EGFR T790M mutations are the dominant mechanism of resistance identified. MET amplification, small cell histologic transformation, and HER2 amplification are uncommon. There were no acquired mutations in other oncogenes identified. This protocol illustrates that post-progression tumor biopsies can be done on a large scale with minimal adverse events. Comprehensive tumor analysis at the time of resistance is important to develop new therapeutic strategies and also to inform patient care in the acquired resistance setting.
Acknowledgments
Role of Funding Source: This study was supported by NIH funding including R01CA121210 (William Pao), R21CA115051 (Vincent Miller) and P01CA129243 (Mark Kris). The sponsor played no role in data collection, analysis or interpretation.
Footnotes
Conflicts of Interest: Mark Kris has consulted for Pfizer, Boehringer Ingelheim, Genentech, and has received grants from Pfizer and Boehringer Ingelheim. Vincent Miller is currently employed by and owns stock in Foundation Medicine. Gregory Riely has consulted for AstraZeneca, Boehringer-Ingelheim, Chugai, Ariad, Tragara, Daiichi, Novartis, Abbott Molecular and Celgene, and has received grants from Infinity Pharmaceuticals, Bristol-Myers Squibb, Novartis, Chugai, Pfizer, Merck and GlaxoSmithKline. William Pao has consulted for MolecularMD, AstraZeneca, Bristol-Myers Squibb, Symphony Evolution and Clovis Oncology. He has received research funding from Enzon, Xcovery, AstraZeneca, and Symphogen. Rights to EGFR T790M testing were licensed on behalf of William Pao, Vincent Miller and others by MSKCC to MolecularMD. Helena Yu, Maria Arcila, Natasha Rekhtman, Camelia Sima, Maureen Zakowski, Marc Ladanyi have no conflicts of interest.
References
- 1.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The lancet oncology. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 3.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 4.Janne PA, Wang X, Socinski MA, Crawford J, Stinchcombe TE, Gu L, et al. Randomized Phase II Trial of Erlotinib Alone or With Carboplatin and Paclitaxel in Patients Who Were Never or Light Former Smokers With Advanced Lung Adenocarcinoma: CALGB 30406 Trial. J Clin Oncol. 2012;30(17):2063–9. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–5. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 8.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 11.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR mutant lung cancers that lack the second-site EGFR T790M mutation. Cancer discovery. 2012;2(10):922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin Y-L, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci. 2012;109(31):E2127–33. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxnard GR, Arcila ME, Sima C, Riely GJ, Chmielecki J, Kris MG, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clinical Cancer Research. 2011;17(6):1616–22. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hata AKN, Katakami N, Yoshioka H, Takeshita J, Tanaka K, Nanjo S, et al. Rebiopsy of non-small cell lung cancer patients with acquired resistance to EGFR-TKI: Comparison between T790M mutation-positive and -negative populations. J Clin Oncol. 2012:30. doi: 10.1002/cncr.28364. (suppl; abstr 7528) [DOI] [PubMed] [Google Scholar]
- 15.Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, Zakowski MF, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clinical Cancer Research. 2011;17:1169–80. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:225–35. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan Q, Pao W, Ladanyi M. Rapid Polymerase Chain Reaction-Based Detection of Epidermal Growth Factor Receptor Gene Mutations in Lung Adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arcila ME, Nafa K, Oxnard GR, Miller VA, Ladanyi M. Improved Detection of the EGFR T790M Mutation in Lung Cancer Patients with Acquired Resistance to EGFR TKIs Using a Locked Nucleic Acid-based Approach. J Mol Diagn. 2009;11 Abstracts 655. [Google Scholar]
- 20.Bean J, Brennan C, Shih JH, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cappuzzo F, Varella-Garcia M, Shigematsu H, Domenichini I, Bartolini S, Ceresoli GL, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor-positive non-small-cell lung cancer patients. Journal of clinical oncology. 2005;23:5007–18. doi: 10.1200/JCO.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. Journal of clinical oncology. 2005;23:6838–45. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 23.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clinical cancer research. 2006;12:6494–501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 24.Bean J, Riely GJ, Balak M, Marks JL, Ladanyi M, Miller VA, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clinical cancer research. 2008;14:7519–25. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS medicine. 2007;4:1669–79. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suda K, Murakami I, Katayama T, Tomizawa K, Osada H, Sekido Y, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clinical cancer research. 2010;16:5489–98. doi: 10.1158/1078-0432.CCR-10-1371. [DOI] [PubMed] [Google Scholar]
- 27.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clinical cancer research. 2006;12:839–44. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 29.Harichand-Herdt S, Ramalingam SS. Gender-associated differences in lung cancer: clinical characteristics and treatment outcomes in women. Seminars in oncology. 2009;36:572–80. doi: 10.1053/j.seminoncol.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Radzikowska E, Glaz P, Roszkowski K. Lung cancer in women: age, smoking, histology, performance status, stage, initial treatment and survival. Population-based study of 20 561 cases. Annals of oncology. 2002;13:1087–93. doi: 10.1093/annonc/mdf187. [DOI] [PubMed] [Google Scholar]
- 31.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Science translational medicine. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaft JE, Oxnard GR, Sima CS, Kris MG, Miller VA, Riely GJ. Disease flare after tyrosine kinase inhibitor discontinuation in patients with EGFR-mutant lung cancer and acquired resistance to erlotinib or gefitinib: implications for clinical trial design. Clinical cancer research. 2011;17:6298–303. doi: 10.1158/1078-0432.CCR-11-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riely GJ, Kris MG, Zhao B, Akhurst T, Milton DT, Moore E, et al. Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clinical cancer research. 2007;13:5150–5. doi: 10.1158/1078-0432.CCR-07-0560. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg SB, Oxnard GR, Digumarthy S, Muzikansky A, Jackman DM, Lennes IT, et al. Chemotherapy with erlotinib or chemotherapy alone in advanced NSCLC with acquired resistance to EGFR tyrosine kinase inhibitors (TKI) J Clin Oncol. 2012;30 doi: 10.1634/theoncologist.2013-0168. (suppl; abstr 7524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oxnard GR, Lo P, Jackman DM, Butaney M, Heon S, Johnson BE, et al. Delay of chemotherapy through use of post-progression erlotinib in patients with EGFR-mutant lung cancer. J Clin Oncol. 2012;30 (suppl; abstr 7547) [Google Scholar]
- 36.Faehling M, Eckert R, Kamp TG, Kuom S, Spengler W. Treatment with EGFR tyrosine kinase inhibitors beyond progression in long-term responders to erlotinib in advanced non-small cell lung cancer: A case-control study of overall survival. J Clin Oncol. 2012;30 doi: 10.1016/j.lungcan.2013.02.010. (suppl; abstr 7572) [DOI] [PubMed] [Google Scholar]
- 37.Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer research. 2008;68:9479–87. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 38.Yano S, Yamada T, Takeuchi S, Tachibana K, Minami Y, Yatabe Y, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. Journal of thoracic oncology. 2011;6:2011–7. doi: 10.1097/JTO.0b013e31823ab0dd. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramoise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nature genetics. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, et al. Reactivation of ERK Signaling Causes Resistance to EGFR Kinase Inhibitors. Cancer discovery. 2012;2(10):934–947. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]