An E2 enzyme Ubc11 is required for ubiquitination of Slp1/Cdc20 and spindle checkpoint silencing in fission yeast (original) (raw)
Abstract
For ordered mitotic progression, various proteins have to be regulated by an ubiquitin ligase, the anaphase-promoting complex or cyclosome (APC/C) with appropriate timing. Recent studies have implied that the activity of APC/C also contributes to release of mitotic checkpoint complexes (MCCs) from its target Cdc20 in the process of silencing the spindle assembly checkpoint (SAC). Here we describe a temperature-sensitive mutant (ubc11-P93L) in which cell cycle progression is arrested at mitosis. The mutant grows normally at the restrictive temperature when SAC is inactivated, suggesting that the arrest is not due to abnormal spindle assembly, but rather due to prolonged activation of SAC. Supporting this notion, MCCs remain bound to APC/C even when SAC is satisfied. The ubc11+ gene encodes one of the two E2 enzymes required for progression through mitosis in fission yeast. Remarkably, Slp1 (a fission yeast homolog of Cdc20), which is degraded in an APC/C-dependent manner, stays stable throughout the cell cycle in the ubc11-P93L mutant lacking the functional SAC. Other APC/C substrates, in contrast, were degraded on schedule. We have also found that a loss of Ubc4, the other E2 required for progression through mitosis, does not affect the stability of Slp1. We propose that each of the two E2 enzymes is responsible for collaborating with APC/C for a specific set of substrates, and that Ubc11 is responsible for regulating Slp1 with APC/C for silencing the SAC.
Keywords: APC/C, SAC, ubiquitin, mitosis, Ubc11, Slp1, Mad2
Introduction
Proper mitotic progression is a collective phenomenon, which can be archived by an orderly sequence of intricate reactions among various regulators. It is remarkable that most of the individual enzymatic activities are strictly limited to only when required. The quantitative control by ubiquitin-dependent proteasomal degradation is an essential one of such regulatory mechanisms, and quite a few proteins are targeted by an identical ubiquitin ligase, the anaphase-promoting complex or cyclosome (APC/C). To date, it has been revealed that in order to deal with the orchestrated proteolysis, APC/C sequentially alters its substrate specificity during mitosis,1,2 and it has been suggested that at least three factors contribute to this algorithm: multiple coactivators, inhibitors and mitotic checkpoint.3
Previous studies have indicated that APC/C-mediated ubiquitination requires its coactivators, which play a key role in substrate recognition. Cdc20 (fission yeast Slp1) and Cdh1 (fission yeast Ste9/Srw1) are responsible for destruction of mitotic regulators.4,5 In general, the difference between the two is the timing of interaction with APC/C: the former is assigned to activate from early mitosis to metaphase/anaphase transition, whereas the latter after that.6,7 This coactivator switching produces diversity of the substrate specificity,8 and accordingly, main targets of APC/CCdc20 are anaphase inhibitor securin and B-type cyclin. Eukaryotic cells are equipped with an APC/CCdc20 inhibitory mechanism, termed the spindle assembly checkpoint (SAC), which blocks disruption of these substrates when a kinetochore is unattached to the mitotic spindle. The core components of SAC include Mad1, Mad2, Mad3/BubR1, Bub1, Bub3 and Mps1, all of which are well conserved evolutionarily.9 SAC is therefore responsible for preventing premature sister chromatid separation and mitotic exit. During the SAC activation10,11 mitotic checkpoint complexes (MCCs) composed of Mad2, Mad3/BubR1 and Bub3 directly bind APC/CCdc20.12-16
A serious question still remains controversial: how can Cdc20 be released from MCCs and activate APC/C? It has been proposed that a cognate E2 of APC/C, UBE2C/UbcH10 (fission yeast Ubc11)-dependent Cdc20 ubiquitination and the subsequent conformational change causes its dissociation from MCCs.17 By contrast, recent studies have provided evidence that the above-mentioned modification is just a degradation signal.18-20 However, they also reached an agreement that APC/C-mediated ubiquitination reactions are involved in MCCs disassembly.
We have previously proposed that p31comet plays a key role in silencing the SAC.21 Its homologs, however, have been identified only in higher eukaryotes, suggesting the existence of more conserved regulators. Unlike metazoan,22 yeast SAC genes are non-essential.12,23-30 It is therefore conceivable that a defect in the checkpoint silencing mechanism that perturbs cell cycle progression would be suppressed by inactivation of SAC. Based on this assumption, a screen for mutants defective in SAC silencing was performed, and we isolated mutants whose temperature sensitivities are dependent on expression of Mad2. Here we report one of the mutants identified through the screen. The mutation was found on the ubc11+ gene encoding one of the two E2 enzymes collaborating with APC/C for ubiquitination at mitosis. Although SAC is satisfied in the mutant, the onset of anaphase is delayed with accumulation of Slp1, but not Cdc13 (a fission yeast homolog of cyclin B) or Cut2 (a fission yeast homolog of securin). In a temperature-sensitive mutant of the ubc4+ gene encoding the other E2 enzyme, Cdc13 and Cut2, but not Slp1, accumulated. Based on these results, we propose that E2s can determine the substrate specificity for APC/C, and that the MCCs disassembly, the vital process of silencing the SAC, can be induced by the Ubc11-dependent ubiquitination.
Results
Isolation of the ubc11-P93L mutant
As previous studies have shown, constitutive activation of the SAC leads to mitotic arrest, yet the SAC components are non-essential in fission yeast.26,27,29,30 Taking this fact as advantage, we designed a genetic screen for mutants defective in silencing the SAC with an assumption that perturbation of mitotic progression by a defect in silencing SAC can be suppressed by loss of the functional SAC. A strain conditionally expressing Mad2 was mutagenized, and the survivors were screened for mutants whose temperature sensitivity was dependent on expression of Mad2. In this study, we focused on one of the mutants identified through the screen.
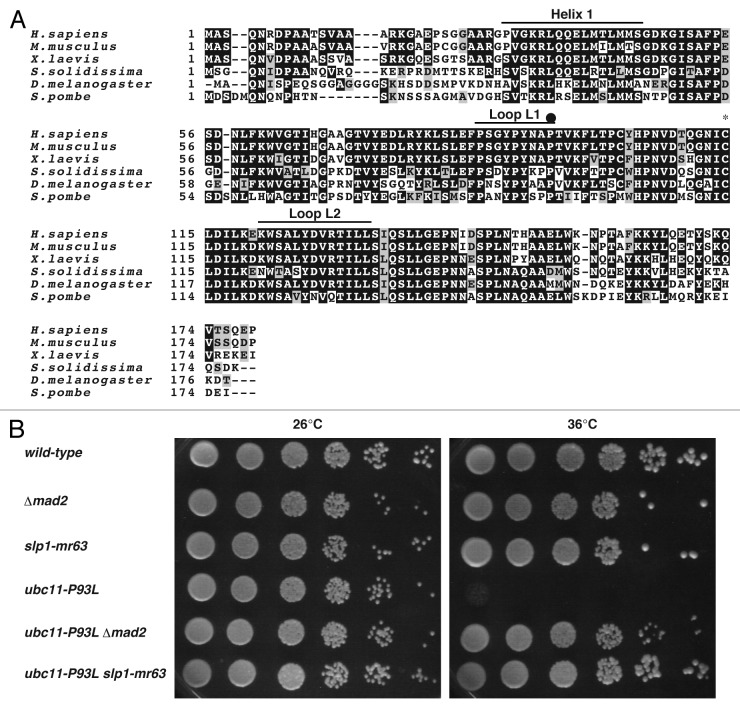
By using the temperature sensitivity in the presence of the functional SAC as a selection marker, a genomic DNA fragment complementing the temperature sensitivity was isolated from a fission yeast genomic DNA library.31,32 The integration mapping proved that this fragment originated from the ubc11 locus encoding a cognate ubiquitin-conjugating enzyme of APC/C.33 Sequencing of the mutated gene identified an amino acid residue substitution, the 93rd proline by leucine, and thereby this allele was designated ubc11-P93L (Fig. 1A). The temperature sensitivity could be suppressed by deletion of the mad2 gene as well as by introduction of slp1-mr63,34 an allele defective in binding to Mad2 (Fig. 1B), suggesting that physical interaction between Slp1 and Mad2 might cause the growth defect in the ubc11-P93L mutant.
Figure 1. Isolation of the ubc11-P93L mutant. (A) Full-length amino acid sequences of human UbcH10/UBE2C, mouse Ube2C, frog UBCx, clam E2-C, fruitfly Vihar and fission yeast Ubc11 are aligned. A closed circle indicates the mutation site of Ubc11P93L, where prorine is replaced with leucine. A cysteine residue in UBC domain for thiolester formation with ubiquitin is marked by an asterisk. (B) A wild-type strain, a strain deleted for mad2+, Δ_mad2_, slp1-mr63, ubc11-P93L, ubc11-P93L Δ_mad2_, ubc11-P93L slp1-mr63 were spotted on YEA media and were incubated at 26°C or 36°C for 3 d.
Biochemical analysis of Ubc11P93L in vivo
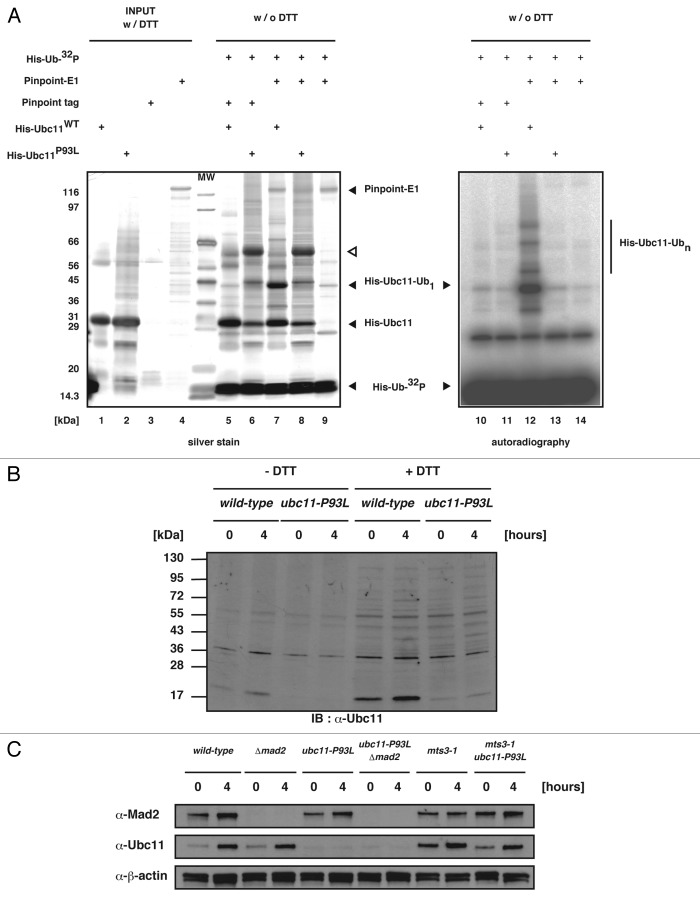
In ubiquitination, E2s are well known to play two key roles: primarily receive an activated ubiquitin from E1 ubiquitin-activating enzyme and subsequently catalyze substrate ubiquitination through interaction with E3 ubiquitin-protein ligases.35 To examine whether Ubc11P93L mutant protein maintains such general functions, we performed an in vitro ubiquitin transfer assay. Each component was prepared from E.coli as a recombinant protein (Fig. 2A). In the positive control reaction using wild-type Ubc11 (Ubc11WT), bands at approximately 45 kDa and upper molecular weight could be detected (Fig. 2A, lane 12). Because these bands were sensitive to boiling in the presence of DTT (data not shown), they were protein complexes consisting of Ubc11 and ubiquitins linked via the thiolester bond. In the case of Ubc11P93L, by contrast, there seemed to be no difference between negative controls, suggesting that the mutant was defective in transfer of activated ubiquitins from E1 (Fig. 2A, lane 13).
Figure 2. Characterization of Ubc11P93L protein. (A) The reaction mixtures for the in vitro ubiquitin transfer assay were run on SDS-PAGE and silver-stained (left panel) or dried and exposed to the X-ray film (right panel). (B and C) Western blot was performed with extracts prepared from cells incubated at 26°C or 36°C for 4 h. The genotype of each strain is indicated on the top.
We noticed that the His-tagged Ubc11P93L mutant proteins prepared from E.coli run on SDS-PAGE slower when incubated without DTT. The position on the gel (shown with the open triangle in Fig. 2A) suggested that they may form a dimer. Western blot analysis of cell extracts, however, revealed that the Ubc11P93L mutant protein existed as a monomer (Fig. 2B). We speculated that the mutant protein would be protected by a cellar factor from dimerization. In addition, western blot analysis revealed that Ubc11P93L became extremely unstable in yeast cells (Fig. 2C). Taken together, these results indicated that the level of the Ubc11-dependnet ubiquitination would be extremely low in the ubc11-P93L mutant cells.
Mitotic arrest in the ubc11-P93L mutant
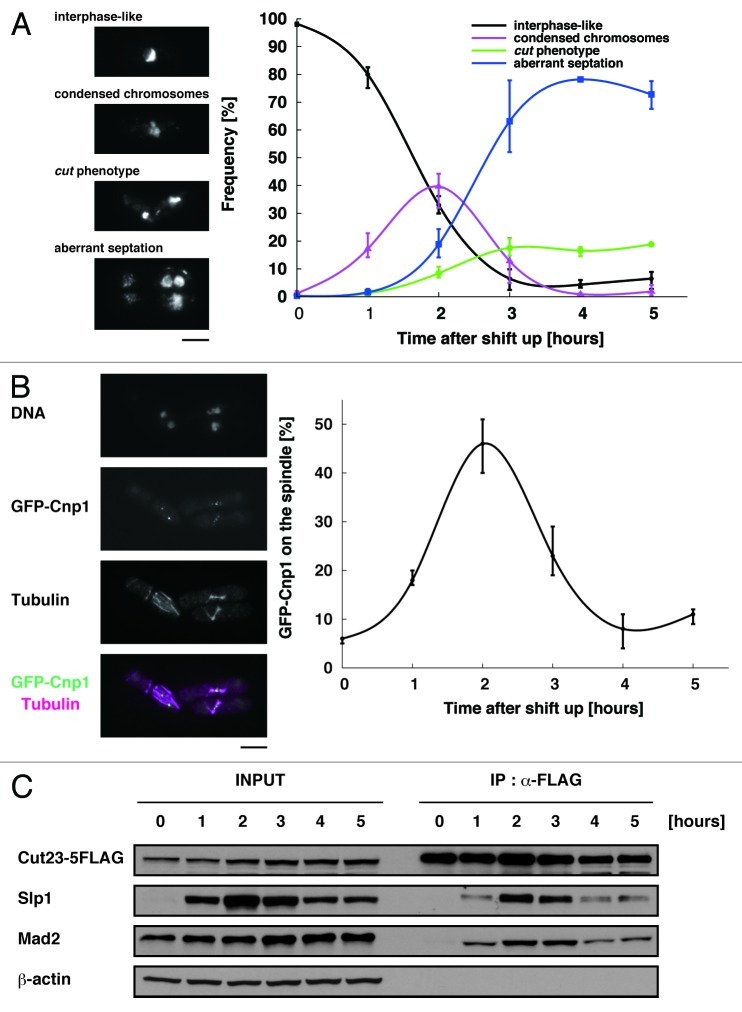
In order to determine at which stage of the cell cycle the ubc11-P93L mutant was arrested, we first examined the nuclear morphology. Two hours after the shift to the restrictive temperature, the mutant cells exhibited condensed chromosomes (Fig. 3A) and short spindle (Fig. 3B), indicating that they were arrested around metaphase. The arrest was not tight as the mutant cells thereafter exhibited cut (cell untimely torn; undivided nucleus is intersected by the septum and torn into two parts by the subsequent cytokinesis) or aberrant septation36 (Fig. 3A).
Figure 3. Mitotic arrest in the ubc11-P93L mutant. (A) The frequency of respective phenotypes of nuclear morphology: interphase-like normal phenotype, condensed chromosomes, the cut phenotype and aberrant septation are shown at each time point after the shift to the restrictive temperature. (B) The ubc11-P93L mutant cells were incubated for up to 5 h at the restrictive temperature. The sample taken at 2 h after the shift was processed for indirect immunofluorescent staining with the aniti-tubulin antibody (magenta). DNA was visualized by staining with DAPI. The position of the centromere (Cnp1-GFP, green) was also examined. The scale bar indicates 5 μm. The frequency of the cells with Cnp1-GFP on the spindle was also shown at each time point after the shift to the restrictive temperature. (C) Western blot (INPUT) and immunoprecipitation with the anti-FLAG antibody (IP: α-FLAG) were performed with extracts prepared from the ubc11-P93L cells incubated at 26°C or 36°C for up to 5 h.
We attempted to further analyze which events the defect of Ubc11 could influence, and first investigated whether kinetochore-spindle attachment was established properly. GFP-Cnp1 signals, an indicator of the position of centromere/kinetochore, were found as multiple foci no more than six, which overlapped with microtubules in 45% of the ubc11-P93L mutants shifted to the restrictive temperature for 2 h (Fig. 3B), suggesting that SAC was satisfied with establishment of kinetochore-spindle attachment in these cells.
Finally, we examined whether APC/CMCC was disassembled. Co-immunoprecipitation was performed with cell extracts prepared from a strain expressing a component of APC/C (Cut23) tagged with FLAG epitope to precipitate APC/C. As shown in Figure 3C, both Slp1 and Mad2 were bound to APC/C in cells shifted to the restrictive temperature for 2 to 3 h. The level of Slp1 both in cell extracts and immunoprecipitates with APC/C thereafter decreased, probably due to a residual activity of Ubc11 or other E2 enzymes with a low activity/specificity.
We were particularly interested in the phenotypes observed at 2 to 3 h after the shift. They would suggest that a loss of Ubc11 caused a significant delay in mitotic progression. While kinetochore-spindle attachment was established, SAC was not silenced. Considering together with the fact that the temperature sensitivity was suppressed by loss of SAC, we speculated that the delay in the mutant might be caused a defect in silencing SAC.
Specific requirement of Ubc11 for destruction of Slp1
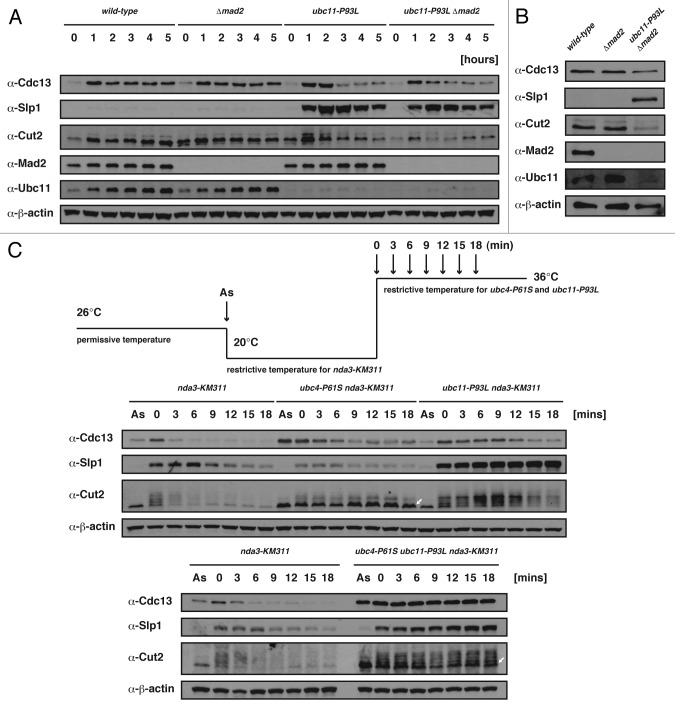
A previous study proposed that APC/C cooperates with at least two E2s, fission yeast Ubc11 and Ubc4, each of which has an essential and incompatible role in polyubiquitin-chain formation.37 The B-type cyclin/Cdc13 and securin/Cut2 cause a mitotic arrest if not properly degraded in the APC/C-dependent manner.38-41 We therefore attempted to examine how the ubc11-P93L mutation affects the stability of proteins normally degraded in the APC/C-dependent manner. The cellular levels of Slp1, Cdc13 and Cut2 were first monitored in the ubc11-P93L mutant. As shown in Figure 4A, the level of Slp1 began to increase and reached a peak 2 h after the shift to the restrictive temperature. The other two proteins, Cdc13 and Cut2, transiently accumulated after the shift, but were degraded much more rapidly than Slp1. Remarkably, Slp1 also accumulated in the double mutant (ubc11-P93L Δ_mad2_) that could grow at the restrictive temperature. We also examined the stability of the three proteins in the double mutant grown asynchronously at 36°C for 10 h and found that Slp1, but not the other two proteins, abnormally accumulated (Fig. 4B). These results suggested that Cdc13 and Cut2 could be ubiquitinated and thereafter degraded in the ubc11-P93L mutant, whereas Slp1 could not.
Figure 4. Stability of Slp1 in the ubc11-P93L mutant. (A) Western blot was performed with extracts prepared from cells incubated at 26°C or 36°C for up to 5 h. The genotype of each strain is indicated on the top. (B) Western blot was performed with extracts prepared from cells incubated at 36°C for 10 h. The genotype of each strain is indicated on the top. (C) Each strain was arrested by the shift to the restrictive temperature for the nda3-KM311 mutation (20°C) for 6 h and released to 36°C at time 0. Cell extracts were prepared at indicated time points and processed for western blotting with each antibody. The genotype of each strain is indicated on the top. Extracts prepared from an asynchronous culture (As) were also examined in the same way.
To further understand the relationship between the three substrates and two E2s, we have examined the levels of Cdc13, Slp1 and Cut2 at the transition from prophase to anaphase. The cold-sensitive nda3-KM311 mutation defective in the spindle formation42 was introduced into each of the ubc11-P93L mutant, ubc4-P61S mutant37 and ubc4-P61S ubc11-P93L double mutant, respectively. In the nda3-KM311 single mutant after incubation at its restrictive temperature (20°C), the three proteins accumulated. Upon the shift from 20°C to 36°C, they were mostly degraded within 9 min (Fig. 4C). The level of Cdc13, which gradually decreased in each of the ubc4-P61S nda3-KM311 and ubc11-P93L nda3-KM311 double mutant, was stably maintained in the triple mutant (ubc4-P61S ubc11-P93L nda3-KM311). The result would suggest that degradation of Cdc13 requires either one of the two E2s (though it proceeds more rapidly in the presence of the two E2s). While the level of Slp1 decreased in the ubc4-P61S nda3-KM311 double mutant, it was stable in both the double mutant (ubc11-P93L nda3-KM311) and the triple mutant, reinforcing that Ubc11 is largely responsible for degradation of Slp1. In the single nda3-KM311 mutant at time point 0, the band representing Cut2, which was observed as a ladder,43,44 rapidly disappeared after the shift to 36°C. In the ubc4-P61S nda3-KM311 double mutant, Cut2 was stable largely as a single band (indicated with a white arrow in Fig. 4C). The Cut2-ladder persisted in the ubc11-P93L nda3-KM311 double mutant, though it was degraded to some extent at later time points. In the triple mutant, both of the two forms, the ladder and the single band, of Cut2 remained stable, suggesting that each of the two E2 would take a separate responsibility for degradation of Cut2. The analysis demonstrated the substrate specific roles of the two E2s and the specific requirement of Ubc11 for degradation of Slp1.
Ubc11-dependent ubiquitination of Slp1
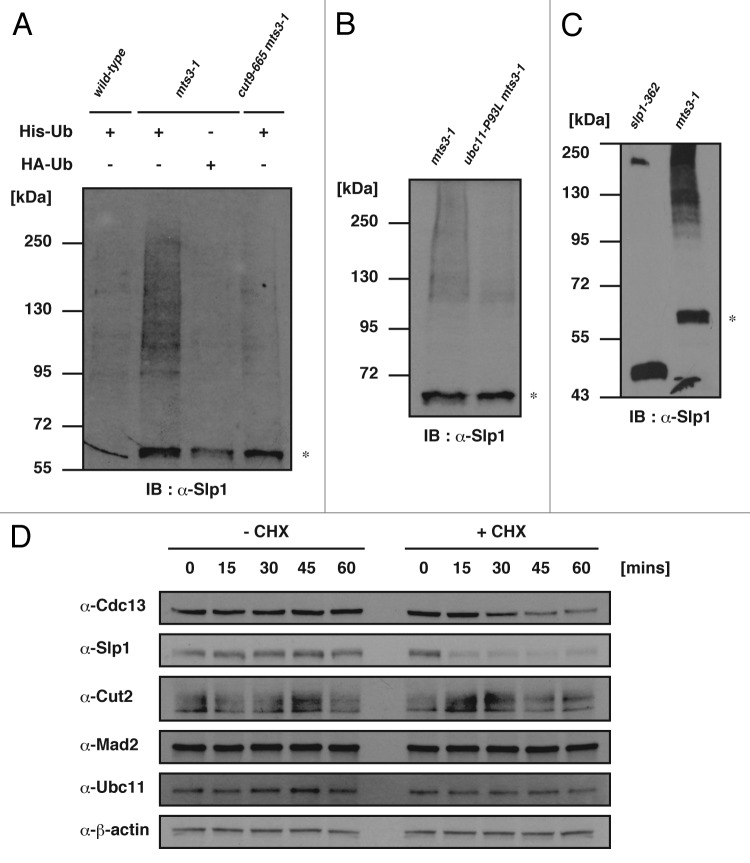
Although previous studies showed that Cdc13 and Cut2 could be ubiquitinated dependently on APC/C,45-47 no evidence was presented for ubiquitination of Slp1 in fission yeast. We therefore attempted to test if Slp1 was ubiquitinated by following an assay previously reported.48 His6-tagged ubiquitin (His-Ub) was ectopically expressed in the following three strains, the wild-type, mts3-1 mutant defective in 26 S proteasome function49 and cut9–66550 mts3-1 double mutant defective in APC/C and 26 S proteasome function. Four hours after the shift to the restrictive temperature (36°C) for the mts3-1 and cut9–665 mutations, His-Ub and its conjugates were purified from the respective cell extracts with the metal beads, and the presence/status of Slp1 was examined by immunoblot with the anti-Slp1 antibody. The mts3-1 mutant expressing HA-tagged ubiquitin (HA-Ub) was also examined as a negative control. As shown in Figure 5A, in the extracts of the mts3-1 mutant expressing His-Ub, we detected a smearing band above 95 kDa. This band could not be detected in the double mutant (cut9–665 mts3-1) expressing His-Ub, suggesting it was produced dependently on the functional APC/C. We also found it was dependent on Ubc11 (Fig. 5B). In order to examine the identity of the band appeared at the position slightly above 55 kDa (indicated by the asterisk in Fig. 5A and B), we prepared cell extracts from the slp1–362 mutant,51 and the metal beads-bound fraction was analyzed by immunoblot with the anti-Slp1 antibody. The mutant expresses a truncated Slp1 protein with the molecular weight of approximately 43 kDa due to a single nonsense mutation, which is defective in interaction with APC/C.52,53 As shown in Figure 5C, in the cell extracts prepared from the slp1–362 mutant, we detected a band at the position of 43 kDa, but not around 55 kDa in the metal beads-bound fraction, indicating that the asterisk in Figure 5A and B represented the wild type Slp1. Furthermore, the smearing band above 95 kDa was not detectable in the metal beads-bound fraction of extracts prepared from the slp1–362 mutant. We speculated that the Slp1–362 mutant protein was not a good substrate due to a lack of interaction with APC/C. Taken together, these results would suggest that Slp1 can be ubiquitinated in an APC/C- and Ubc11-dependent manner and subsequently degraded by the 26 S proteasome.
Figure 5. Ubiquitination of Slp1. (A and B) Cell extracts were prepared from cultures first incubated at 26°C and then shifted up to 36°C for 4 h. Ubiquitinated proteins were purified and analyzed by immunoblotting with the Slp1 antibody. Note that the band likely representing nonubiquitinated Slp1, indicated by asterisk, can also be detected in the pull-down samples, likely due to a His-residue cluster within the Slp1 protein. (C) The cell extracts were prepared and processed as in (A). (D) The nda3-KM311 mutant cells were first incubated at 20°C for 6 h. Cycloheximide was added to the media at time 0 and continuously incubated for up to 60 min. Cell extracts were processed for immunoblot at each time point.
Previous studies provided evidence that APC/C-mediated ubiqutination and the following proteasomal degradation of Cdc20 occurs even during the SAC activation.18,54 To test whether this is also the case for fission yeast Slp1, its stability was examined in the nda3-KM311 mutant cells arrested at the restrictive temperature due to SAC. In the presence of 100 μg/ml cycloheximide, an inhibitor of protein synthesis, the level of Slp1 rapidly decreased, while other proteins, including Cdc13 and Cut2, were more stable (Fig. 5D), indicating that as well as its homologs, Slp1 could be quantitatively regulated via the ubiquitin-dependent proteasomal proteolysis during the SAC activation.
Discussion
To date it has been proposed that APC/C cooperates with two ubiquitin-conjugating enzymes, Ubc11 and Ubc4 in fission yeast, both of which are individually required for polyubiquitin-chain formation at different steps; the former for initiation and the latter for elongation. Based on this prevailing model, a defect of either of the two E2s can be sufficient for stabilization of APC/C substrates including B-type cyclin/Cdc13, securin/Cut2 and Cdc20/Slp1. In the present study, however, we reported that a loss of Ubc11 could inhibit the degradation of Slp1 among the three substrates. These results therefore imply an additional feature of the two E2 enzymes: an active selection of substrates for APC/C.
What makes the difference between Ubc11- and Ubc4-mediated ubiquitination? Valid speculations can be led from researches about ubiquitin topology. According to them, UbcH10 (a homolog of Ubc11 in higher eukaryotes) preferentially forms polyubiquitin-chain linked through K11, while such bias there cannot be observed in Ubc4/UbcH5-mediated ubiquitination.55,56 X-ray crystallography indeed revealed that the K11-linked, K48-linked and K63-linked polyubiquitin-chains adopt distinct structures, respectively.57 Given this, our experimental data can be interpreted that Ubc4 also has the ability to ubiquitinate Slp1, but the chain cannot become targeted for proteasomal proteolysis. On the other hand, a recent study has suggested that multiple monoubiquitination can also become a degradation signal, at least for Cyclin B1.58
A previous report intriguingly provided evidence that the N-terminal extension unique to UbcH10, a striking difference between the two E2s, restricts both polyubiquitination and multiubiquitination.59 Ectopic expression of UbcH10 lacking the N-terminal extension overrode an arrest imposed by SAC, suggesting that the N-terminal extension of UbcH10 might play a role in restricting ubiquitination activity when SAC is activated. In any case, it is probable that the competition and/or collaboration between the two E2s can result in producing diversity of ubiquitination kinetics and apparent substrate specificity.
Finally, we demonstrated that the defect of Ubc11 retained Slp1 bound to Mad2 even when the SAC was satisfied. Whether the ubiquitination of Slp1/Cdc20 triggers MCCs disassembly still remains controversial among various studies, though it has been generally accepted that APC/C-dependent ubiquitination reactions is required for MCCs disassembly.17-19,60,61 Our findings would suggest that it is the Ubc11-mediated ubiquitination that can trigger MCCs disassembly.
Materials and Methods
Yeast strains, media and transformation
The strains used in this study are derivatives of Schizosaccaharomyces pombe h− 972 and h+ 975.62 The Δ_mad2_, slp1-mr63, mts3-1, GFP-cnp1, nda3-KM311, cut9-665, slp1-362, his5-303 and lys1-131 are our laboratory stock.34,42,49-51,63,64 The mutant ubc4-P61S was a generous gift from Dr Hiroaki Seino.37 Yeast transformations were performed by lithium acetate methods.65,66 Total RNAs were prepared as previously described.67 EMM was used as a minimal medium, and YEA62 as a rich medium except for nda3-KM311 mutant strains, which were grown in YPAD medium. For spot assays, exponentially growing cultures washed twice with distilled water were serially diluted by the factor of 5 and spotted onto agar plates.
Isolation of ubc11-P93L mutant
A fission yeast strain, RBC115 (h− leu1-32 mad2::ura4::nmt1-mad2),68 was constructed by replacing the ura4+ gene inserted at the mad2+ locus with the open reading frame of mad2+ franked by the nmt1 promoter and terminator. RBC115 was chemically mutagenized as described previously,69 and the survivors were screened as the following two criteria: (1) to exhibit a temperature sensitivity only when expression of Mad2 was turned on; and (2) to be arrested at mitosis after the shift to the restrictive temperature. Twenty-eight mutants, including the ubc11-P93L mutant, were isolated among approximately 200,000 survivors.
FLAG tagging of Cut23
An about 0.4-kbp DNA fragment of C-terminus region of cut23+ was amplified by PCR. The resulting _Pvu_II-_Bgl_II DNA fragment was digested with appropriate restriction enzymes and then inserted into the _Pvu_II-BamHI site of pFA6a-5FLAG-kanMX6 (generously provided by Dr Jun-ichi Nakayama). An about 1.2-kbp DNA fragment downstream of the stop codon of cut23+ was also amplified by PCR. The resulting _Sac_I-_Spe_I DNA fragment was digested with appropriate restriction enzymes and then inserted into the _Sac_I-_Spe_I site of pFA6a-3HA-His3MX6.70 Both of the resultant plasmids were digested with _Pvu_II and _Bgl_II, and an about 0.8-kbp DNA fragment encoding a part of C-terminus region of FLAG-tagged Cut23 and yADH terminator from the former was inserted into the _Pvu_II-_Bgl_II site of the latter. The resulting plasmid, pFA6a-Cut23-5FLAG-His3MX6, was digested with _Bam_HI and introduced into the wild-type his5-303 strain. His+ transformants were isolated. The gene replacement was confirmed by PCR and immunoblotting.
Strains expressing tagged ubuiquitin
For construction of strains expressing tagged ubiquitin, an integration plasmid containing a part of C-terminus region of lys1+ gene as a selection marker, pYAT1 was employed. The DNA fragment encoding the His-tag was obtained from pREP1(6His-Ubi), and those encoding tif51 promoter, HA-tag or ubiquitin were obtained from Ptif51(HA-Ubi)-LEU2. These two plasmids were generously provided by Dr Kenji Kitamura. The _Kpn_I-_Sac_I DNA fragments encoding tif51 promoter, tagged ubiquitin and the nmt1 terminator were inserted into the _Kpn_I-_Sac_I site of pYAT1. The resulting plasmids were introduced into the wild-type lys1-131 strain and Lys+ transformants were isolated.
Ubiquitin transfer assay
mRNA of Ptr3, a fission yeast ubiquitin activating enzyme E1, was reverse-transcribed by use of ReverTra Ace (TOYOBO), and then the resulting cDNA was amplified by PCR. The resulting _Bam_HI-_Not_I DNA fragment was digested with appropriate restriction enzymes and then inserted into _Bam_HI-_Not_I site of PinPointXa-1 vector (Promega) in-frame with biotinylated purification protein tag sequence. Ubc11 mRNAs were also reverse transcribed by use of ReverTra Ace, and then the resulting cDNA was amplified by PCR. The resulting _Eco_RI-_Xho_I DNA fragment was digested with combinations of appropriate restriction enzymes and then inserted into _Eco_RI-_Xho_I site of pET30a vector (Promega) in-frame with Histidine tag sequence. To obtain 32P-labeled ubiquitin protein, protein kinase A phosphorylation site (Arg-Arg-Ala-Ser-Val) was introduced into pET30a in-frame with 6× Histidine tag sequence (pET30a KT). Yeast ubiquitin was amplified by PCR using the plasmid Ptif51(HA-Ubi)-LEU2 as a template. The resulting DNA fragment was digested with combinations of appropriate restriction enzymes, and then cloned into pET30a KT in-frame with 6× Histidine and PKA phosphorylation site. All the fusion proteins were, respectively, expressed in E.coli BL21(DE3) strain and directly purified with the following resin according to the manufacturer’s protocol: the biotinylated purification tagged Ptr3 and the biotylated purification tagged protein with SoftLink soft release avidin resin (Promega), or the His-tagged Ubc11 and His-PKA site tagged ubiquitin with TALON Metal Affinity Resin (Clontech). In vitro phosphorylation reaction was performed by cAMP-dependent kinase-catalytic subunit [20 mM TRIS-HCl, 100 mM NaCl, 12 mM MgCl2, 1 mM DTT, 25% (v/v) glycerol, pH 7.4, 2.4 units/μl cAMP-dependent kinase-catalytic subunit (Promega), 1.5 μCi/μl (γ-32P)ATP, 0.5 μg/μl His-Ubc11 purified protein] at 25°C for 30 min. The labeling mixture was directly loaded onto BioSpin-P6 column (Bio-Rad) pre-equilibrated with incubation buffer [20 mM TRIS-HCl, 100 mM NaCl, 12 mM MgCl2, 1 mM DTT, 25 %(v/v) glycerol, pH 7.4] and the flow-through fraction was collected and calibrated with autoradiography. Two hundred cpm/protein μg were used for thioester formation reaction. Thioester formation reactions contained 0.5 μg of 32P-labeled His ubiquitin, 0.1 μg of Pinpoint-Ptr3 or PinPoint tag protein and 0.5 μg of His-Ubc11WT or Ubc11P93L in 4 mM TRIS-HCl, 0.5 mM MgCl2, 0.2 mM ATP, pH 7.6) at 25°C, for 30 min. Reactions were terminated by incubating the mixtures for 10 min at 25°C in 50 mM TRIS-HCl pH6.8, 2% SDS, 10% glycerol and 0.5% BPB or boiling the mixtures for 3 min in 50 mM TRIS-HCl pH6.8, 2% SDS, 10% glycerol, 1 mM DTT and 0.5% BPB. The duplicated gels were stained by silver-staining protocol or take the autoradiogram on X-ray film.
Fluorescent microscopy
Indirect immunofluorescent staining was performed as previously described,30 except that Alexa Fluor 594 Goat Anti-mouse (1:1,000; Molecular Probes) was employed as a secondary antibody. Images were acquired with a laser-scanning microscope (Leica) and processed with IP lab (BD) and Photoshop version 12.0.4 (Adobe).
Proteins analysis
Cells were collected, washed once with PBS, rapidly frozen by liquid nitrogen and kept at −80°C. Cells were thawed, washed once with below-mentioned buffers and thereafter suspended in such buffers, including protease/proteasome inhibitors [1 mM PMSF, 10 μM MG132 and 1% (v/v) of the final concentration of protease inhibitor cocktail (nacalai tesque)]. Cells were lysed by glass beads and centrifuged at 13,000 rpm for 15 min. Supernatants were used for analyses. For immunoprecipitation, modified HB buffer [25 mM TRIS-HCl, (pH 7.5), 15 mM EGTA, 15 mM MgCl2, 60 mM β-glycerophosphate, 15 mM ρ-nitrophenylphosphate, 0.5 mM Na3VO4, 10 mM N-ethylmaleimide, 0.1% NP-40, 0.1 mM NaF] was used. Soluble proteins were incubated with Anti-FLAG M2 Affinity Gel (Sigma), followed by SDS-PAGE. Cell lysates were manipulated at 4°C. For investigation of protein stabilities, after wash with modified HB buffer cells were suspended in modified HB buffer containing Laemmli sample buffer and boiled for 5 min before cell disruption. Cell extracts were manipulated at room temperature. Detection of ubiquitinated Slp1 was performed as described previously,48 except that tagged ubiquitins were overproduced from tif51 promoter71 integrated at the lys1 locus, and thereby cells were cultured in rich media. Antibodies were used as follows: anti-Slp1 (1:500), anti-Mad2 (1:50), anti-Cdc13 (1:200; Santa Cruz Biotechnology), anti-Cut2 (1:3,000; generously provided from Dr Mitsuhiro Yanagida), anti-Ubc11 (1:200; generously provided by Dr Fumiaki Yamao), anti-FLAG M2 (1:1,000; Sigma) and anti-β-actin (1:8,000; Abcam).
Acknowledgments
The authors greatly appreciate Drs Andrea Baines, Kenji Kitamura, Taro Nakamura, Jun-ichi Nakayama, Hiroaki Seino, Chikashi Shimoda, Fumiaki Yamao, Mitsuhiro Yanagida and National BioResource Project for reagents, Drs Hiro Yamano, Tadashi Uemura, Osamu Chisaka, Jun Takeda and Tatsuki Kunoh for discussion and Ms Kyoko Matsui for technical assistance. This work was supported by a grant from Ministry of Education, Culture, Sports, Science and Technology of Japan to T.M.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
References
- 1.Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–43. doi: 10.1016/S1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 2.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 4.Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–93. doi: 10.1016/S0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 5.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–3. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 6.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–71. doi: 10.1016/S1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 7.Kramer ER, Gieffers C, Hölzl G, Hengstschläger M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–10. doi: 10.1016/S0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 8.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–65. [PMC free article] [PubMed] [Google Scholar]
- 9.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 10.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–10. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–8. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–82. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–67. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–13. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 17.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–5. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–20. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia L, Li B, Warrington RT, Hao X, Wang S, Yu H. Defining pathways of spindle checkpoint silencing: functional redundancy between Cdc20 ubiquitination and p31(comet) Mol Biol Cell. 2011;22:4227–35. doi: 10.1091/mbc.E11-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13:1234–43. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habu T, Kim SH, Weinstein J, Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21:6419–28. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/S0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 23.Hoyt MA, Totis L, Roberts BTS. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–17. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 24.Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–91. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–20. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–70. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernard P, Hardwick K, Javerzat JP. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J Cell Biol. 1998;143:1775–87. doi: 10.1083/jcb.143.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–18. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol Cell Biol. 2002;22:2728–42. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikui AE, Furuya K, Yanagida M, Matsumoto T. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J Cell Sci. 2002;115:1603–10. doi: 10.1242/jcs.115.8.1603. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, et al. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–69. doi: 10.1128/MCB.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura T, Nakamura-Kubo M, Hirata A, Shimoda C. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1(+)-encoding syntaxin-like protein. Mol Biol Cell. 2001;12:3955–72. doi: 10.1091/mbc.12.12.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osaka F, Seino H, Seno T, Yamao F. A ubiquitin-conjugating enzyme in fission yeast that is essential for the onset of anaphase in mitosis. Mol Cell Biol. 1997;17:3388–97. doi: 10.1128/mcb.17.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–7. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 35.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagida M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 1998;8:144–9. doi: 10.1016/S0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]
- 37.Seino H, Kishi T, Nishitani H, Yamao F. Two ubiquitin-conjugating enzymes, UbcP1/Ubc4 and UbcP4/Ubc11, have distinct functions for ubiquitination of mitotic cyclin. Mol Cell Biol. 2003;23:3497–505. doi: 10.1128/MCB.23.10.3497-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–8. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 39.Yamano H, Gannon J, Hunt T. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 1996;15:5268–79. [PMC free article] [PubMed] [Google Scholar]
- 40.Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–28. [PMC free article] [PubMed] [Google Scholar]
- 41.Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, et al. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–87. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–58. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 43.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature. 1996;381:438–41. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 44.Aoki K, Nakaseko Y, Kinoshita K, Goshima G, Yanagida M. CDC2 phosphorylation of the fission yeast dis1 ensures accurate chromosome segregation. Curr Biol. 2006;16:1627–35. doi: 10.1016/j.cub.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, et al. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature. 1996;384:276–9. doi: 10.1038/384276a0. [DOI] [PubMed] [Google Scholar]
- 46.Berry LD, Feoktistova A, Wright MD, Gould KL. The schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function. Mol Cell Biol. 1999;19:2535–46. doi: 10.1128/mcb.19.4.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon HJ, Feoktistova A, Wolfe BA, Jennings JL, Link AJ, Gould KL. Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr Biol. 2002;12:2048–54. doi: 10.1016/S0960-9822(02)01331-3. [DOI] [PubMed] [Google Scholar]
- 48.Takayama Y, Mamnun YM, Trickey M, Dhut S, Masuda F, Yamano H, et al. Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell. 2010;18:385–96. doi: 10.1016/j.devcel.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon C, McGurk G, Wallace M, Hastie ND. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem. 1996;271:5704–11. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- 50.Hirano T, Funahashi S, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–9. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto T. A fission yeast homolog of CDC20/p55CDC/Fizzy is required for recovery from DNA damage and genetically interacts with p34cdc2. Mol Cell Biol. 1997;17:742–50. doi: 10.1128/mcb.17.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada HY, Matsumoto S, Matsumoto T. High dosage expression of a zinc finger protein, Grt1, suppresses a mutant of fission yeast slp1(+), a homolog of CDC20/p55CDC/Fizzy. J Cell Sci. 2000;113:3989–99. doi: 10.1242/jcs.113.22.3989. [DOI] [PubMed] [Google Scholar]
- 53.Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, Chen JS, et al. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol Cell. 2007;28:871–85. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–51. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–10. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 56.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–65. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–84. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–76. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–56. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foster SA, Morgan DO. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol Cell. 2012;47:921–32. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19:1116–23. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 63.Cottarel G. The Saccharomyces cerevisiae HIS3 and LYS2 genes complement the Schizosaccharomyces pombe his5-303 and lys1-131 mutations, respectively: new selectable markers and new multi-purpose multicopy shuttle vectors, pSP3 and pSP4. Curr Genet. 1995;28:380–3. doi: 10.1007/BF00326437. [DOI] [PubMed] [Google Scholar]
- 64.Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell. 2008;19:682–90. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–9. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen R, Sprague GF, Jr., Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci USA. 1983;80:3035–9. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ito D, Saito Y, Matsumoto T. Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1. Proc Natl Acad Sci USA. 2012;109:209–14. doi: 10.1073/pnas.1114647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsumoto T, Beach D. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell. 1991;66:347–60. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- 70.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 71.Matsuyama A, Shirai A, Yoshida M. A series of promoters for constitutive expression of heterologous genes in fission yeast. Yeast. 2008;25:371–6. doi: 10.1002/yea.1593. [DOI] [PubMed] [Google Scholar]