The Vam6 and Gtr1–Gtr2 pathway activates TORC1 in response to amino acids in fission yeast (original) (raw)
Abstract
The Rag family of GTPases has been implicated in the TORC1 activation in Drosophila and in mammalian cells in response to amino acids. We have investigated the role of the Rag GTPases Gtr1 and Gtr2 in TORC1 regulation in Schizosaccharomyces pombe. Fission yeast Gtr1 and Gtr2 are non-essential proteins that enhance cell growth in the presence of amino acids in the medium. The function of Gtr1 and Gtr2 in nutrient signaling is further supported by the observation that even in rich medium the deletion of either gene results in the promotion of mating, meiosis and sporulation, consistent with the downregulation of TORC1. We show that Gtr1 and Gtr2 colocalize with TORC1 in vacuoles, where TORC1 is presumably activated. Epistasis analyses indicated that Gtr1 and Gtr2 function downstream of Vam6 and upstream of TORC1 in response to amino acid signals. Our data demonstrate the existence of an evolutionarily conserved pathway with the Vam6 and Gtr1–Gtr2 pathway activating TORC1, which in turns stimulates cell growth and inhibits sexual differentiation.
Key words: S. pombe, Cell growth, TOR, Gtr1, Gtr2, Vam6
Introduction
Cell survival depends on continuous sensing of the nutritional environment and adjusting cell growth, cell division and cell differentiation accordingly. In the presence of nutrients, Schizosaccharomyces pombe cells grow and divide, but when nutrients (mainly nitrogen) are limited they arrest in G1 and undergo sexual differentiation. The target of rapamycin (TOR) pathway is an evolutionarily conserved signal transduction pathway that couples nutrient availability with the diverse cellular responses that ultimately drive cell growth and proliferation, and inhibit sexual differentiation.
A broad range of signals regulates the activity of TOR to control cell growth. TOR is a serine/threonine protein kinase that is structurally and functionally conserved from yeasts to mammals. TOR exists in two distinct complexes TORC1 and TORC2 (Loewith et al., 2002; Sarbassov et al., 2004). TORC1 contains Raptor and positively regulates cell growth and size by promoting anabolic processes, such as protein synthesis (Fingar et al., 2002; Hay and Sonenberg, 2004), and by inhibiting catabolic processes, such as autophagy (Blommaart et al., 1995; Noda and Ohsumi, 1998; Shigemitsu et al., 1999). In contrast, TORC2, which contains Rictor, regulates Akt and also affects the actin cytoskeleton (Jacinto et al., 2004; Sarbassov et al., 2005). In mammalian cells, mTOR (for mammalian TOR) is a crucial player in the TSC1-TSC2–Rheb–mTOR signaling pathway, which regulates cell growth in response to growth factors, nutrients and energy conditions. TORC1 is activated by the GTPase Rheb, which is negatively regulated by the TSC1-TSC2 tuberous sclerosis complex (Long et al., 2005; Smith et al., 2005). Unlike higher eukaryotes, which contain a single TOR protein, S. pombe and Saccharomyces cerevisiae have two: Tor1 and Tor2.
In contrast to S. cerevisiae, the TSC1-TSC2–Rheb–TOR pathway is conserved in S. pombe, providing an excellent model to study TOR pathway regulation. In S. pombe, Tor2 forms part of the TORC1 complex and is essential for cell growth and the repression of sexual differentiation, meiosis and sporulation (Alvarez and Moreno, 2006; Matsuo et al., 2007; Uritani et al., 2006; Weisman et al., 2007), whereas Tor1 is not essential for growth and is included in the TORC2 complex (Alvarez and Moreno, 2006; Hayashi et al., 2007; Matsuo et al., 2007).
Amino acids are potent activators of TORC1 (Hara et al., 1998). The Rag proteins are GTPases with a canonical N-terminal Ras-like GTPase domain and a unique C-terminal RagA conserved region. Two complementary studies have reported that the conserved Rag GTPases act as upstream regulators of TORC1 and have important roles in coupling amino-acid-derived signals to TORC1 in both Drosophila and mammalian cells (Kim et al., 2008; Sancak et al., 2008). In mammals, there are four Rag proteins (RagA, RagB, RagC and RagD). RagA and RagB are very similar to each other and are orthologues of budding yeast Gtr1p, whereas RagC and RagD are similar to each other and are orthologs of yeast Gtr2p (Bun-Ya et al., 1992; Hirose et al., 1998). Rag and Gtr proteins function in heterodimeric complexes that contain one Gtr1-like GTPase and one Gtr2-like GTPase (Nakashima et al., 1999; Sekiguchi et al., 2001), and the two GTPases bind different forms of guanine nucleotides; one binds GTP and the other binds GDP. Only when RagA or RagB is bound to GTP and RagC or RagD is bound to GDP is the heterodimer fully active to stimulate TORC1. In addition, RagA and RagB have a dominant role over RagC and RagD in TORC1 activation (Binda et al., 2009; Li and Guan, 2009). The active Rag heterodimer can directly bind Raptor (Sancak et al., 2008), which is a key subunit in TORC1. This interaction between Rag and Raptor depends on the GTP-binding status of RagA or RagB. The Rag might activate TORC1 by transporting this complex to the vicinity of Rheb in mammalian cells, although Rag proteins do not directly stimulate the kinase activity of mammalian TORC1 (Sancak et al., 2008). This proposed mechanism of activation, through the amino-acid-induced subcellular localization, is not conserved in budding yeast because the subcellular localizations of both TORC1 components and Gtr proteins are not affected by amino acids (Binda et al., 2009).
In S. cerevisiae Vam6 is a GTP-exchange factor (GEF) (Wurmser et al., 2000) that forms part of the HOPS complex (Starai et al., 2008), which is involved in vacuolar fusion (Price et al., 2000) and required for autophagy (Kinchen et al., 2008). Recently, Vam6 has been reported to control the activity of TORC1 by activating Gtr1. Vam6 colocalizes with the TORC1 complex and the Rag proteins at the membrane of the vacuole and functions as a GEF of Gtr1 (Binda et al., 2009). In S. pombe Vam6 has been described as a protein required for entry into and the maintenance of the G0 status (Sajiki et al., 2009). The vam6 mutant has numerous small vesicles, possibly owing to a reduction in vacuolar fusion, but the role of this GEF remains unclear and no relationship with TORC1 or Gtr1–Gtr2 has been described previously.
Here, we show that Rag proteins in S. pombe induce cellular growth and repress sexual differentiation by activating the TORC1 complex in response to the presence of amino acids in the medium. We also provide evidence that Vam6 activates the Gtr1–Gtr2 complex.
Results
Rag proteins activate TORC1 in S. pombe
The multiprotein mTOR kinase complex is the central component of a pathway that promotes growth in response to different signals, including insulin, energy levels and amino acids. In Drosophila and mammalian cells, Rag proteins are mediators of the amino acid signaling to mTOR to promote cell growth (http://www.pombase.org/). Loss of Gtr1 or Gtr2 resulted in the inability of the cells to grow properly, and they divided with a doubling time longer than that of wild-type cells (Fig. 1A). The _gtr1_Δ and _gtr2_Δ mutants were unable to sense the availability of amino acids and did not increase their rate of growth (measured as doubling time) when amino acids were added to the medium (Fig. 1A). Therefore, fission yeast Rag proteins are non-essential proteins, but they are important for the enhancement of cell growth in response to amino acids.
Fig. 1.
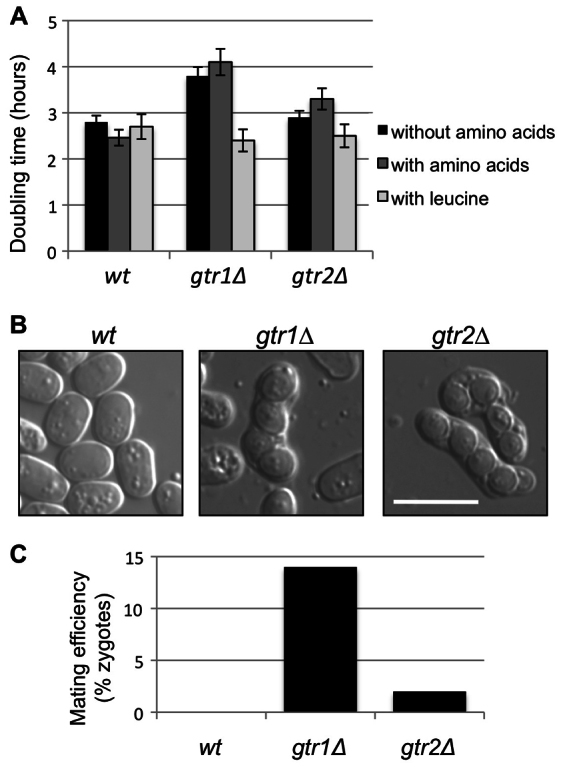
Role of Gtr1 and Gtr2 in cell growth and sexual differentiation. (A) Wild-type (wt), _gtr1_Δ and _gtr2_Δ cells were grown in Edinburgh minimal medium (EMM) at 30°C, and then transferred into fresh EMM and EMM supplemented with amino acids. At the same time, _leu1-32, gtr1_Δ leu1-32 and _gtr2_Δ leu1-32 cells were grown in EMM supplemented with leucine. This experiment was performed three times and the number of cells per ml was counted every 2 hours to calculate the doubling time. (B) _gtr1_Δ and _gtr2_Δ cells of opposite mating types were incubated on YES plates for 2 days at 25°C. The _gtr1_Δ and _gtr2_Δ mutant cells were able to mate and form spores in rich medium. Scale bar: 10 μm. (C) Mating efficiency in rich medium was determined by counting the number of zygotes observed in YES plates.
Leucine uptake is reduced in tsc1 or tsc2 mutants in which Tor2 is hyperactive (Weisman et al., 2005; Weisman et al., 2007), indicating that the TORC1 complex inhibits leucine uptake. If Gtr1 and Gtr2 are mediating the activation of Tor2 in response to amino acids, in cells lacking Gtr1 (or Gtr2) Tor2 should be less active and leucine uptake should increase. We deleted the gtr1 and gtr2 genes in a leu1-32 mutant and analyzed the growth rate (measuring the doubling time) in the presence of leucine. As shown in Fig. 1A, the growth rate of _leu1-32 gtr1_Δ and _leu1-32 gtr2_Δ mutants improved considerably in the presence of leucine, suggesting that Gtr1 and Gtr2 regulate TORC1 positively.
Another important phenotype related to Tor2 activity is the inhibition of sexual differentiation. Inactivation of Tor2 in rich medium leads to cell cycle exit, G1 arrest, sexual differentiation, meiosis and sporulation (Alvarez and Moreno, 2006; Hayashi et al., 2007; Urano et al., 2007; Uritani et al., 2006). When _gtr1_Δ and _gtr2_Δ haploid cells of opposite mating types were grown together in rich medium, zygotes were observed and quantified (Fig. 1B and Fig. 1C, respectively), indicating that Gtr2, and in particular Gtr1, inhibit sexual differentiation in rich medium. Collectively, these observations support the idea that Gtr1 and Gtr2 activate TORC1 in S. pombe.
Gtr1 and Gtr2 form heterodimers that localize to the vacuole membrane
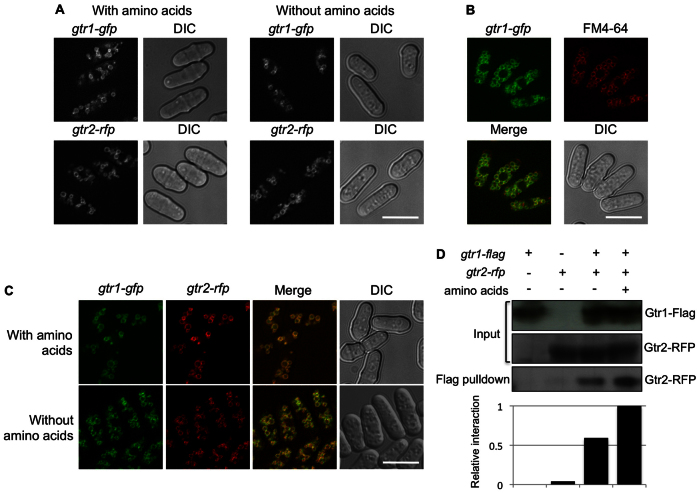
Mammalian Rag proteins localize to the lysosomal surface independently of the availability of amino acids (Sancak et al., 2008). In S. cerevisiae, Gtr1 and Gtr2 have been reported to localize to the vacuole membrane (Dubouloz et al., 2005). Therefore, the localization of Rag proteins to the membrane of the vacuoles, in yeast, or to the lysosome, the corresponding organelle in higher eukaryotes, highlights the role of the TOR pathway in autophagy. We looked at Gtr1 and Gtr2 localization in the absence and presence of amino acids in S. pombe. To accomplish this, we tagged the chromosome copies of gtr1 and gtr2 with sequences encoding GFP and RFP (designated gtr1-gfp and gtr2-rfp), respectively. Exponentially growing wild-type cells revealed the localization of Gtr1–GFP and Gtr2–RFP to intracellular membrane structures that could have been vacuolar membranes (Fig. 2A). The observed pattern was identical, regardless of the presence or not of amino acids in the medium. To confirm the localization to the vacuole membrane, we stained the gtr1-gfp cells with the lipophilic vacuolar membrane fluorescent dye FM4-64. As shown in Fig. 2B, Gtr1–GFP colocalized with FM4-64 staining, indicating that Gtr1–GFP is concentrated at the membranes of vacuoles in S. pombe.
Fig. 2.
Subcellular localization of Gtr1 and Gtr2. (A) gtr1-gfp and gtr2-rfp cells were grown in EMM at 30°C in the presence and in the absence of amino acids. Gtr1–GFP and Gtr2–RFP localized to structures similar to the vacuolar membranes, independently of the presence of amino acids. (B) gtr1-gfp cells were grown in EMM at 30°C, and FM4-64 staining was used to identify vacuoles and corroborate that Gtr1–GFP localized to the vacuole membranes. (C) Gtr1–GFP and Gtr2–RFP colocalized to the vacuolar surface in the presence and in the absence of amino acids. (D) gtr1-flag, gtr2-rfp and gtr1-flag gtr2-rfp cells were grown in EMM at 30°C and transferred into EMM in the presence and in the absence of amino acids. Exponentially growing cells were collected after 1 hour and cell lysates were pulled down using anti-Flag M2 beads. Gtr1–Flag physically interacted with Gtr2–RFP and this interaction increased in the presence of amino acids. The relative interaction between Gtr1–Flag and Gtr2–RFP was estimated by densitometry using ImageJ software and normalized to the amount of Gtr2–RFP pulled down with anti-Flag antibodies in the presence of amino acids, which was set as 1. Scale bars: 10 μm.
In mammalian cells, RagA and RagB can form heterodimers with RagC and RagD (Sekiguchi et al., 2001). In budding yeast, Gtr1 can form a homodimer as well as a heterodimer with Gtr2 (Nakashima et al., 1999). To check whether Gtr1 and Gtr2 form heterodimers in vivo, we tagged Gtr2 with RFP in a gtr1-gfp background. The localization Gtr1–GFP and Gtr2–RFP was identical and independent of the availability of amino acids (Fig. 2C), suggesting that they interact in vivo. To corroborate this interaction biochemically, we immunoprecipitated Gtr1 tagged with the Flag epitope in cells growing in the absence of amino acids and at 1 hour after adding amino acids. We observed that Gtr2–RFP co-precipitated with Gtr1 (Fig. 2D) and that the Gtr1–Gtr2 interaction was stronger in cells growing in the presence of amino acids, indicating that the formation of the heterodimer is stimulated by amino acids.
TORC1 also localizes to the vacuole membrane
The activation of the mTORC1 pathway by amino acids is correlated with the movement of mTORC1 from an undefined location in the cytoplasm to the surface of the lysosomes where the Rag proteins reside (Sancak et al., 2008). Amino acids promote the translocation of mTORC1 in a Rag-dependent fashion to the surface of lysosomes, where mTORC1 can find its well-known activator Rheb (Sancak et al., 2008; Sancak et al., 2010). In budding yeast, Tor1 is located diffusely in the cytoplasm and is more concentrated near the vacuolar membrane (Sturgill et al., 2008). This localization of Tor1 is not altered by the presence of amino acids. Thus, the proposed mechanism of TORC1 activation by the amino acid-induced subcellular localization of TORC1 may not be conserved in S. cerevisiae.
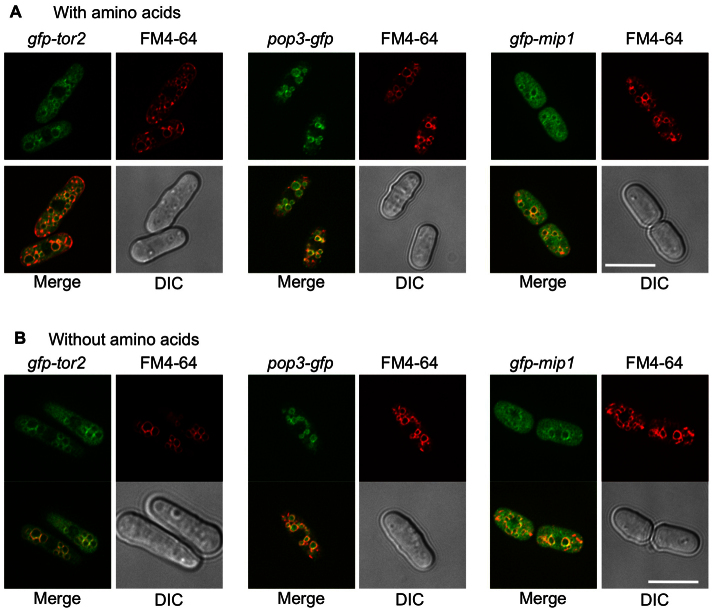
In S. pombe, TORC1 contains Tor2, Mip1 (the Raptor ortholog) and Pop3 (also known as Wat1; the Lst8 ortholog) (Alvarez and Moreno, 2006; Matsuo et al., 2007). In order to analyze whether these TORC1 components alter their subcellular localization in an amino-acid-dependent manner, we tagged Tor2, Mip1 and Pop3 with GFP. The localization of GFP–Tor2 in cytoplasmic structures has been reported previously (Hayashi et al., 2007) but no vacuolar membrane localization has been described. Pop3–GFP was expressed under its own promoter, whereas GFP–Tor2 and GFP–Mip1 were expressed under the nmt1 promoter in the presence of thiamine. Under these conditions, the level of overexpression was diminished; cells grew normally and mating was not suppressed, indicating that TORC1 was exerting physiological activity. As shown in Fig. 3, GFP–Tor2, GFP–Mip1 and Pop3–GFP showed similar GFP signals that colocalized with FM4-64 staining. Thus, the three components of the TORC1 complex showed vacuolar membrane localization, independently of the presence or not of amino acids in the medium.
Fig. 3.
Subcellular localization of TORC1. gfp-tor2, pop3-gfp and gfp-mip1 cells were grown in EMM containing thiamine (EMMT) at 30°C and transferred into EMMT with (A) and without amino acids (B). Cells were stained with FM4-64 to observe the vacuolar membranes. The three subunits of the TORC1 complex (GFP–Tor2, Pop3–GFP and GFP–Mip1) localized to the vacuolar membranes. Scale bars: 10 μm.
The Gtr1–Gtr2 heterodimer interacts with the TORC1 complex
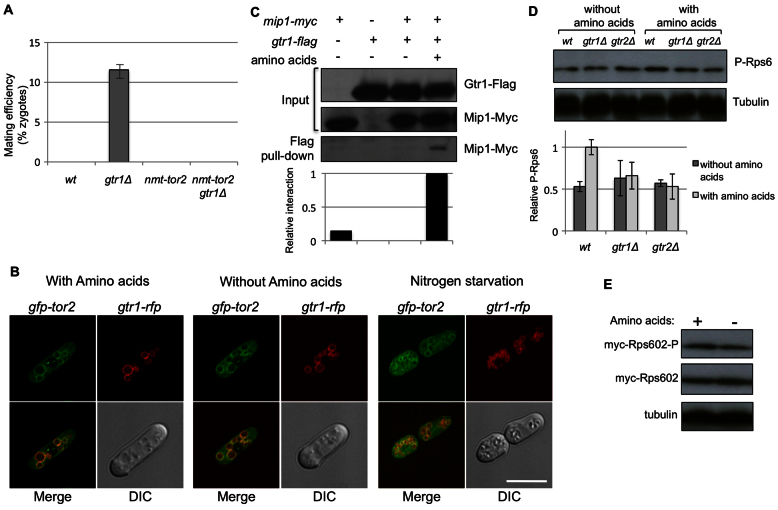
Cells lacking either Gtr1 or Gtr2 undergo mating and sexual differentiation in rich medium (Fig. 1B), as do Tor2 mutants. Moreover, overexpression of Tor2 suppresses mating (Alvarez and Moreno, 2006). To test whether Tor2 and Gtr1 act in the same pathway to impair sexual differentiation, we counted the number of zygotes in _gtr1_Δ cells (_gtr1_Δ), in cells overexpressing Tor2 (nmt-tor2) and in _gtr1_Δ cells overexpressing Tor2 (_gtr1_Δ nmt-tor2). In rich medium, _gtr1_Δ cells mate and nmt-tor2 cells are sterile (Fig. 4A). The double mutant _gtr1_Δ nmt-tor2 was unable to mate and form zygotes (Fig. 4A), indicating that the overexpression of Tor2 suppresses the phenotype of the gtr1 deletion.
Fig. 4.
TORC1 and Gtr1 interact in vivo. (A) Wild-type (wt), _gtr1_Δ, nmt-tor2 and _nmt-tor2 gtr1_Δ cells of opposite mating types were grown on YES plates at 25°C for 2 days. Overexpression of Tor2 rescued the mating derepression observed in _gtr1_Δ cells. (B) Double-mutant gfp-tor2 gtr1-rfp cells were grown EMM containing thiamine (EMMT) at 30°C and transferred into fresh EMMT, EMMT supplemented with amino acids and EMMT without nitrogen for 1 hour. GFP–Tor2 and Gtr1–RFP colocalized on the vacuolar membrane, regardless of the presence of amino acids or nitrogen in the medium. Scale bar: 10 μm. (C) mip1-myc, gtr1-flag and mip1-myc gtr1-flag cells were grown in EMM and transferred into EMM in the presence or absence of amino acids for 1 hour. Cell lysates (Input) and Flag pull-down fractions were subjected to SDS-PAGE and immunoblots were incubated with anti-Myc and anti-Flag antibodies, as indicated. Gtr1–Flag interacted physically with Mip1–Myc (the Raptor ortholog) in an amino-acid-dependent manner. The relative interaction between Gtr1–Flag and Mip1–Myc was estimated by densitometry using ImageJ software and normalized to the Mip1–Myc pulled down in the presence of amino acids, which was set as 1. (D) Wild-type, _gtr1_Δ and _gtr2_Δ cells were grown in EMM and transferred into EMM with and without amino acids for 90 minutes. Cell extracts were subjected to SDS-PAGE and immunoblotted to detect phosphorylated Rps6 (P-Rps6) as a measurement of TORC1 activity. The relative amount of phosphorylated Rps6 was estimated by densitometry and normalized to tubulin expression. The amount of phosphorylated Rps6 in wild-type cells growing in the presence of amino acids was set as 1. (E) _rps601_Δ myc-rps602 cells were grown in the presence and in the absence of amino acids. The amount of Myc–Rps6 was examined with the anti-Myc antibody. Rps6 phosphorylation (Rps602-P) was used as a measurement of TORC1 activity. Tubulin was used as a loading control.
Because Gtr1, Gtr2 and TORC1 localize to the vacuolar membrane, we analyzed whether GFP–Tor2 and Gtr1–RFP colocalized in this organelle. As shown in Fig. 4B, most of the GFP–Tor2 colocalized with Gtr1–RFP independently of the presence of amino acids or nitrogen. To confirm further that Gtr1 forms a complex with the TORC1 complex, Gtr1 was tagged with the Flag epitope and Mip1 (Raptor) with the Myc epitope. As shown in (Fig. 4C), Mip1–Myc was pulled down with Gtr1–Flag only in the presence of amino acids. Together, these findings suggest that in fission yeast cells the Gtr1–Gtr2 heterodimer and TORC1 are located in the vacuolar membrane independently of the presence of amino acids. However, only when amino acids are present in the medium does the Gtr1–Gtr2 heterodimer interact physically with TORC1 and activate the TOR pathway to induce cell growth and repress sexual differentiation.
TORC1 activity can be monitored by measuring ribosomal protein S6 (Rps6) phosphorylation (Nakashima et al., 2010). We analyzed Rps6 phosphorylation in wild-type cells and in _gtr1_Δ and _gtr2_Δ mutant cells growing in the presence and in the absence of amino acids. Wild-type cells were able to sense the availability of amino acids in the medium by activating TORC1 and the amount of Rps6 phosphorylation observed was higher in the presence than in the absence of amino acids (Fig. 4D). It has been reported that expression of Myc–Rps6 does not alter the expression level under nitrogen starvation conditions (Nakashima et al., 2010), and hence we used the same cells to monitor the total amount of Myc–Rps6 in cells growing in the presence and absence of amino acids. Similar to the presence of nitrogen availability, the presence of amino acids did not alter Rps6 expression levels (Fig. 4E). Cells lacking gtr1 or gtr2 were unable to sense the presence of amino acids and no further increases in Rps6 phosphorylation were observed after the addition of amino acids (Fig. 4D), showing that Gtr1–Gtr2 indeed has an important role in TORC1 activation in response to amino acids in fission yeast and supporting the notion that the function of the Rag family GTPase is conserved in eukaryotes (Binda et al., 2009; Kim et al., 2008; Sancak et al., 2008).
Vam6, Gtr1–Gtr2 and Tor2 act in the same pathway
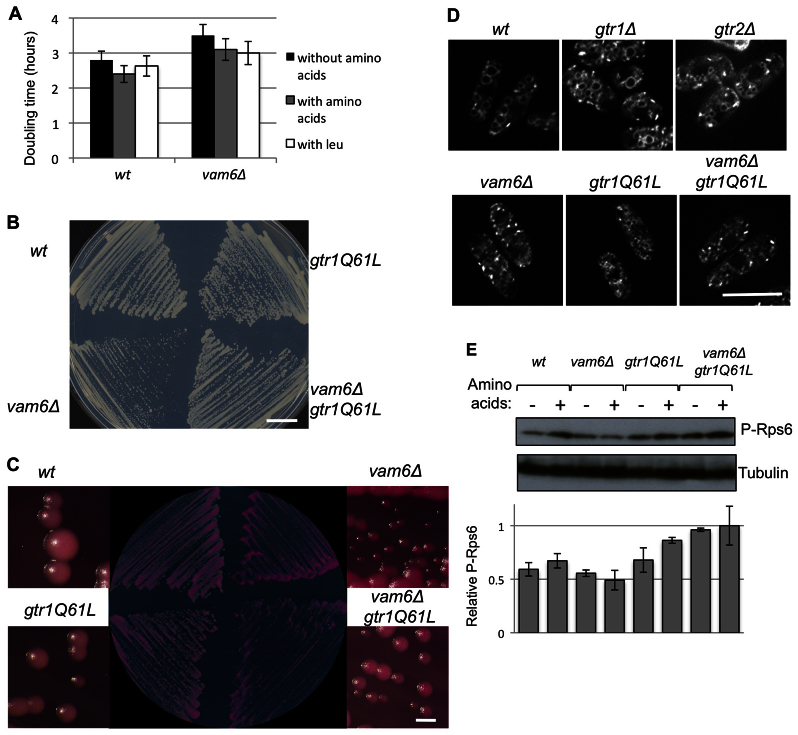
Recently Vam6 has been described to control the activity of TORC1 by activating Gtr1 in S. cerevisiae (Binda et al., 2009). Vam6 colocalizes with the TORC1 complex and the Rag proteins in the membrane of the vacuole and functions as a GEF of Gtr1 (Binda et al., 2009). We analyzed the vam6 ortholog in S. pombe to see whether it was able to act as the GEF that regulates Gtr activity. First, we deleted vam6 and observed a clear defect in cell growth, with a longer doubling time than that of wild-type cells for cells growing in the absence of amino acids. The doubling time of _vam6_Δ was shorter in the presence of amino acids, indicating that these cells were still able to respond, at least partially, to the presence of amino acids (Fig. 5A) and that Vam6 has an important role in regulating cell growth in S. pombe but is not essential for responding to the availability of amino acids.
Fig. 5.
_vam6_Δ cells show a defect in cell growth. (A) Wild-type (wt) and _vam6_Δ mutant cells were grown exponentially in EMM, in EMM plus amino acids and in EMM plus leucine. Cell numbers were counted every 2 hours to establish the doubling time in each medium. Wild-type, _vam6_Δ, gtr1Q61L and _vam6_Δ gtr1Q61L cells were grown on EMMT (EMM supplemented with thiamine) plates (B) and on plates supplemented with Phloxine B (C) in the presence of thiamine to induce expression of low levels of _gtr1Q61L. vam6_Δ cells formed small colonies (B) containing a high number of dead cells, generating dark red colonies (Phloxine B positive) (C). The viability of _vam6_Δ cells expressing the active form of Gtr1 (gtr1Q61L) increased; these cells formed larger colonies (B and C) and the number of dead cells decreased (C). (D) Wild-type, _gtr1_Δ, _gtr2_Δ, _vam6_Δ, gtr1Q61L and _vam6_Δ gtr1Q61L cells were grown in EMMT (i.e. in the presence of thiamine) at 30°C and membrane vacuoles were stained with FM6-64. _vam6_Δ cells showed a defect in membrane fusion; FM4-64 was unable to stain membrane vacuoles in _vam6_Δ cells, and the fluorescence appeared as small dots corresponding to endosomal vesicles. _gtr1_Δ, _gtr2_Δ or gtr1Q61L cells did not show this phenotype and FM4-64 was able to stain the vacuolar membrane correctly. The same defect of _vam6_Δ cells in membrane fusion was observed in _vam6_Δ gtr1Q61L, indicating that the active form of Gtr1 is unable to restore the vacuolar structure of _vam6_Δ. (E) Wild-type, _vam6_Δ, gtr1Q61L and _vam6_Δ gtr1Q61L cells were grown at 30°C in EMMT and transferred to EMMT in the presence and in the absence of amino acids (supplemented with thiamine in both cases). Cells were collected after 90 minutes and the extracts were subjected to SDS-PAGE and immunoblotting to detect Rps6 phosphorylation (P-Rps6) as a measurement of TORC1 activity. The relative amount of phosphorylated Rps6 was estimated by densitometry and normalized to tubulin expression. The amount of phosphorylated Rps6 in _vam6_Δ gtr1Q61L cells growing in the presence of amino acids was normalized to 1. In the presence of amino acids the amount of phosphorylated Rps6 increased in wild-type cells. The vam6 mutant was unable to respond to amino acids and the activity of TORC1 was low. By contrast, in cells expressing gtr1Q61L (the active form of Gtr1) Rps6 phosphorylation increased. The mutant vam6 showed high Rps6 phosphorylation when Gtr1Q61L was expressed, indicating that the constitutively active form of Gtr1 is able to restore TORC1 activity in _vam6_Δ cells. Scale bars: 1 cm (B); 1 mm (C); 10 μm (D).
As an indirect assay for Tor2 activity, we compared the growth rate in medium with and without leucine. To determine whether Vam6 was able to activate Gtr1 or Gtr2, we measured the cell growth of the leu1-32 strain. Interestingly, in the presence of leucine the deletion of vam6 improved growth (shorter doubling time) (Fig. 5A); this shortening in doubling time was similar to that observed in _gtr1_Δ and _gtr2_Δ cells (Fig. 1A), indicating that these mutants were able to increase their rate of cell growth in the presence of leucine, probably because TORC1 activity is lower and induces leucine uptake. These data suggest that Vam6 activates TORC1 through Gtr1–Gtr2.
Vam6 acts upstream of Gtr1 in S. pombe
Vam6 is required for entry into and the maintenance of quiescence after nitrogen starvation (Sajiki et al., 2009). Our findings show that Vam6 is also important for cell growth (Fig. 5A). To investigate whether Vam6 acts as a GEF of Gtr1, we mutated the conserved Q61 residue in Gtr1 to a leucine residue, which is expected to result in a constitutively active GTP-bound form of Gtr1 (Nakashima et al., 1999). We introduced Gtr1Q61L in a _vam6_Δ background and found that the double mutant was able to grow normally (Fig. 5B), indicating that constitutively active Gtr1 rescues the cell growth defect of the _vam6_Δ mutant. Viability was also assayed by growing the cells on plates containing Phloxine B, a vital dye that accumulates in dead cells and turns the colonies dark pink. The colonies of the _vam6_Δ mutant were dark pink, indicating that some of the cells were dying. However, the colonies of _vam6_Δ cells expressing Gtr1Q61L had a lighter pink color, similar to that of wild-type cells (Fig. 5C), corroborating the notion that the active form of Gtr1 rescues the lethality of _vam6_Δ cells. These results suggest that Vam6 functions upstream of Gtr1, possibly by acting as a GEF.
Vam6 is part of the HOPS complex (Starai et al., 2008), which is required for vacuolar fusion (Price et al., 2000). To check whether this was also the case in S. pombe, we used FM4-64 to stain vacuolar membranes (Fig. 5D). If the vacuolar fusion process diminishes, staining with FM4-64 remains visible only as small dots in the cytoplasm because the endosomes are unable to fuse; however, if normal vacuolar fusion occurs, the vacuolar membranes are stained with the FM4-64 dye. FM4-64 stained only small vesicles in the cytoplasm of _vam6_Δ cells, confirming a defect in vacuolar fusion in these cells. However, _gtr1_Δ, _gtr2_Δ and gtr1Q61L cells did not show any defects and vacuolar membranes were stained completely, whereas the _gtr1Q61L vam6_Δ double mutant showed small vesicles after FM4-64 staining, the same phenotype observed in _vam6_Δ cells, indicating that gtr1Q61L is unable to restore the vacuolar fusion defect of the vam6 deletion (Fig. 5D).
We measured TORC1 activity (as Rps6 phosphorylation) in wild-type and in _vam6_Δ mutant cells growing in the presence and in the absence of amino acids (Fig. 5E). _vam6_Δ mutant cells showed similar Rps6 phosphorylation levels to that of wild-type cells in the absence of amino acids. However, in contrast to wild-type cells, _vam6_Δ cells did not show an increase in Rps6 phosphorylation in the presence of amino acids. In the gtr1Q61L cells (i.e. with the active form of Gtr1), high levels of Rps6 phosphorylation were observed independently of the availability of amino acids, indicating a constitutively high activity of the TORC1 complex. When the active form of Gtr1 was expressed in a _vam6_Δ background, Rps6 phosphorylation was higher than in _vam6_Δ cells. These results suggest that TORC1 is not properly activated in _vam6_Δ cells and that these cells are unable to respond to the presence of amino acids; however, a constitutively active form of Gtr1 is able to restore TORC1 activity (Fig. 5E).
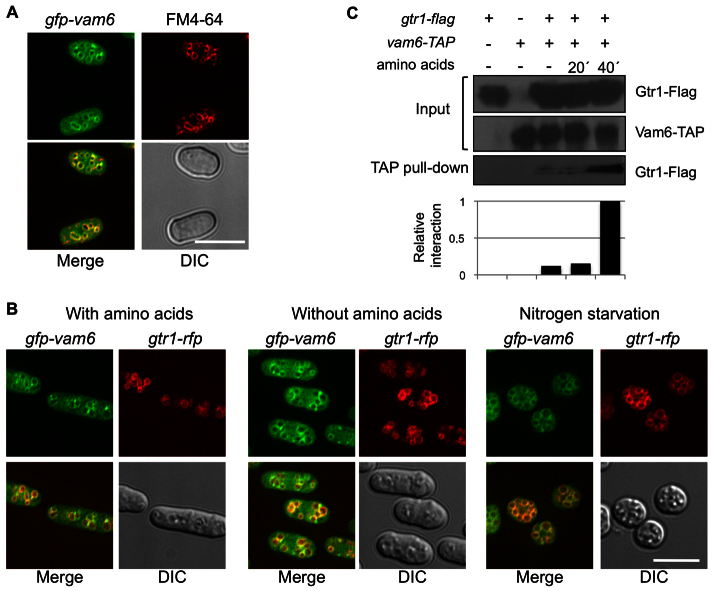
Vam6 was tagged with the GFP epitope in order to study its subcellular localization under different nutritional conditions. As shown in Fig. 6A, Vam6–GFP colocalized with the FM4-64 staining of vacuolar membranes. As reported above, Gtr1 could be acting downstream of Vam6, because a constitutively active form of Gtr1 (Gtr1Q61L) is able to rescue the growth defect of cells lacking Vam6. Accordingly, we examined the possible colocalization of Gtr1 and Vam6 under different nutritional conditions. Vam6–GFP colocalized with Gtr1–RFP to vacuolar membranes, regardless of the presence of amino acids or nitrogen starvation (Fig. 6B). To confirm that Vam6 did interact physically with Gtr1 we tagged Gtr1 with the Flag epitope and Vam6 with the TAP epitope. After pulling down Vam6–TAP with IgG–Sepharose, we were only able to detect Gtr1–Flag in cells grown in the presence of amino acids (Fig. 6C). This finding indicates that the Vam6–Gtr1 interaction is dependent upon the presence of amino acids in the medium.
Fig. 6.
Vam6 and Gtr1 interact in vivo. (A) gfp-vam6 cells were grown in EMM with thiamine at 30°C and vacuolar membranes were stained with FM4-64. GFP–Vam6 localized to the vacuolar membranes in growing cells. (B) gfp-vam6 gtr1-rfp cells were grown at 30°C in EMM and transferred to EMM, EMM supplemented with amino acids and EMM without nitrogen (always in the presence of thiamine). GFP–Vam6 and Gtr1–RPF colocalized to the vacuolar membranes in all conditions. (C) gtr1-flag, vam6-tap and gtr1-flag vam6-tap cells were grown in EMM and transferred into EMM supplemented with amino acids. Cells were collected after 20 and 40 minutes. Cell lysates (Input) and TAP pulldown fractions were subjected to SDS-PAGE and immunoblots were incubated with anti-TAP and anti-Flag antibodies, as indicated. The interaction between Gtr1–Flag and Vam6–TAP was estimated by densitometry using ImageJ software and the Gtr1–Flag pulled down in the presence of amino acids was normalized to 1. Gtr1–Flag physically interacted with Vam6–TAP in an amino-acid-dependent manner. Scale bars: 10 μm.
Discussion
The TOR pathway couples the nutritional environment with cell growth and proliferation. In fission yeast, TORC1 senses the presence of nitrogen in order to promote cell growth and inhibit sexual differentiation. In this paper, we describe the pathway of Vam6, Gtr1 and Gtr2 and TORC1 that responds to amino acid signaling in S. pombe.
In fission yeast there are two Gtr proteins: Gtr1 and Gtr2. Here, we show that they are non-essential proteins important for sensing the presence of amino acids in the medium by activating TORC1. In the absence of amino acids, cell growth is compromised in _gtr1_Δ and _gtr2_Δ mutants, probably because the TORC1 pathway is not properly regulated and sexual differentiation is induced. In Drosophila and mammalian cells, Rag proteins are activators of TOR. Here, we show that the fission yeast orthologs Gtr1 and Gtr2 also activate TORC1. Like tor2 mutants, cells lacking Gtr1 or Gtr2 show an increase in cellular growth in the presence of leucine and derepression of sexual differentiation. This phenotype is suppressed by Tor2 overexpression, suggesting that Tor2 is acting downstream from Gtr1 and Gtr2.
Mammalian Rag proteins localize to the lysosomal surface (Sancak et al., 2008). As in S. cerevisiae, S. pombe Gtr1, Gtr2 and TORC1 are localized to the vacuolar membrane, independently of the presence or the absence of amino acids (Binda et al., 2009; Dubouloz et al., 2005). Moreover, we describe that the formation of the Gtr1–Gtr2 heterodimer in fission yeast is stimulated by the presence of amino acids in the medium. Amino acids also stimulate the interaction between Gtr1 and Mip1 (Raptor), a component of the TORC1 complex. Thus, the Gtr1–Gtr2 heterodimer and the TORC1 complex are located to the vacuolar membrane and only when amino acids are present does the Gtr1–Gtr2 heterodimer form and associate with the TORC1 complex to activate Tor2 and promote cell growth.
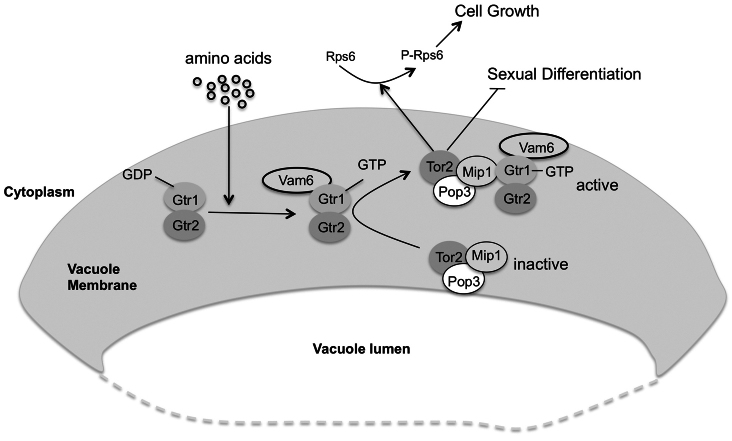
Previous studies have shown that Vam6 controls TORC1 activity by activating Gtr1 in S. cerevisiae (Binda et al., 2009). Here, we studied the role of Vam6 in S. pombe. Although Vam6 is not an essential protein the vam6 mutant grows very slowly. The growth of cells lacking Gtr1, Gtr2 or Vam6 improved in the presence of leucine, suggesting that, as in S. cerevisiae, Vam6 acts in the same pathway. Vam6 is essential for proper TORC1 activation; however, cells lacking Vam6 were able to shorten their doubling time in the presence of amino acids, suggesting that another pathway can stimulate cell growth in the presence of amino acids. Interestingly, a constitutively active version of Gtr1, where the conserved Q61 is mutated into a leucine residue, was able to rescue the growth defect phenotype of the _vam6_Δ mutant (Fig. 5B,C) and restore TORC1 activity (Fig. 5E). This result suggests that fission yeast Vam6 acts upstream of the Gtr1–Gtr2 heterodimer, as in the case of S. cerevisiae where Vam6 has been described as a GEF of Gtr1. Additionally, we show that Vam6 is localized to the vacuole membrane, where it interacts physically with Gtr1 in an amino-acid-dependent manner. In conclusion, we propose a model in which all the components of the Vam6, Gtr1 and Gtr2 and TORC1 pathway reside on the vacuolar surface, although only under conditions of stimulation by amino acids do these proteins interact physically (Fig. 7). This physical interaction activates Tor2, promoting cell growth and repressing sexual differentiation.
Fig. 7.
Hypothetical model showing how Vam6, Gtr1–Gtr2 and TORC1 respond to amino acids. Amino acids activate Tor2 by means of Vam6 and the Rag proteins to induce translation and promote cell growth. In the presence of amino acids, Gtr1 interacts strongly with Gtr2 and this heterodimer binds Vam6. Vam6 would activate Gtr1 and this, in turn, would bind to the TORC1 complex by means of Mip1. The active TORC1 complex induces Rps6 phosphorylation and, hence, induces cell growth and inhibit sexual differentiation. This activation would occur in the vacuolar membrane, where all the components would interact strongly in an amino-acid-dependent manner.
Materials and Methods
Fission yeast strains and media
All S. pombe strains used in this study are listed in supplementary material Table S1. Standard methods were used for growth, transformation and genetic manipulations (Sazer and Sherwood, 1990). Gene-tagged versions (gtr1-gfp, gtr1-rfp, gtr2-gfp, gfp-tor2, pop3-gfp, gfp-mip1, mip1-myc, gtr1-flag, gfp-vam6 and vam6-TAP) were generated by a PCR-based method (Bahler et al., 1998). Except where specifically indicated, all experiments in liquid culture were performed in Edinburgh minimal medium (EMM) containing the required supplements (except when mentioned), starting with a cell density of 2–4(×106) cells per ml, corresponding to the mid-exponential phase of growth. When cells were grown at 30°C in the presence of amino acids, EMM was supplemented with 225 mg/l of histidine and lysine hydrochloride. In the presence of leucine, we used leu1-32 auxotrophic strains, which were grown in EMM supplemented with 225 mg/l of leucine.
Protein extraction, immunopurification and western blotting
Protein extracts were obtained using trichloroacetic acid (TCA) extraction, as described previously (Foiani et al., 1994). For immunoprecipitation, exponentially growing cells (approximately 5×108 cells) were lysed in 250 μl of HB buffer [25 mM MOPS pH 7.2, 60 mM β-glycerophosphate, 15 mM MgCl2, 1mM DTT, 1% Triton X-100, 5 mM EGTA and 15 mM p-nitrophenylphosphate, supplemented with complete protease inhibitor cocktail tablets (Roche)]. Extracts were incubated with 20 μl of anti-Flag M2 affinity gel (Sigma-Aldrich) for 2 hours with shaking. For western blotting, 75–100 μg of total protein extract was run on SDS-PAGE (7.5% gels), transferred onto a nitrocellulose filter (Amersham), and probed with anti-TAP-peroxidase antiperoxidase soluble complex PAP (Sigma), anti-Flag-M2–peroxidase (Sigma-Aldrich), mouse anti-Myc 9E10 (a gift from Karim Labib, The Paterson Institute for Cancer Research, Manchester, UK), rabbit anti-RFP (MBL), and mouse anti-tubulin (a gift of Keith Gull, Sir William Dunn School of Pathology, University of Oxford, UK) primary antibodies, and horseradish-peroxidase-conjugated anti-mouse-IgG (NA 931, Amersham) and anti-rabbit-IgG (NA 943, Amersham) secondary antibodies. Immunoblots were developed using the enhanced chemiluminescence procedure (ECL kit, Amersham). TORC1 activity was monitored by measuring ribosomal protein S6 (Rps6) phosphorylation. Protein extracts were obtained using TCA extraction and the phosphorylation of Rps6 was detected by western blotting using the anti-phosphorylated Akt substrate (PAS) antibody (Cell Signaling Technology).
Epifluorescence microscopy
Epifluorescence microscopy was carried out using an Olympus IX71 fluorescence microscope improved with Delta Vision equipment from Applied Precision. To visualize vacuoles, FM4-64 (Biotium) was used to stain the vacuole membrane (Vida and Emr, 1995). Deconvolution option from Delta Vision and ImageJ software were used to process the images. Yeast colony pictures were taken using a Leica MZ75 binocular microscope equipped with an Olympus Camedia digital camera.
Acknowledgements
We thank Karim Labib, Keith Gull and Fuyu Tamanoi for antibodies and strains, the IBMCC Microscopy Unit for their help with the Deltavision microscope and Nicholas Skinner for corrections to the manuscript.
Footnotes
Funding
This work is supported by grants from Ministerio de Economía y Competitividad [grant numbers BFU2008-01808, BFU2011-28274, Consolider CSD2007-00015] and Junta de Castilla y León Grupo de Excelencia [grant number GR 265] to S.M. N.V. is supported by a postdoctoral grant from the Carlos III Institute, Ministerio de Sanidad.
References
- Alvarez B., Moreno S. (2006). Fission yeast Tor2 promotes cell growth and represses cell differentiation. _J. Cell Sci._119, 4475-4485 [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. _Yeast_14, 943-951 [DOI] [PubMed] [Google Scholar]
- Binda M., Peli-Gulli M. P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., De Virgilio C. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. _Mol. Cell_35, 563-573 [DOI] [PubMed] [Google Scholar]
- Blommaart E. F., Luiken J. J., Blommaart P. J., van Woerkom G. M., Meijer A. J. (1995). Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. _J. Biol. Chem._270, 2320-2326 [DOI] [PubMed] [Google Scholar]
- Bun-Ya M., Harashima S., Oshima Y. (1992). Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. _Mol. Cell. Biol._12, 2958-2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R. (2006a). Cell growth control: little eukaryotes make big contributions. _Oncogene_25, 6392-6415 [DOI] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R. (2006b). The TOR signalling network from yeast to man. _Int. J. Biochem. Cell Biol._38, 1476-1481 [DOI] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C. (2005). The TOR and EGO protein complexes orchestrate microautophagy in yeast. _Mol. Cell_19, 15-26 [DOI] [PubMed] [Google Scholar]
- Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. (2002). Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. _Genes Dev._16, 1472-1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. (1994). The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. _Mol. Cell. Biol._14, 923-933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., Avruch J. (1998). Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. _J. Biol. Chem._273, 14484-14494 [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. (2004). Upstream and downstream of mTOR. _Genes Dev._18, 1926-1945 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., Yanagida M. (2007). Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. _Genes Cells_12, 1357-1370 [DOI] [PubMed] [Google Scholar]
- Hirose E., Nakashima N., Sekiguchi T., Nishimoto T. (1998). RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. _J. Cell Sci._111, 11-21 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Ruegg M. A., Hall A., Hall M. N. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. _Nat. Cell Biol._6, 1122-1128 [DOI] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008). Regulation of TORC1 by Rag GTPases in nutrient response. _Nat. Cell Biol._10, 935-945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O., Ravichandran K. S. (2008). A pathway for phagosome maturation during engulfment of apoptotic cells. _Nat. Cell Biol._10, 556-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Guan K. L. (2009). Amino acid signaling to TOR activation: Vam6 functioning as a Gtr1 GEF. _Mol. Cell_35, 543-545 [DOI] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. _Mol. Cell_10, 457-468 [DOI] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. (2005). Rheb binds and regulates the mTOR kinase. _Curr. Biol._15, 702-713 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Otsubo Y., Urano J., Tamanoi F., Yamamoto M. (2007). Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. _Mol. Cell. Biol._27, 3154-3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Sato T., Tamanoi F. (2010). Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. _J. Cell Sci._123, 777-786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N., Noguchi E., Nishimoto T. (1999). Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through tr2p. _Genetics_152, 853-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. (1998). Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. _J. Biol. Chem._273, 3963-3966 [DOI] [PubMed] [Google Scholar]
- Price A., Wickner W., Ungermann C. (2000). Proteins needed for vesicle budding from the Golgi complex are also required for the docking step of homotypic vacuole fusion. _J. Cell Biol._148, 1223-1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajiki K., Hatanaka M., Nakamura T., Takeda K., Shimanuki M., Yoshida T., Hanyu Y., Hayashi T., Nakaseko Y., Yanagida M. (2009). Genetic control of cellular quiescence in S. pombe. _J. Cell Sci._122, 1418-1429 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. _Science_320, 1496-1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. _Cell_141, 290-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. _Curr. Biol._14, 1296-1302 [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. _Science_307, 1098-1101 [DOI] [PubMed] [Google Scholar]
- Sazer S., Sherwood S. W. (1990). Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. _J. Cell Sci._97, 509-516 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T., Hirose E., Nakashima N., I. i M., Nishimoto T. (2001). Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. _J. Biol. Chem._276, 7246-7257 [DOI] [PubMed] [Google Scholar]
- Shigemitsu K., Tsujishita Y., Hara K., Nanahoshi M., Avruch J., Yonezawa K. (1999). Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. _J. Biol. Chem._274, 1058-1065 [DOI] [PubMed] [Google Scholar]
- Smith E. M., Finn S. G., Tee A. R., Browne G. J., Proud C. G. (2005). The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. _J. Biol. Chem._280, 18717-18727 [DOI] [PubMed] [Google Scholar]
- Starai V. J., Hickey C. M., Wickner W. (2008). HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. _Mol. Biol. Cell_19, 2500-2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Cohen A., Diefenbacher M., Trautwein M., Martin D. E., Hall M. N. (2008). TOR1 and TOR2 have distinct locations in live cells. _Eukaryot. Cell_7, 1819-1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J., Sato T., Matsuo T., Otsubo Y., Yamamoto M., Tamanoi F. (2007). Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. _Proc. Natl. Acad. Sci. USA_104, 3514-3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani M., Hidaka H., Hotta Y., Ueno M., Ushimaru T., Toda T. (2006). Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. _Genes Cells_11, 1367-1379 [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. _J. Cell Biol._128, 779-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., Roitburg I., Nahari T., Kupiec M. (2005). Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. _Genetics_169, 539-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., Roitburg I., Schonbrun M., Harari R., Kupiec M. (2007). Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. _Genetics_175, 1153-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. (2000). New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. _J. Cell Biol._151, 551-562 [DOI] [PMC free article] [PubMed] [Google Scholar]