Intestinal inflammation targets cancer-inducing activity of the microbiota (original) (raw)
. Author manuscript; available in PMC: 2013 May 6.
Published in final edited form as: Science. 2012 Aug 16;338(6103):120–123. doi: 10.1126/science.1224820
Abstract
Inflammation alters host physiology to promote cancer, as seen in colitis-associated colorectal cancer (CRC). Here we identify the intestinal microbiota as a target of inflammation that impacts the progression of CRC. High-throughput sequencing revealed that inflammation modifies gut microbial composition in colitis-susceptible interleukin-10-deficient (_Il10_−/−) mice. Monocolonization with the commensal Escherichia coli NC101 promoted invasive carcinoma in azoxymethane (AOM)-treated _Il10_−/− mice. Deletion of the polyketide synthase (pks) genotoxic island from E. coli NC101 decreased tumor multiplicity and invasion in AOM/_Il10_−/− mice, without altering intestinal inflammation. Mucosa-associated pks+ E. coli were found in a significantly high percentage of inflammatory bowel disease (IBD) and CRC patients. This suggests that in mice, colitis can promote tumorigenesis by altering microbial composition and inducing the expansion of microorganisms with genotoxic capabilities.
Chronic inflammation is a well-established risk factor for several cancers, including colorectal cancer (CRC) (1). Although the mechanism by which chronic intestinal inflammation leads to CRC is still unclear, numerous experimental studies suggest that inflammatory cells and their associated mediators such as interleukin-6 (IL-6), tumor necrosis factor-α (TNF- α), IL-23, and reactive oxygen species form a microenvironment favoring the development of CRC, presumably by enhancing DNA damage in epithelial cells (2–4).
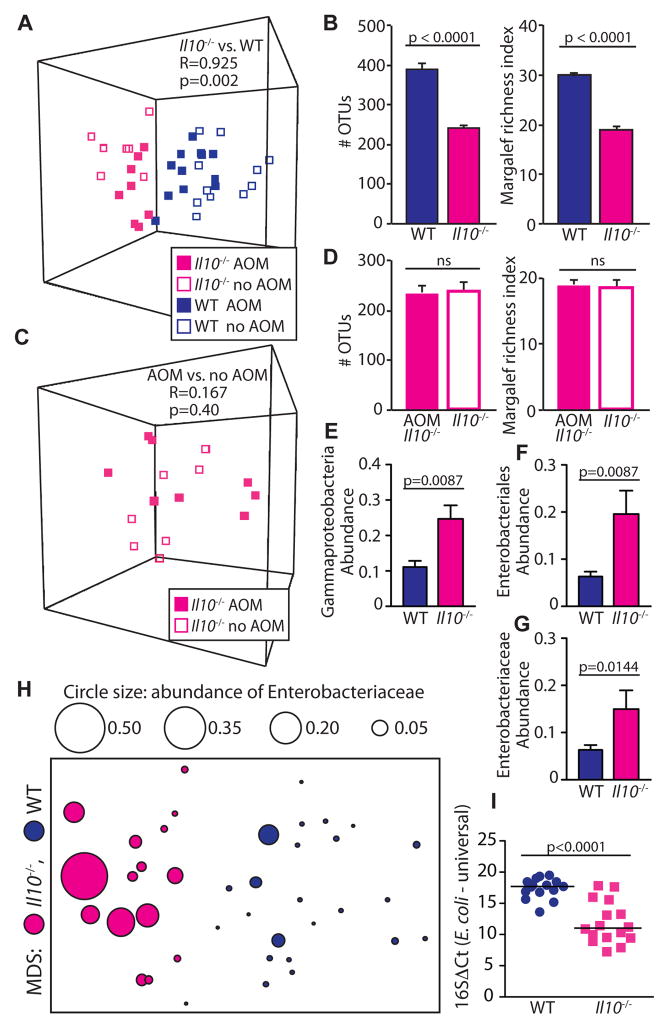
In the colon, trillions of commensal bacteria, termed the microbiota, are in close proximity to a single layer of epithelial cells. A critical question is whether these microorganisms actively participate in the process of carcinogenesis. We have previously shown that microbial status modulates development of colitis-associated CRC using the colitis-susceptible _Il10_−/− mouse strain (5). To evaluate the effect of inflammation and carcinogenesis on the colonic microbiota, we utilized Illumina HiSeq2000 sequencing targeting the hypervariable V6 region of the 16S rRNA gene in mucosal biopsies and stool samples of _Il10_−/− and WT mice, in the presence and absence of the colon-specific carcinogen AOM. Germ-free (GF) _Il10_−/− and control WT adult mice were transferred to specific pathogen free (SPF) conditions for 20 weeks. During this timeframe, 100% of _Il10_−/− mice develop colitis; with the addition of AOM, 60–80% of mice develop colon tumors (5). WT mice develop neither colitis nor tumors (5). We first compared the luminal microbiota between all _Il10_−/− (colitis/cancer) and WT mice (healthy control) and found that the microbiota of _Il10_−/− mice clustered apart from those of WT controls (Fig 1A, Fig. S1, ANOSIM (Analysis of Similarity) R=.925, p=0.002). The altered microbiota of _Il10_−/− mice showed reduced richness compared to WT controls (Fig 1B, p<0.0001). Analysis of mucosal biopsies revealed the colonic-adherent microbiota of AOM/_l10_−/− mice with colitis/cancer clustered apart from healthy AOM/WT controls (Fig S2A), with alterations in microbial evenness but not richness (Fig S2B, p=0.023). To determine the impact of AOM on the microbiota in the context of inflammation, we compared the luminal microbiota of _Il10_−/− mice with colitis to AOM/_Il10_−/− mice with colitis/cancer and found that AOM treatment had no significant effect on luminal microbial composition or richness in _Il10_−/− mice (Fig 1C, D). These data suggest that inflammation rather than cancer is associated with the observed microbial shifts.
Fig 1.
Inflammation alters fecal microbial community structure. (A–B) Luminal microbiota of _Il10_−/− vs. WT mice. (A) Operational taxonomic unit (OTU) abundances were standardized by total, square root transformed, and assembled into a Bray Curtis similarity matrix to generate a multidimensional scaling (MDS) plot, where in these plots each symbol represents the microbiota of an individual mouse analyzed by Illumina sequencing of 16S V6 region. _Il10_−/− vs. WT comparison by ANOSIM (Analysis of Similarity), R=1 is maximum dissimilarity. (B) Richness, mean + SEM of cage means, 5–6 cages/group, 2–4 mice per cage, t test. (C–D) Luminal microbiota of AOM/_Il10_−/− vs. _Il10_−/− (C) MDS plot, AOM/_Il10_−/− vs. _Il10_−/−comparison by ANOSIM. (D) Richness, mean + SEM of cage means, 2–3 cages/group, 2–4 mice per cage, t test. (E–G) Standardized transformed abundance, median + SEM of cage means, 5–6 cages/group, 2–4 mice per cage, Mann Whitney U. (H) MDS plot depicting luminal microbiota of _Il10_−/− vs. WT, overlaid with Enterobacteriaceae abundance depicted by circle size. (I) E. coli ΔCt relative to total bacteria (16S). Each symbol depicts one mouse, line at median, Mann Whitney U.
We then hypothesized that inflammation-induced changes in microbial composition includes the expansion of bacteria within the Proteobacteria phylum, as several members have been associated with colitis and CRC (6–9). Analysis of phylum-level distribution revealed that inflammation in _Il10_−/− mice was associated with significantly increased levels of luminal Verrucomicrobia, Bacteroidetes, and Proteobacteria compared to WT controls (Fig S3). Although Verrucomicrobia significantly differed between groups, this phylum is not well characterized, restricting detailed molecular analysis. Within Proteobacteria, however, the Gammaproteobacteria class, Enterobacteriales order, and Enterobacteriaceae family were all significantly more abundant in _Il10_−/− mice (Fig 1E–H). Because E. coli are members of the family Enterobacteriaceae and adherent-invasive E. coli have been associated with human IBD and CRC (8, 10–13), we determined by quantitative PCR if E. coli was more abundant in the context of inflammation in _Il10_−/− mice. We found that relative to WT mice, the luminal microbiota of _Il10_−/− mice exhibited a ~100-fold increase in E. coli (Fig 1I). AOM treatment did not affect E. coli abundance (Fig S4A). Total bacterial loads between WT and _Il10_−/− mice did not differ (Fig S4B), nor did levels of the common commensal Lactobacillus (Fig S4C).
To determine the causative effect of commensal E. coli on CRC, we administered AOM to GF _Il10_−/− mice mono-associated with either the commensal murine adherent-invasive E. coli NC101 or the human commensal Enterococcus faecalis OG1RF, both of which cause aggressive colitis in _Il10_−/− mice (14). As expected, both E. coli NC101 and E. faecalis mono-associated, AOM-treated _Il10_−/− mice developed severe colitis (Fig 2A). Despite similar levels of colitis, 80% of E. coli mono-associated mice developed invasive mucinous adenocarcinoma, whereas E. faecalis mono-associated mice rarely developed tumors (Fig 2B–D). Colonic cytokines involved in inflammation and carcinogenesis including Il6, Tnfa, Ifng, Il1b, Il18, Il17 and Il23 were not significantly different between AOM-treated E. coli and E. faecalis mono-associated mice (Fig 2E, S5, Table S1–2). In addition, infiltrating CD3+ T cells, F4/80+ macrophages, and Ly6B.2+ monocytes and neutrophils were similar between AOM-treated E. coli and E. faecalis mono-associated mice (Fig. S6). These observations demonstrate that in addition to inflammation, bacteria-specific factors may be required for the development of colitis-associated CRC.
Fig 2.
Colonization of germ-free mice Il10−/− mice with E. coli (E.c.) or E. faecalis (E.f.) differentially impacts tumorigenesis without affecting inflammation. (A) Histologic inflammation scores, t test. (B) Macroscopic tumor counts; two tailed Mann Whitney. (C) Percent of mice with invasive adenocarcinoma; Fisher’s exact test. (D) Representative H&E histology. (A–D): Mean + SEM, 7–12/group, single experiment. (E) Colonic cytokine mRNA expression relative to GF _Il10_−/−; Mean + SEM, 4/group, t test.
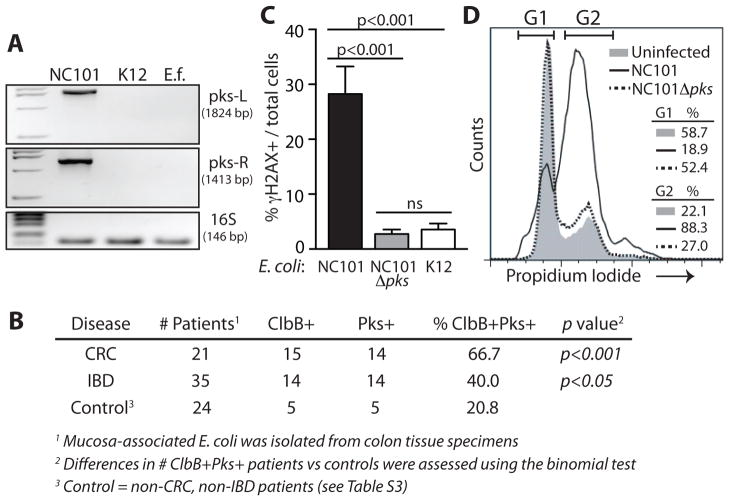
We hypothesized that E. coli NC101 has carcinogenic capabilities not shared by E. faecalis. Several members of the family Enterobacteriaceae, including select E. coli strains of B2 phylotype, harbor a ~54kb polyketide synthases (pks) pathogenicity island that encodes multi-enzymatic machinery for synthesizing a peptide-polyketide hybrid genotoxin named Colibactin (15–18). A bioinformatics BLAST search of the E. coli NC101 genome (accession NZ_AEFA00000000) revealed the presence of pks and the absence of other known E. coli genotoxins Cif, CNF and CDT. Using PCR and sequencing (15), we detected the pks island in E. coli NC101, but not E. faecalis or non-colitogenic E. coli K12 (Fig 3A). To determine if E. coli pks is associated with human CRC or IBD, we screened mucosa-associated E. coli strains isolated from colorectal tissue specimens of 35 patients with IBD, 21 with CRC, and 24 non-IBD/non-CRC controls (11). CRC specimens could not be obtained from IBD-associated CRC patients as these patients typically undergo colectomy upon diagnosis of dysplasia. Although 5 of the 24 (20.8%) non-IBD/non-CRC controls harbored _pks_+ E. coli, the genotoxic island was detected in 14 of 35 (40%, p<0.05) IBD patients and in 14 of 21 (66.7%, p<0.001) CRC patients (Figure 3B, Table S3). This suggests pks+ bacteria are associated with chronic intestinal inflammation and CRC and may impact carcinogenesis.
Fig 3.
_Pks_+ E. coli strains are associated with CRC and DNA damage. (A–B) PCR screen for pks (A) using primers targeting the left (1824bp) and right (1413bp) ends of the pks island in E. coli strains NC101 and K12, and E. faecalis (E.f.), and (B) using primers targeting the right end and ClbB gene of the pks island in mucosa-associated E. coli isolated from human colorectal tissue specimens, Binomial test *p<0.05, ***p<0.001. (C–D) NC101 pks induces (C) γH2AX (MOI 20, 4hr) in IEC-6 cells; Mean + SEM, ANOVA + Tukey, and (D) G2 cell cycle arrest (MOI 100, 24hr). A, C–D are representative of 3 experiments.
To functionally link pks with the development of CRC, we created an isogenic pks-deficient E. coli NC101 strain (NC101Δ_pks). Absence of pks did not affect bacterial growth in vitro (Fig. S7) nor did it impair colonization capacity in vivo (109–1010 per 200g stool pellet, 4–6 mice/group). As pks from strains of extraintestinal pathogenic E. coli can elicit mammalian DNA damage (15, 16), we tested the ability of E. coli NC101 pks to induce a DNA damage response. We infected the non-transformed rat intestinal epithelial cell line IEC-6 with NC101 or NC101Δ_pks and assessed levels of phosphorylated histone H2AX (γH2AX), a surrogate marker of DNA damage (19–21). WT NC101 induced γH2AX in ~30% of cells, whereas NC101Δ_pks_ induced γH2AX in <5%, a level equivalent to that induced by non-colitogenic E. coli K12 (Fig 3C). Consistent with these results, we observed that WT NC101 induced a 3-fold increase in the percent of cells arrested in G2/M phase relative to untreated and NC101Δ_pks_ infected cells (Fig 3D). These experiments demonstrate that pks alone has the capacity to induce DNA damage and indicate that the genotoxic island does not block the initiation of DNA damage response. This led us to hypothesize that pks would also promote tumorigenesis in vivo.
To test this hypothesis, we mono-associated GF Il10_−/− mice with E. coli NC101 or NC101Δ_pks, with or without AOM treatment, and assessed inflammation and tumorigenesis. The absence of pks did not affect the severity of colonic inflammation in Il10_−/− mice with colitis (12wk no AOM), or colitis/cancer (14wk+AOM and 18wk+AOM) (Fig 4A). Similarly, colon tissue pro-inflammatory cytokine transcripts and immune cell infiltration were not significantly different between mice mono-associated with NC101 vs. NC101Δ_pks (Fig S8, S9; Table S4). Importantly though, at both 14wk+AOM and 18wk+AOM, the absence of pks was associated with significantly reduced neoplastic lesions (Fig 4B). At 14wk+AOM, high grade dysplasia (HGD) or invasive carcinomas were present in 5/8 mice mono-associated with NC101, whereas only 1/8 NC101Δ_pks_ mono-associated mice developed HGD. At 18wk+AOM, the absence of pks did not affect mouse survival or tumor size, however, macroscopic tumor burden and carcinoma invasion were significantly decreased (Fig 4C–F, S10). In addition, all 9 NC101 mono-associated mice developed invasive carcinoma, with 4/9 fully invading the muscularis propria and serosa. In contrast, 0/9 NC101Δ_pks_ mono-associated mice exhibited full invasion. This likely suggests that the presence of E. coli pks accelerates progression from dysplasia to invasive carcinoma. In the absence of AOM, GF _Il10_−/− mice colonized with NC101 for 21 weeks developed only mild dysplasia (Fig. S11A). GF WT mice mono-associated with E. coli NC101 and treated with AOM developed neither inflammation nor dysplasia/tumors (Fig. S11B), suggesting that this bacterium is not carcinogenic in the absence of inflammation. Together these data indicate that the absence of pks reduces the tumorigenic potential of E. coli NC101 without altering colonic inflammation.
Fig 4.
Deletion of pks reduces the tumorigenicity, but not inflammatory potential, of E. coli NC101 in AOM/_Il10_−/− mice. (A) Histologic inflammation and (B) neoplasia scores in 3 cohorts of mice at weeks 12, 14+AOM, 18+AOM, (C–F) 18wks+AOM: (C) Histologic invasion score, (D) Macroscopic tumor number, (E) Mean macroscopic tumor diameter in each mouse (F) Representative H&E histology. (A–F) Each symbol represents data from one mouse, line at mean, pairwise comparisons by t test. (G) Cells/crypt with ≥4 γH2AX foci, and γH2AX IHC (400x). Each symbol represents data from mouse, ANOVA + Tukey, line at mean, arrowheads indicate γfoci+ cells.
To evaluate the impact of pks on host DNA damage in vivo, we measured colonocyte γH2AX+ nuclear foci (γ-foci) in AOM/Il10_−/− mice mono-associated with NC101 vs. NC101Δ_pks for 14 weeks (19–21). The abundance of γ-foci+ colonocytes/crypt was significantly reduced in AOM/Il10_−/− mice mono-associated with E. coli NC101Δ_pks vs. E. coli NC101 (Fig 4G). We detected a 5-fold reduction in γ-foci+ colonocytes/crypt in E. coli NC101 mono-associated AOM/WT mice relative to E. coli NC101-associated AOM/_Il10_−/− mice (Fig 4G). This suggests that both host inflammation and _E. coli_-derived pks act in concert to create a host microenvironment that promotes DNA damage and tumorigenesis in AOM/_Il10_−/− mice.
Although the etiology of colitis associated CRC is multifactorial, this work indicates that chronic inflammation targets the intestinal microbiota and can induce the expansion of microbes, including E. coli, that influence CRC in mice. The carcinogenic effect of E. coli NC101 pks clearly demonstrates that genotoxic microorganisms promote CRC in the presence of the carcinogen AOM in _Il10_−/− mice. It remains to be seen if NC101 and other _pks_-harboring bacteria have similar effects in other models of colitis-associated CRC. An increased prevalence of pks+ E. coli in IBD and CRC patients may suggest a cancer-promoting role in human CRC. We propose a model in which inflammation creates an environment that supports carcinogenesis through its effects on both the host and the microbiota. In this two-hit model, inflammation targets the microbiota to foster the expansion of bacteria with genotoxic potential, such as _pks_+ bacteria. In parallel, inflammation creates an opportunity for _pks_+ bacteria to adhere to the colonic mucosa by decreasing protective mucins and antimicrobial peptide production (22, 23), a process prevented by natural barrier function present in non-inflamed WT mice. The genotoxic effect of pks requires bacteria-host cell contact (15, 16), thus an environment in which bacteria can more readily access the epithelium could result in increased delivery of the pks product Colibactin to epithelial host cells. This would explain the lack of cancer in pks+ E. coli -associated WT mice. Although other microbes likely participate in the progression of CRC, our findings highlight the complex effects of inflammation on both microbial composition/activity and the host’s ability to protect itself from a dysbiotic microbiota.
Supplementary Material
Acknowledgments
We thank M. Bower and S. Tonkonogy of the National Gnotobiotic Rodent Resource Center at University of North Carolina and North Carolina State University for assistance with gnotobiotic mice (NIH P40 R018603). Histology was performed at the CGIBD histology core (P30 DK034987). This work was supported by funding from NIH T32 DK007737 (JCA), R01 DK73338 (CJ), R01 DK47700 (CJ), R01 CA136887 (TOK), R01 DK53347-11 (KWS/RBS), the American Institute for Cancer Research (CJ), NY Presbyterian/Weill Cornell Medical College (KWS), Crohn’s and Colitis foundation UK (BJC), the National Institute of Health Research Specialist Biomedical Research Center in Microbial Disease (JMR), the North West Cancer Research Fund UK (BJC), and the Canadian Institutes of Health Research MOP#114872 (AS). All data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Illumina sequencing data are deposited in NCBI’s sequence read archive (SRA) and are accessible under SRA055272. Inflammation PCR array data are deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE39085. Carrying on the good fight for Mathieu Jobin, Lyne Gauthier, and Janine Drysdale.
Footnotes
All Supplementary Materials have been supplied as a separate file, ArthurSOM.pdf. This file is 30 pages, 25.6MB, and includes the following:
Materials and Methods
References #1–25 from main text and #26–39 supplemental
References and Notes
- 1.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 2.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 3.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danese S, Mantovani A. Inflammatory bowel disease and intestinal cancer: a paradigm of the Yin-Yang interplay between inflammation and cancer. Oncogene. 2010;29:3313–3323. doi: 10.1038/onc.2010.109. [DOI] [PubMed] [Google Scholar]
- 5.Uronis JM, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luperchio SA, Schauer DB. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 2001;3:333–340. doi: 10.1016/s1286-4579(01)01387-9. [DOI] [PubMed] [Google Scholar]
- 8.Shen XJ, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. gutmicrobes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marshall BJ. The 1995 Albert Lasker Medical Research Award. Helicobacter pylori. The etiologic agent for peptic ulcer. JAMA. 1995;274:1064–1066. doi: 10.1001/jama.274.13.1064. [DOI] [PubMed] [Google Scholar]
- 10.Swidsinski A, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 11.Martin HM, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 12.Darfeuille-Michaud A, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 13.Masseret E, et al. Genetically related Escherichia coli strains associated with Crohn’s disease. Gut. 2001;48:320–325. doi: 10.1136/gut.48.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SC, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 16.Cuevas-Ramos G, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homburg S, Oswald E, Hacker J, Dobrindt U. Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli. FEMS Microbiol Lett. 2007;275:255–262. doi: 10.1111/j.1574-6968.2007.00889.x. [DOI] [PubMed] [Google Scholar]
- 18.Putze J, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 20.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inaba Y, et al. Expression of the antimicrobial peptide alpha-defensin/cryptdins in intestinal crypts decreases at the initial phase of intestinal inflammation in a model of inflammatory bowel disease, IL-10-deficient mice. Inflamm Bowel Dis. 2010;16:1488–1495. doi: 10.1002/ibd.21253. [DOI] [PubMed] [Google Scholar]
- 23.Schwerbrock NMJ, et al. Interleukin 10-deficient mice exhibit defective colonic Muc2 synthesis before and after induction of colitis by commensal bacteria. Inflamm Bowel Dis. 2004;10:811–823. doi: 10.1097/00054725-200411000-00016. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes JM, Campbell BJ. Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol Med. 2002;8:10–16. doi: 10.1016/s1471-4914(01)02194-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.