PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells (original) (raw)
Abstract
We have previously shown that α-thalassemia mental retardation X-linked (ATRX) and histone H3.3 are key regulators of telomeric chromatin in mouse embryonic stem cells. The function of ATRX and H3.3 in the maintenance of telomere chromatin integrity is further demonstrated by recent studies that show the strong association of ATRX/H3.3 mutations with alternative lengthening of telomeres in telomerase-negative human cancer cells. Here, we demonstrate that ATRX and H3.3 co-localize with the telomeric DNA and associated proteins within the promyelocytic leukemia (PML) bodies in mouse ES cells. The assembly of these telomere-associated PML bodies is most prominent at S phase. RNA interference (RNAi)-mediated knockdown of PML expression induces the disassembly of these nuclear bodies and a telomere dysfunction phenotype in mouse ES cells. Loss of function of PML bodies in mouse ES cells also disrupts binding of ATRX/H3.3 and proper establishment of histone methylation pattern at the telomere. Our study demonstrates that PML bodies act as epigenetic regulators by serving as platforms for the assembly of the telomeric chromatin to ensure a faithful inheritance of epigenetic information at the telomere.
INTRODUCTION
Telomeres are specialized structures that protect the chromosome ends. Telomere maintenance is essential for the unlimited proliferative potential of human cells. Many human tumor cells have acquired an indefinite replicative capacity by maintaining their telomeres through increased telomerase expression and activity. However, in 10–20% of tumors, a recombination-mediated Alternative Lengthening of Telomeres (ALT) mechanism is used (1–5). Hallmarks of ALT cancer cells include the highly heterogeneous length of their telomeres, ranging from very short to longer than 100 kb (3,6) and the presence of extra-chromosomal telomeric DNA (7,8).
Mammalian telomeres contain DNA and histone modifications characteristic of repressive chromatin. Knockout studies in mice have demonstrated the importance of DNA hypermethylation, H3K9me3 (H3 Lys9 trimethylation) and H4K20me3 (H4 Lys20 trimethylation) for negative regulation of telomere length and repression of telomeric recombination (9–11). However, unlike the somatic cell counterparts, pluripotent mouse embryonic stem (ES) cells comprise much lower H3K9me3 and H4K20me3 levels at telomeres (12,13). On induction of differentiation, these repressive chromatin marks are rapidly increased at telomeres, suggesting that the telomeric chromatin state, in this case, histone modification pattern in ES cells, is less ‘heterochromatic’ in nature and undergoes a dynamic configuration change during differentiation (12,13). It is interesting that a low density of H3K9me3 and H4K20me3 is also found at telomeres in induced pluripotent stem cells, highlighting the importance of the maintenance of a unique telomeric chromatin state in pluripotent cell types (14). This change in telomeric chromatin state during cellular differentiation also occurs in vivo in mice as demonstrated by the transition of transcriptionally active subtelomeric regions in ES cells to repressive chromatin in somatic tissues (15).
The telomere in mouse ES cells is enriched with ATRX (α-thalassemia mental retardation X-linked) and histone variant H3.3 (12,13). ATRX is a member of SWI2/SNF2 family of helicase/ATPases (16,17) that plays a role in controlling DNA methylation at ribosomal repeats, subtelomeres and chromosome Y satellite repeats (18). H3.3 is a H3 variant commonly, but not exclusively, associated with active chromatin (19,20). In mouse ES cells, the depletion of either ATRX or H3.3 induces a telomeric dysfunction phenotype (12,13), providing evidence for their function in maintaining telomeric chromatin integrity. Recent studies have shown that ATRX and its interacting partner Death domain associated protein (DAXX) (Death associated domain 6) act as chaperones that deposit H3.3 at the pericentric and telomeric DNA (21–23). The loss of ATRX/DAXX function results in an increase in pericentric satellite and telomeric transcription, providing evidence for the function of ATRX/DAXX/H3.3 as chromatin regulators at the repressive heterochromatin. Importantly, recent studies have also reported a common mutation in ATRX/DAXX/H3.3 in human ALT-positive cancers including the pancreatic neuroendocrine tumors, pediatric glioblastoma multiforme, neuroblastoma and several other cancers of the central nervous system (24–32), further implicating the role of ATRX/DAXX/H3.3 in the maintenance of telomere chromatin integrity.
Promyelocytic leukemia (PML) nuclear bodies are spheres of 0.1–1.0 μm in diameter found in the nucleus. They regulate cellular processes including gene transcription, tumor suppression, DNA replication and repair (33–35). Consistent with their multi-faceted role in cellular function, PML bodies contain a variety of regulatory proteins including PML, SP100, p53, pRb, HP-1, ATRX, DAXX and SUMO-1. In human ALT cancer cells, a distinct kind of PML bodies known as the ALT-associated PML bodies (APBs) is found. In these structures, telomeric DNA, telomere binding proteins (e.g. TERF-1, TERF-2) and DNA repair proteins (e.g. RAD50, Mre11, NBS1) co-localize with the PML protein (8,36). Although it is unclear as to whether APBs directly drive ALT activity, there is evidence that they promote ALT activity. The disassembly of APBs, as a result of RNAi-mediated knockdown of either PML or Mre11/Rad50/Nbs1 (MRN) proteins, leads to inhibition of telomere elongation in ALT cells (37–40).
Here, we show that ATRX, DAXX and H3.3 co-localize with telomeres at the PML bodies in pluripotent mouse ES cells. Other proteins found within these bodies include and DNA repair proteins including NBS1 and Mre11. The assembly of these telomere-associated PML bodies is associated with the pluripotent state of mouse ES cells, as their presence is not found in human and mouse somatic cell types. RNAi-mediated depletion of PML expression in mouse ES cells results in the disassembly of these telomere-associated PML bodies, and consequently loss of ATRX and H3.3 binding at the telomeres. These changes are accompanied by increased levels of repressive chromatin marks including H3K9me3 and H4K20me3 at the telomeres, indicating a disruption to the maintenance of the unique and less ‘heterochromatic’ histone methylation modification pattern at the telomeres in ES cells. Our study provides evidence that PML bodies play an important role in maintaining telomeric chromatin integrity. Considering the link between telomere function and chromosome stability (35), this study further reveals the function of PML bodies in regulating genome integrity (41,42), in this case, ensuring the faithful inheritance of epigenetic states at telomeres.
MATERIALS AND METHODS
Cell culture
Human HT1080 fibrosarcoma and mouse NIH3T3 transformed fibroblast were cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% FCS. Mouse ES129.1, ES-W9.5, EGRA2 and EGRA3 cells were cultured in DMEM with 15% heat-inactivated FCS and 103 units/ml of leukemia inhibitory factor (LIF) and 0.1 mM β-mercaptoethanol.
Induction of cellular differentiation of mouse ES cells
ES129.1 cells were subjected to cellular differentiation by LIF withdrawal and retinoid acid treatment. Before differentiation, >90% of cells expressed the stem-cell surface marker POU5F1 (also known as OCT4). After 3 days of differentiation, <50% expressed POU5F1. By 6 days, POU5F1 was barely detectable.
Appending the MYC epitope to the N-terminus of H3.3A
A standard replacement vector was used to target modified sequence to one of the endogenous alleles of the H3f3a (H3 histone, family 3A) gene. A neomycin positive selection cassette flanked by loxP sites was contained in intron 2–3, which was later removed by transient exposure to Cre recombinase. The first 15 residues of the resulting protein produced by the modified H3f3a locus were M-EQKLISEEDL-ARTK. The only other sequence change at the locus was a single loxP site contained within intron 2–3 (M.T., L.H.W. and J.R.M., unpublished data).
Immunofluorescence and FISH analysis
Cells were treated with microtubule-depolymerizing agent Colcemid at 37°C for 1 h and harvested for immunofluorescence analysis (43). Cells were subjected to hypotonic treatment in 0.075 M KCl, cytospun on slides and incubated in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris–HCl, pH 7.2, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 0.1% (v/v) Triton X-100 and protease inhibitor). Slides were blocked in KCM buffer containing 1% bovine serum albumin (BSA), incubated with relevant primary and secondary antibodies at 37°C for 1 h. After each round of antibody incubation, slides were washed three times in KCM buffer. Slides were then fixed in KCM with 4% formaldehyde and mounted in mounting medium. Images were collected using a fluorescence microscope linked to a CCD camera system. For telomere-fluorescence in situ hybridization (FISH), slides were fixed in cold methanol followed by dehydration in an ethanol series before denaturation and hybridization with Cy3-conjugated telomere DNA probe (Dako).
Antibodies
Primary antibodies used include rabbit polyclonal antisera against TERF-1 (44), TERF-2 (Novus), ATRX (H300; Santa Cruz Biotechnologies); H3K9me3 (Millipore), H4K20me3 (Abcam/Millipore), DAXX (Millipore/Merck), Mre11 (Millipore/Merck); mouse monoclonal antisera against MYC tag (Millipore), POU5F1 (Santa Cruz Biotechnologies), PCNA (Proliferating Cell Nuclear Antigen; Millipore/Merck) 53BP1 (Millipore/Merck) and NBS1 (Millipore/Merck) and PML (Millipore/Merck).
Cell cycle synchronization and FACS analysis
Cells were treated with 2 mM thymidine for 12 h, released from cell cycle blockage by washing with warm medium and incubated in medium at 37°C for various time points. For fluorescence-activated cell sorting (FACS) analysis, cells were harvested, washed in 1× phosphate buffered saline (PBS), fixed with ice-cold ethanol, stained with propidium iodide and analyzed on a LSR II Becton Dickinson Flow Cytometry Analyzer.
RNAi-mediated knockdown and real-time PCR analysis
Transfections of small interfering RNA (siRNA) oligonucleotides were performed using Lipofectamine 2000 (Invitrogen). These oligonucleotides have been pre-validated to confirm their targeting specificity to messenger RNAs of PML, NBS1 and Mre11. A set of control scrambled siRNA oligonucleotides was also used. Cells were harvested 24–48 h after RNAi-mediated knockdown for the assessment of the levels of endogenous protein expression by real-time polymerase chain reaction (PCR) analysis using the SYBR system (Applied Biosystem). As internal controls, specific primers corresponding to house-keeping gene Actin were used in real-time PCR analysis. Changes in expression levels were calculated according to the manufacturer’s instruction.
CO-FISH
Cells were incubated for 16–20 h in fresh medium containing BrdU (10 µg/ml). An hour before harvesting, Colcemid was added to the media to accumulate mitotic cells. Cells were harvested and resuspended in 0.075 M KCl (pre-warmed to 37°C). Ice-cold methanol-acetic acid (3:1 ratio) was added to cell suspension. The cell suspension was spun (5 min at 1000 rpm) and washed twice in methanol-acetic acid. Cells were dropped onto slides and allowed to dry overnight. Slides were rehydrated in 1× PBS for 5 min at room temperature, incubated with 0.5 µg/ml RNase A (in PBS, DNase free) for 10 min at 37°C and stained with 0.5 µg/ml Hoechst 33258 in 2× SSC for 15 min at room temperature. Subsequently, slides were placed in a shallow plastic tray, covered with 2× SSC and exposed to 365 nm ultraviolet light at room temperature for 45 min. The BrdU-substituted DNA strands were digested with at least 10 U/µl of Exonuclease III at room temperature for 30 min. Slides were washed in 1× in PBS, dehydrated in ethanol series 70%, 95%, 100% and air dried. FISH was performed as described above, except that slides were not subjected to DNA denaturation.
Chromatin immunoprecipitation assays (CHIP)
Cells were fixed in 0.8% paraformaldehyde in 1× PBS, washed extensively in 1× PBS, lysed in ice-cold Lysis buffer [1% sodium dodecyl sulphate (SDS), 10 mM EDTA, 50 mM Tris–HCl, pH 8.0, and protease inhibitors; sonicated chromatin products of ∼100–300 bp] and diluted 10× in Dilution buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, protease inhibitors and 1 mg/ml BSA). ChIP was performed with the relevant antibody and captured with Protein-A/G Sepharose. DNA-protein complex was washed with 1× Wash buffer I (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton X-100 and 2 mM EDTA), 2× Wash buffer II (20 mM Tris–HCl, pH 8.0, 250 mM NaCl, 0.1% SDS, 1% Triton X-100 and 2 mM EDTA) and eluted in 1% SDS and 100 mM NaHCO3. Eluate fraction was de-crosslinked by high-salt treatment (200 mM NaCl) at 60°C followed by proteinase K treatment at 50°C. DNA extracted was subjected to dot blot analysis.
Dot blot analysis
Fifty nanograms of DNA was diluted with 0.6 M NaCl, denatured (by heating DNA at 100°C for 5 min and cooled on ice for 5 min), transferred onto Hybond N+ nitrocellulose membrane and rinsed in 0.5 M NH2SO4. The membrane was incubated in 0.4 N NaOH at RT for 5 min, followed by neutralization with 2× SSC twice. In subsequent, the membrane was subjected to Southern blot analysis with either γ-32p ATP-end-labeled (TTAGGG)4 telomere probe. Signal intensities were analyzed with Typhoon PhosphoImager System and ImageQuant software.
Sequences of siRNA oligonucleotides
- PML set 1 GCAGUGGCAUGAGGAACUAUU
- GAUUCAGCAGUGGCATGAGGAACUAGGCAC
- PML set 2 GCGCGACUACCAGGAAAUAUU
- CAGCGCGACUACCAGGAAAUAGCUGGCCAG
- PML set 3 CCUCAAGAUUGACAAUGAAUU
- CUUUGACCUCAAGAUUGACAAUGAAACCCA
- PML set 4 CGGUGAACCGGGAAAGCAAUU
- UUGCUUUCCCGGUUCACCGCG
- MreII set 1 GAGGCUUUGAACCUUUCAAUU
- UUGAAAGGUUCAAAGCCUCUU
- MreII set 2 GGGCACAACAUCUAGCAAAUU
- UUUGCUAGAUGUUGUGCCCUU
- NBS1 set 1 GCCCUUGGUUGUUUGUUCUUU
- AGAACAAACAACCAAGGGCUU
- NBS1 set 2 CCAGAAAUCUAUGUGUAAAUU
- UUUACACAUAGAUUUCUGGUU
Primer sets used for real-time PCR analysis
- Actin for1 TCCCTGGAGAAGAGCTACGA and
- Actin rev1 AGCACTGTGTTGGCGTACAG
- Actin for2 CATGTTTGAGACCTTCAACA and
- Actin rev2 GTGAGGATCTTCATGAGGTA
- PML exon1/2 for GACTATGAACACAGCCAAAGCC and
- PML exon1/2 rev GCCTTGCAGATGGGGCACTGC
- PML exon5/6 for GCCTGGAGCACACCCTGTACC and
- PML exon5/6 rev GGAGCTTGTGGCCAACCTGTC
- Mre11 exon17/18 for CCAATTCCAGGGCTGATCAA and
- Mre11 exon17/18 rev CTCTGGGACATCCGTTTGCT
- Mre11a exon9/11 for GTTCTGGCTAACCACCCAAA and
- Mre11a exon9/11 rev CCACCCGTAGTCGGATAAGA
- Nbs1 exon11 for CCTGCCGGACCCTCACT and
- Nbs1 exon11 rev CCAGGGTCGCATTCTGAGA
- Nbs1 exon2/3 for GGAGTACGTTGTTGGGAGGA and
- Nbs1 exon2/3 rev CTGTCTTCAACGTGCAGGAA
- Nbs1 exon6/7 for TAAATGCCAAGCAGCACAAG and
- Nbs1 exon6/7 rev ACATCAACAACGCAGGTTCC
RESULTS
Localization of telomeric complex at PML bodies during S phase
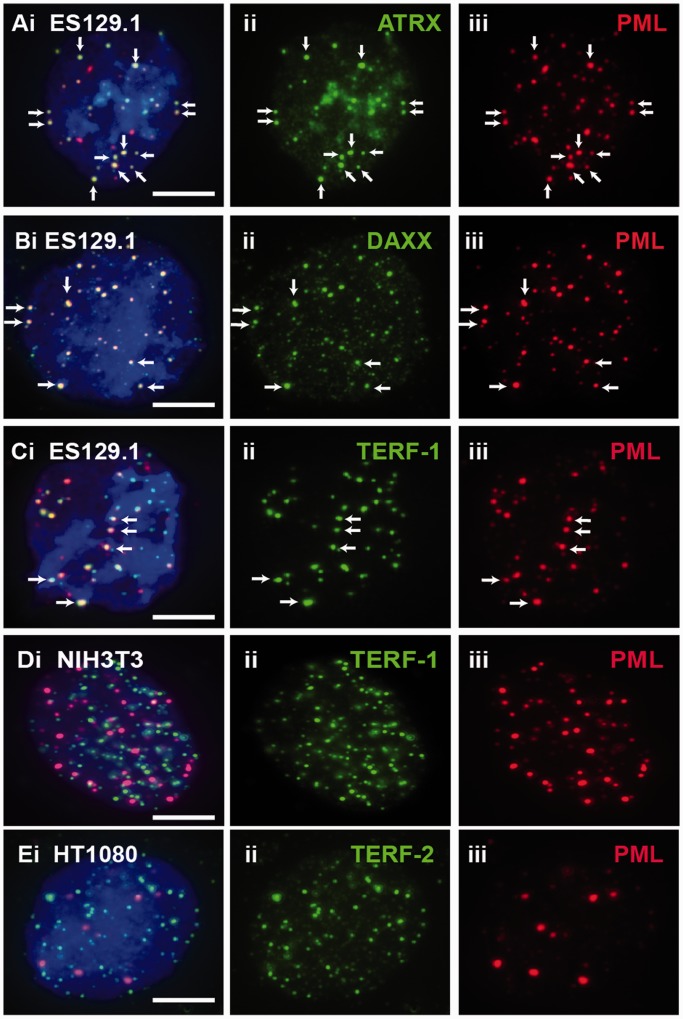
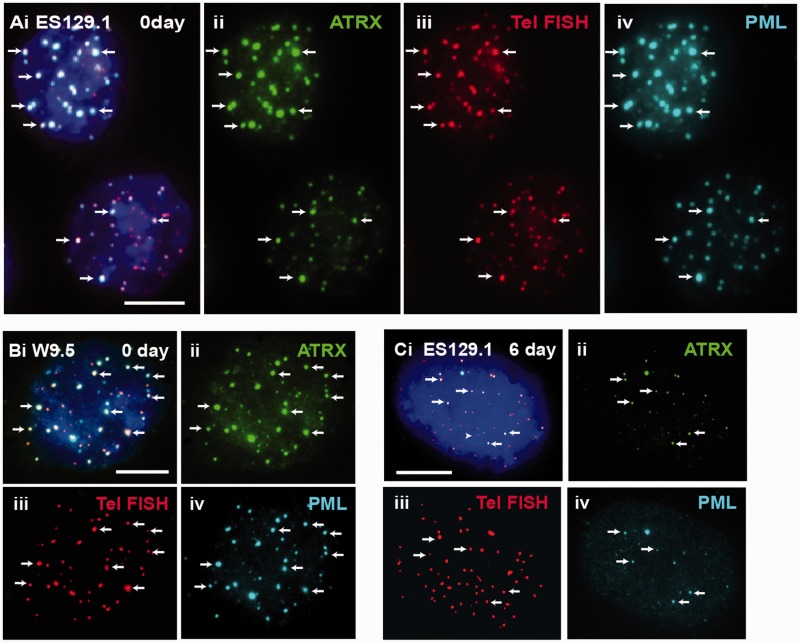
In this study, our aim was to determine the platform involved in the establishment of epigenetic information at the telomeres, in particular, the recruitment of ATRX, DAXX and deposition of H3.3 at the telomeres during S phase in mouse ES cells (12,21–23,45). PML bodies have been implicated in many cellular pathways such as transcriptional regulation, DNA replication and repair, chromatin organization and tumor suppression. Here, we studied the role of PML bodies as sites for telomeric chromatin assembly in mouse ES cells. In somatic cells, localization of ATRX and DAXX at the PML nuclear bodies has been well documented (21,45–47). In mouse ES cells, ATRX and DAXX also localized at the PML bodies (Figure 1A and B). However, beside ATRX and DAXX, telomeres also localized at the PML bodies in these cells (Figure 1C). The localization of telomeres at the PML bodies was confirmed by combined immunofluorescence and telomere FISH analysis (Figure 2A and B). Beside ES cells, we also observed localization of the telomeres at PML bodies in pluripotent mouse embryonic germ (EG) cell lines—EGRA2 and EGRA3 (data not shown); these pluripotent cells also show enriched association of ATRX and H3.3 at telomeres, as reported in our previous studies (12,13). The assembly of telomere-associated PML bodies could potentially be linked to the pluripotent state of the ES cells because telomeres in non-pluripotent somatic cell types such as NIH3T3 and HT1080 were not associated with the PML bodies (Figure 1D and E). To investigate this possibility, we determined the assembly of telomere-associated PML bodies in mouse ES129.1 cells subjected to cellular differentiation. Before induction of cellular differentiation by LIF withdrawal and retinoid acid treatment (12,13), >90% of cells expressed the stem-cell surface marker POU5F1 (data not shown). After 3 days, only ∼50% expressed POU5F1, and it was barely detectable after 6 days. The presence of telomere-associated PML bodies remained relatively prominent in the first 3 days of differentiation (Table 1), with 91.43% of cells showing ≥5 positive ATRX/telomeres co-staining foci at PML bodies per cell. However, after 6 days, this positive staining pattern started to decrease and was reduced to only 14.29–28.57% after 9–12 days (Figure 2C and Table 1). This loss in telomere-associated PML bodies was not simply a consequence of a change in cellular growth rate, as these cells were still actively dividing after differentiation. These observations indicate that the assembly of telomere-associated PML body is linked to a differentiation-state–specific process.
Figure 1.
ATRX, DAXX and telomeres localized at PML bodies in mouse ES cells. (A–C) In ES129.1 cells, beside ATRX (Aii) and DAXX (Bii), telomeres (Cii; TERF-1 staining) also localized at the PML bodies (A-Ciii; arrows). The co-localization is indicated by the presence of overlapped signals. (D and E) Telomeres of NIH3T3 (Dii; TERF-1 staining) and HT1080 (Eii; TERF-2 staining) cells did not localize at PML bodies (D-Eiii). Part ‘i’ is the merged figures and ‘ii and iii’ the split images of ‘i’. The arrows show some ‘examples’ of co-localized foci. (Scale bars: 5 μm).
Figure 2.
ATRX co-localized with telomeres at PML bodies in mouse ES cells. (A and B) ES129.1 and W9.5 cells showed clear co-staining of ATRX (A-B ii) with telomere FISH signals (A–B iii) at the PML bodies (A and B iv). (C) The co-staining pattern of ATRX (ii) with telomeres (iii) at the PML bodies (iv) started to decrease after 6 days of differentiation. Part ‘i’ is the merged figures and ‘ii, iii and iv’ the split images of ‘i’. The arrows show some ‘examples’ of co-localized foci of ATRX/telomeres at PML bodies. (Scale bars: 5 μm).
Table 1.
Localization of telomere at PML bodies during ES cell differentiation
| Time period of differentiation (days) | 0 | 3 | 6 | 9 | 12 |
|---|---|---|---|---|---|
| Number of cells with ≥5 positive foci of telomere-associated PML bodies (total of 35 cells scored) | 32 | 34 | 27 | 10 | 5 |
| Percentage of cell population with ≥5 positive foci of telomere-associated PML bodies (%) | 91.43 | 97.14 | 77.14 | 28.57 | 14.29 |
Considering the role of ATRX and DAXX as chaperones that load H3.3 into chromatin (21), we propose to study the function of these telomere-associated PML bodies as a platform that facilitated the recruitment of ATRX/DAXX complex and the nucleosomal deposition of H3.3 at the telomeres. There is evidence that the number of PML bodies is increased during DNA replication at S phase (48). Moreover, the co-localization of ATRX and H3.3 with the telomeres in mouse ES cells is most prominent during S phase (12,13). To determine if the assembly of the telomere-associated PML bodies is coupled to cell cycle progression, we performed immunofluorescence analysis on ES129.1 cells synchronized using a thymidine G1/S block protocol. The cell cycle progression of cells was assessed by FACS analysis (12,13). Following the release, ES129.1 cells progressed through S phase at 4–6 h after release, entered M phase at 8 h, and re-entered G1 phase at 10 h. The localization of ATRX/telomeres at the PML bodies occurred predominantly at S phase, with 88.57–91.43% of cells showing ≥5 positive co-staining foci of ATRX/telomeres at PML bodies per cell after 4–6 h of release (Table 2). This suggests that the assembly of telomere-associated PML bodies in mouse ES cells is in synchrony with S phase of cell cycle (49). These PML bodies may be required for the establishment of chromatin marks at newly replicated telomeric DNA to ensure a proper maintenance of telomeric chromatin integrity.
Table 2.
Cell cycle analysis of telomeric localisation at PML bodies
| Time period after G1/S release (hours) | 0 | 2 | 4 | 6 | 8 | 10 |
|---|---|---|---|---|---|---|
| Number of cells with <5 positive foci of telomere-associated PML bodies (total of 35 cells scored) | 28 | 13 | 4 | 3 | 22 | 27 |
| Percentage of cell population with <5 positive foci of telomere-associated PML bodies (%) | 80.00 | 37.14 | 11.43 | 8.57 | 62.86 | 77.14 |
| Number of cells with ≥5 positive foci of telomere-associated PML bodies (total of 35 cells scored) | 7 | 22 | 31 | 32 | 13 | 8 |
| Percentage of cell population with ≥5 positive foci of telomere-associated PML bodies (%) | 20.00 | 62.86 | 88.57 | 91.43 | 37.14 | 22.86 |
DNA repair proteins are not required for assembly of telomere-associated PML bodies
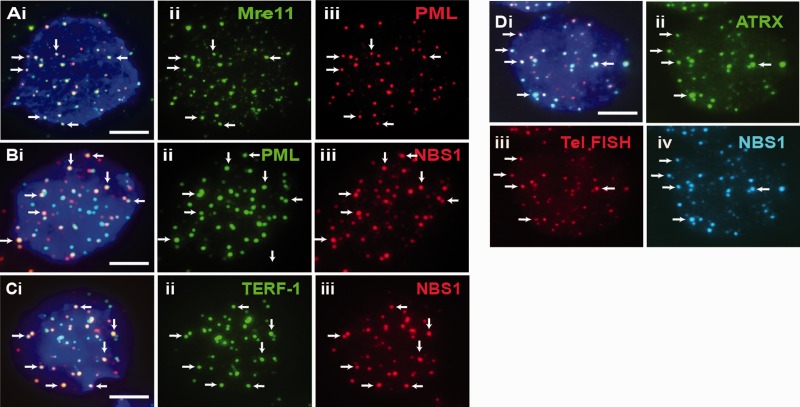
To define the function of PML bodies in maintaining telomeric chromatin integrity, we determined the presence of other proteins at telomere-associated PML bodies. Beside ATRX and DAXX, we detected the presence of DNA repair and recombination proteins NBS1 and Mre11 at the PML bodies and at the telomeres (examples are shown in Figure 3). The co-localization of NBS1 and Mre11 with telomeres at the PML bodies was confirmed by telomere FISH analysis. In human ALT cancer cells, the presence of these DNA repair proteins is vital for APB assembly (37,38). Depletion of the MRN (Mre11/Rad50/NBS1) complex results in the disassembly of APBs and maintenance of telomeric recombination or ALT activity. To determine the requirement of NBS1 and Mre11 in maintaining the assembly of telomere-associated PML bodies in ES cells, we performed RNAi-mediated knockdown of NBS1 and Mre11 expression (Supplementary Table S1a and Figure S1). The FACS analysis showed that transient loss of expression of these proteins did not cause a severe cell cycle block (Supplementary Table S1b). Unlike that of the ALT cancer cells (37,38), loss of function of NBS1 and Mre11 following knockdown had no significant impact on the assembly of telomere-associated PML bodies (Supplementary Table S1c), indicating that they are not required for the assembly of telomere-associated PML bodies in mouse ES cells.
Figure 3.
Proteins that localized at telomeres-associated PML bodies. (A–C) DNA repair protein MRE11 (Aii) was present at PML bodies (Aiii) in ES129.1 cells. NBS1 (B-C iii) was also present at PML bodies (Bii) and telomeres (Cii; TERF-1 staining). (D) NBS1 (iv) co-localized with ATRX (ii) at telomeres (iii) in ES129.1 cells. Part ‘i’ is the merged figures and ‘ii, iii and iv’ the split images of ‘i’. The arrows show some ‘examples’ of co-localized foci. (Scale bars: 5 μm).
Loss of PML bodies induces a telomeric dysfunction phenotype
PML is the most prominent component of PML bodies, and its presence is required for their assembly. To determine the role of PML bodies in maintaining telomeric chromatin integrity, we had established RNAi-mediated knockdown of PML expression in mouse ES129.1 cells. As shown by real-time reverse transcription PCR assay, two of these oligonucleotide sets (set 3 and 4) resulted in 70–80% depletion of PML expression (Supplementary Table S2a). The reduction in PML protein levels was shown by western blot and immunofluorescence analyses in Figure 4A and B. And importantly, the loss of PML expression reduced the assembly of telomere-associated PML bodies. The reduced expression of PML did not significantly impact the cell cycle progression as shown by FACS analysis (Supplementary Table S2b) and positive immunostaining with an antibody against proliferating cell nuclear antigen (PCNA is a cofactor for DNA replication and is expressed in S phase) (Supplementary Figure S2 and Table S2c).
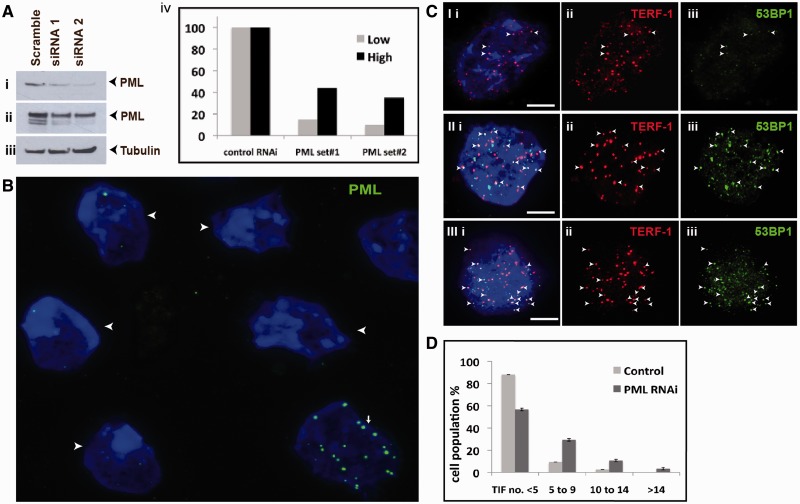
Figure 4.
Effects of loss of PML bodies on telomere function. (A) ES129.1 cells were depleted of PML expression with two sets of siRNA oligonucleotides (set 3 and 4; Supplementary Table S2), followed by western blot analysis with antiserum against PML (i, short exposure; ii, long exposure of blot—the blot was overexposed to detect all PML isoforms) and β-tubulin (iii). Equal loading of protein was achieved by normalization against β-tubulin level. (iv) ‘Low bars’ were graphed according to protein bands in (i), whereas ‘High bars’ according to protein bands in (ii). In PML depleted cells, PML levels (lanes 2 and 3) were reduced by ∼90% 48 h after transfection compared with cells transfected with control siRNA oligonucleotides (lane 1). (B) Two sets of siRNA oligonucleotides (set 3 and 4) were used to deplete PML expression in ES129.1 cells. Immunofluorescence analysis was performed using antiserum against PML. Examples of ES129.1 cells showing the loss of PML bodies are shown by arrowheads. (C) TIFs were detected by co-staining of telomeres (TERF-1 staining; ii) with 53BP1 signals (iii) (see Supplementary Table S3a for scoring data). In ES129.1 cells transfected with control scramble siRNA oligonucleotides, <5 TIFs were detected (I), whereas 48 h of RNAi-mediated knockdown of PML induced the formation of TIFs (11 TIFs in II and 15 TIFs in III, respectively). Arrowheads show examples of TIFs. (**D**) Data were sub-grouped according to number of TIFs (<5, 5–9, 10–14 and >14) per cell. A normal cell contains ∼2–3 TIFs on average, so a threshold of >4 TIFs was used. When depleted of PML expression, percentage of cells with ≥5 TIFs (85 cells were counted) rose by 3–5-folds (from 10–14% to 38–46%).
We have previously shown that RNAi-mediated knockdown of H3.3 and ATRX in mouse ES cells induced telomere-dysfunction phenotypes (12,13). This was indicated by an increase in the number of cells with telomere-induced dysfunctional foci (TIF; a marker for DNA damage at the telomeres) (50). Here, we also assessed the effects of the loss of PML bodies on telomere structural integrity by staining with an antibody against a DNA damage marker 53BP1 (Figure 4C and D). In ES129.1 cells transfected with control scramble siRNA oligonucleotides, only 10–14% of cells showed ≥5 TIFs per cell. In contrast, cells subjected to PML-specific RNAi knockdown showed a >∼4-fold (increased from 10–14% to 38–46%) increase in the population of cells showing ≥5 TIFs per cell (Figure 4C and D and Supplementary Table S3a), demonstrating that telomere functional integrity was impaired following the loss of telomere-associated PML bodies in mouse ES cells.
Loss of PML bodies impacts on establishment of epigenetic marks at telomeres
Considering the link of PML bodies to telomeric recombination in ALT cells (36–40), we performed chromosome orientation (CO)-FISH assay (Supplementary Figure S3) to determine if loss of PML bodies affected the rate of sister chromatin exchange at telomeres (TSCE). In CO-FISH assays, the CO-FISH hybridization normally displays only single telomere-FISH hybridization signal per two sister chromatids. If TSCE occurs, the signal is split and appears on both sister chromatids. No increase in TSCE was observed in ES129.1 subjected to either scramble control or PML-specific RNAi knockdown (Supplementary Table S3b), suggesting that telomere-associated PML bodies in mouse ES cells were not directly involved in telomeric recombination. Our proposal is consistent with a recent work showing that the association of induced ‘long telomeres’ with PML bodies in telomerase overexpressing cells is not accompanied with an increase in TSCE (51), highlighting a distinct role of PML bodies in telomere maintenance. In consideration of the induction of telomeric damage in PML-depleted cells (Figure 4C and D), we propose that PML bodies is involved in maintaining a functional epigenetic state such as the recruitment of ATRX and H3.3 at the telomeres in mouse ES cells (12,13).
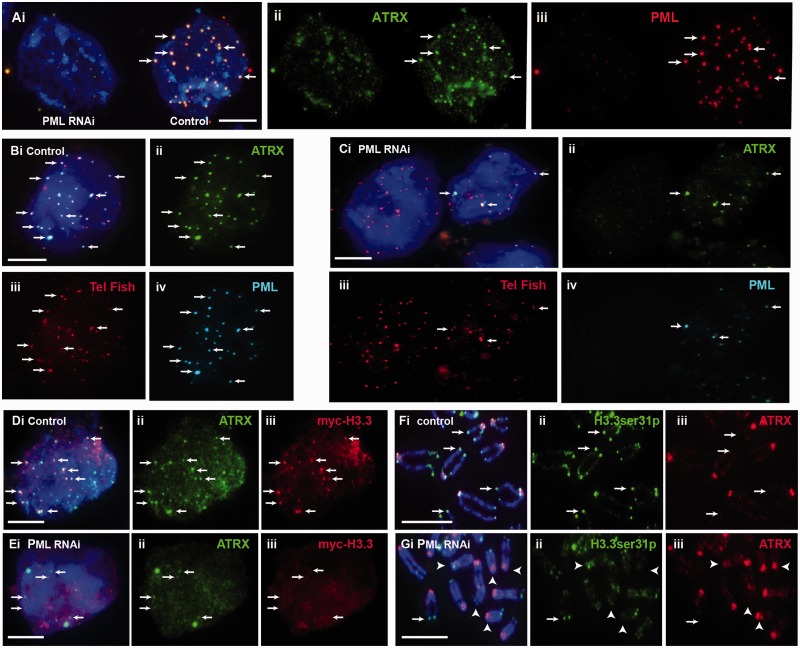
To test this hypothesis, we determined the impact of the loss of PML bodies on the maintenance of epigenetic marks at telomeres. Interestingly, depletion of PML expression and consequential loss of PML bodies led to a reduction in the presence of ATRX at the telomeres (Figure 5A–C). To confirm this, we had specifically assessed co-localization of ATRX and telomeres at PML bodies in PML knockdown cell population during S phase; these cells were blocked at G1/S by thymidine treatment, followed by release into S phase for immunofluorescence analysis. In PML-depleted cells, we found a significant drop in percentage of cell population showing five or more telomere-associated PML bodies, and this loss of assembly of telomere-associated PML bodies resulted in ATRX de-localization from the telomeres during S phase (percentage of cells showing ≥5 positive foci per cell reduced from 82.6 to 42.9%; Table 3). Considering the role of ATRX as chaperon for assembly of H3.3 nucleosomes at the telomeres in mouse ES cells, we also determined if loss of PML bodies also affected H3.3 level at the telomeres. To determine this, we assessed the level of H3.3 phosphorylated at serine 31 (H3.3ser31P) as we have previously shown that telomeres in ES cells are enriched with H3.3ser31P during mitosis (13). Indeed, the reduced association of ATRX at telomeres was also accompanied by a decrease in H3.3ser31P level at the telomeres (Figure 5D and E). Specifically, loss of PML bodies led to a 42% reduction in the level of H3.3ser31P at the telomeres during mitosis (Table 4). These data suggest that PML bodies are required for maintaining ATRX/H3.3 levels at the telomeres.
Figure 5.
Effects of loss of PML bodies on binding of ATRX and H3.3 at telomeres. ES129.1 cells transfected with control scramble siRNA oligonucleotides are indicated as ‘control’, whereas cells subjected to knockdown with PML-specific siRNA oligonucleotides are indicated as ‘PML RNAi’. (A) RNAi knockdown of PML in ES129.1 cells resulted in a loss in the formation of PML bodies (iii; cell on the left), leading to de-localization of ATRX (ii; arrows). (B and C) De-localization of ATRX from telomeres was confirmed by telomere FISH analysis. Compared with scramble control ES129.1 cells (B; control), RNAi knockdown of PML (C; PML RNAi) led to disassembly of PML bodies (Civ), resulting in ATRX de-localization (Cii) from the telomeres (Ciii; telomere FISH) (see Table 3). (D and E) RNAi knockdown of PML and loss of PML bodies (E; PML RNAi) also resulted in a reduction in the co-localization of _myc_-H3.3 (iii) with ATRX (ii) in ES129.1 cells. (F and G) In addition, depletion of PML expression and loss of PML bodies (G; PML RNAi) also resulted in a reduction in the level of H3.3ser31P at the telomeres on metaphase chromosomes (examples of positive H3.3ser31P staining at telomeres are indicated by arrows, whereas examples of the absence of H3.3ser31P signal at telomeres are indicated by arrowheads; see Table 4 for quantification) (Scale bars: 5 μm).
Table 3.
Assembly of telomere-associated PML bodies in PML knockdown cells
| Number of co-localized foci (% of cells) | Control RNAi | PML RNAi | Control RNAi | PML RNAi | Control RNA | PML RNAi |
|---|---|---|---|---|---|---|
| n = 1 | n = 1 | n = 2 | n = 2 | n = 3 | n = 3 | |
| <4 | 17.14 | 57.14 | 14.29 | 60 | 20 | 54.29 |
| 5–9 | 54.29 | 25.71 | 60 | 25.71 | 54.3 | 31.43 |
| 10–14 | 17.14 | 14.29 | 20 | 11.43 | 17.1 | 8.57 |
| >14 | 11.43 | 2.857 | 5.71 | 2.86 | 8.57 | 5.71 |
| % of cell with ≥5 positive foci | 82.86 | 42.86 | 85.71 | 40 | 80 | 45.71 |
| Average % of cells | Control RNAi = 82.6% | PML RNAi = 42.9% |
Table 4.
H3.3ser31P intensities at telomeres in PML knockdown cells
| Experiments | Samples | Centromere signals | H3.3ser31P signals | Ratio of H3.3ser31P/centromere | Fold of change in ratio of H3.3ser31P/centromere |
|---|---|---|---|---|---|
| N = 1 | Control RNAi | 560 | 471 | 0.84 | 0.61 |
| PML RNAi | 688 | 354 | 0.52 | ||
| N = 2 | Control RNAi | 558 | 386 | 0.69 | 0.49 |
| PML RNAi | 766 | 259 | 0.34 | ||
| N = 3 | Control RNAi | 359 | 333 | 0.93 | 0.63 |
| PML RNAi | 386 | 225 | 0.58 | ||
| Average fold of change in ratio of H3.3ser31P/centromere signals | 0.58 (reduced by 42%) |
To further examine the role of PML bodies in maintaining telomeric chromatin state, we performed combined crosslinked chromatin immunoprecipitation (ChIP) and dot blot assays using a telomere DNA probe to assess the changes in telomeric chromatin state (Figure 6 and Supplementary Table S4). Interestingly, beside the reduction in ATRX level at the telomeres (∼0.4-fold or 60%) in cells depleted of PML bodies, there were increases in the levels of H3K9me3 and H4K20me3 at telomeres by ∼2.3- and 3.4-folds, respectively. Interestingly, there was also a slight increase in level of TERF-1 at the telomeres. The enrichment of the repressive histone methylation marks H3K9me3 and H4K20me3 indicated a significant change in the telomeric chromatin properties in cells depleted of PML bodies. These differences in histone methylation profile were unlikely caused by changes in expression of histone modifiers or telomere proteins as shown by western blot analysis (Figure 6). Instead, we propose that these data support our proposal of the importance of PML bodies in ensuring the maintenance of specific epigenetic states at telomeres in mouse ES cells.
Figure 6.
Effects of loss of PML bodies on establishment of chromatin states at telomeres. (A) ES129.1 cells were subjected to RNAi knockdown of PML and western blot analysis with antisera against PML (i, short exposure; ii, long exposure of blot), TERF-1 (iii), β-tubulin (iv), H4K20 monomethylation (v), H4K20 trimethylation (vi, Up, Millipore; and vii, Ab, Abcam), (viii) H3K9 dimethylation, (ix) H3K9 trimethylation and (x) histone H4. No change in expression level was seen in any of these proteins and chromatin marks, except for PML proteins. (B) ChIP analysis (Supplementary Table S4 for data) with antisera against H3K9me3, H4K20me3, ATRX and TERF-1, followed by dot blot analysis by hybridization with a telomere-specific probe. Consistent with the immunofluorescence analysis, a lower level of ATRX is detected at telomeres in PML-depleted cells, whereas, higher levels of H3K9me3 (2.4×) and H4K20me3 (3.4×) were detected at telomeres. In PML-depleted cells, a higher level of TERF-1 (2.5×) was also detected at telomeres.
DISCUSSION
In this study, we examined the platform on which telomeric chromatin state is established, in particular, the assembly of H3.3 nucleosomes by ATRX and DAXX at the telomeres (12,21–23) in mouse ES cells. Our data show that PML bodies act as sites for H3.3 deposition by ATRX and are important for the faithful maintenance of epigenetic information at telomeres in ES cells.
Although PML bodies have been regarded primarily as sites of storage where proteins accumulate and undergo post-translational modifications (35), there are studies showing interaction of specific genomic sites such as the histocompatibility (MHC) class I gene cluster (52) and CMV promoter array (45) with PML bodies. Here, we show that telomeres are associated with PML bodies in mouse ES cells, providing evidence for the propensity for PML bodies to associate with specific genomic loci. The assembly of these telomere-associated PML is linked to the pluripotent state of cells, as these bodies are not found in somatic cells and are reduced in number in differentiated ES cells. It is currently unknown what factor(s) may direct the localization of telomeres to the PML bodies in ES cells. It is, however, tempting to speculate that the unique epigenetic code surrounding the telomeres may direct the formation of PML bodies. Indeed, the hypomethylation status of telomeric DNA in human ALT cancers (53) and mouse DNMT-null cells (10) and the loss of H4K20me3 at telomeres in Suvar-4–20-h knockout cells (11) may promote the assembly of PML bodies surrounding the telomeres in these cells, thus, supporting this possibility. In the same light, the ‘less heterochromatic’ nature of the ES cell telomeric chromatin with lower levels of H3K9me3 and H4K20me3 (13) may drive association of telomeres with PML bodies.
In mouse ES cells, loss of PML bodies following RNAi knockdown of PML induces telomere dysfunctional phenotype and affects binding of ATRX and H3.3 at telomeres. Considering the role of ATRX/DAXX (46) in directing the H3.3 nucleosomal assembly (21,22), the increased telomere dysfunction phenotype in PML-depleted ES cells could be brought about by the disruption in the complex formation between ATRX and DAXX, and a failure in H3.3 deposition at the telomeres.
The depletion of PML expression in mouse ES cells also results in a change in telomeric chromatin properties—as demonstrated by an increase in the level of repressive histone marks including H3K9me3 and H4K20me3 at telomeres. These underlying changes in histone marks indicate a possible disruption to the maintenance of a unique and ‘less heterochromatic’ chromatic state at telomeres in the absence of telomere-associated PML bodies. It is reasonable to suggest that these changes in patterns of histone methylation may contribute instrumentally to the increased telomere dysfunctional phenotype at telomeres in mouse ES cells.
Previous studies in immunodeficiency, centromeric instability and facial dysmorphy (ICF) syndrome (54), a pathological condition caused by mutations in the DNA methyltransferase DNMT3B gene, have provided important clue into how association of PML bodies could contribute to epigenetic modifications. In ICF patient cells, Bloom syndrome protein (BLM) and Topo IIIα within PML bodies resolve the aberrant recombination DNA products formed during replication due to hypomethylation of 1qh and 16qh pericentric satellite DNA, while DAXX, BRCA1 and HP-1 re-establish a repressive and compacted state at these repeats (55). Although it is unclear how changes of epigenetic state at 1qh and 16qh pericentric satellite DNA have promoted the assembly of PML bodies around these loci, these studies clearly demonstrate the involvement of PML bodies as sites for chromatin state re-establishment following DNA replication. In light of this, we propose that PML bodies serve as a fitting platform for recruitment and/or stabilization of chromatin remodeling proteins at telomeres following DNA replication (49). Within the PML bodies, DNA damage processing machinery (e.g. Mre11 and NBS1) could promote telomere replication (49), while ATRX and H3.3 take part in chromatin remodeling by recruiting other chromatin factors such as HP-1 (12). Additionally, the sequestration of telomeric complex within the PML bodies could ensure the assembly of a less ‘heterochromatic’ telomeric chromatin state with lower levels of H3K9me3 and H4K20me3 (13,14) and prevent an aberrant assembly of chromatin marks at telomeres that are not compatible with the pluripotent state of ES cells, This may also explain the presence of high levels of H3K9me3 and H4K20me3 at telomeres in the absence of PML bodies in PML knockdown cells.
It is interesting that the loss of PML bodies also resulted in an increase in TERF-1 level at the telomeres in mouse ES cells. Previous studies have shown that loss of function of ATRX and DAXX induces level of telomeric TERRA transcription (23) and that TERRA RNA binding activity could directly affect association of Shelterin complex and H3K9 methylation at telomeres (56). Considering ATRX/H3.3 recruitment at telomeres is affected in the absence of PML bodies, future studies will be necessary to determine if TERRA transcription is up-regulated in PML knockdown ES cells and if an increase in TERRA transcript level has a part in promoting increased association of TERF1, H3K9 me3 and H4K20me3 at the telomeres in these cells.
Our study builds on the known function of PML bodies as a passive storage domain for proteins or as a reservoir for post-translational modifications; these nuclear bodies bring telomeres into an environment enriched with the appropriate epigenetic modifiers in ES cells. We provide evidence for the important function of PML bodies as active epigenetic regulators in ensuring that epigenetic information on telomeres is properly established and securely inherited on telomere repeats. Our study also provides an important insight into function of PML bodies in the maintenance of genome stability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–3.
FUNDING
National Health and Medical Research Council (NHMRC) of Australia and Victorian Government's Operational Infrastructure Support Programme. L.H.W. receives a Career Development Award from NHMRC. Funding for open access charge: NHMRC Australia.
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
REFERENCES
- 1.Costa A, Daidone MG, Daprai L, Villa R, Cantu S, Pilotti S, Mariani L, Gronchi A, Henson JD, Reddel RR, et al. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. 2006;66:8918–8924. doi: 10.1158/0008-5472.CAN-06-0273. [DOI] [PubMed] [Google Scholar]
- 2.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, McKay MJ, Reddel RR, Royds JA. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 3.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 4.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 5.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 7.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, Reddel RR. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 8.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 11.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–414. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q, Reynolds GE, Innes L, Pedram M, Jones E, Junabi M, Gao DW, Ricoul M, Sabatier L, Van Brocklin H, et al. Telomeric transgenes are silenced in adult mouse tissues and embryo fibroblasts but are expressed in embryonic stem cells. Stem Cells. 2007;25:3085–3092. doi: 10.1634/stemcells.2007-0478. [DOI] [PubMed] [Google Scholar]
- 16.Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, Neuhaus D, Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl Acad. Sci. USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 18.Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 19.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Delbarre E, Jacobsen BM, Reiner AH, Sorensen AL, Kuntziger T, Collas P. Chromatin environment of histone variant H3.3 revealed by quantitative imaging and genome-scale chromatin and DNA immunoprecipitation. Mol. Biol. Cell. 2010;21:1872–1884. doi: 10.1091/mbc.E09-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Wilde RF, Heaphy CM, Maitra A, Meeker AK, Edil BH, Wolfgang CL, Ellison TA, Schulick RD, Molenaar IQ, Valk GD, et al. Loss of ATRX or DAXX expression and concomitant acquisition of the alternative lengthening of telomeres phenotype are late events in a small subset of MEN-1 syndrome pancreatic neuroendocrine tumors. Mod. Pathol. 2012;25:1033–1039. doi: 10.1038/modpathol.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capurso G, Festa S, Valente R, Piciucchi M, Panzuto F, Jensen RT, Delle Fave G. Molecular pathology and genetics of pancreatic endocrine tumours. J. Mol. Endocrinol. 2012;49:R37–R50. doi: 10.1530/JME-12-0069. [DOI] [PubMed] [Google Scholar]
- 32.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 34.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 35.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell. Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 36.Wu G, Jiang X, Lee WH, Chen PL. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 2003;63:2589–2595. [PubMed] [Google Scholar]
- 37.Jiang WQ, Zhong ZH, Henson JD, Neumann AA, Chang AC, Reddel RR. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of the MRE11/RAD50/NBS1 complex. Mol. Cell. Biol. 2005;25:2708–2721. doi: 10.1128/MCB.25.7.2708-2721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–4647. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- 39.Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 2000;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- 40.Yu J, Lan J, Wang C, Wu Q, Zhu Y, Lai X, Sun J, Jin C, Huang H. PML3 interacts with TRF1 and is essential for ALT-associated PML bodies assembly in U2OS cells. Cancer Lett. 2011;291:177–186. doi: 10.1016/j.canlet.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Torok D, Ching RW, Bazett-Jones DP. PML nuclear bodies as sites of epigenetic regulation. Front. Biosci. 2009;14:1325–1336. doi: 10.2741/3311. [DOI] [PubMed] [Google Scholar]
- 42.Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. PML bodies: a meeting place for genomic loci? J. Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- 43.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- 45.Newhart A, Rafalska-Metcalf IU, Yang T, Negorev DG, Janicki SM. Single cell analysis of Daxx and ATRX-dependent transcriptional repression. J. Cell Sci. 2012;125(Pt 22):5489–5501. doi: 10.1242/jcs.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl Acad. Sci. USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 2004;117:3807–3820. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- 48.Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J. Cell Biol. 2006;175:55–66. doi: 10.1083/jcb.200604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 50.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 53.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl Acad. Sci. USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ, Megarbane A, Sarda P, Sirma H, Moncla A, et al. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 2006;119:2518–2531. doi: 10.1242/jcs.02965. [DOI] [PubMed] [Google Scholar]
- 56.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data