Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 17.
Published in final edited form as: Adv Drug Deliv Rev. 2009 Apr 17;61(6):428–437. doi: 10.1016/j.addr.2009.03.009
Abstract
Proteins bind the surfaces of nanoparticles, and biological materials in general, immediately upon introduction of the materials into a physiological environment. The further biological response of the body is influenced by the nanoparticle–protein complex. The nanoparticle's composition and surface chemistry dictate the extent and specificity of protein binding. Protein binding is one of the key elements that affects biodistribution of the nanoparticles throughout the body. Here we review recent research on nanoparticle physicochemical properties important for protein binding, techniques for isolation and identification of nanoparticle-bound proteins, and how these proteins can influence particle biodistribution and biocompatibility. Understanding the nanoparticle-protein complex is necessary for control and manipulation of protein binding, and allows for improved engineering of nanoparticles with favorable bioavailability and biodistribution.
Keywords: Nanoparticles, Protein binding, Immunology, Biodistribution, Biocompatibility
1. Introduction
Nanotechnology has recently gained attention as one of the critical research endeavors of the 21st century [1]. According to the National Nanotechnology Initiative, nanotechnology is the research and development of nanosystems, such as nanoparticles, on the scale of 1–100 nm [1]. Particles of this size are potentially useful tools for medicine and biology, as they are of commensurate size to important biological components (e.g. DNA, proteins, cell membranes) and thus able to interact in a sophisticated and controlled way at the cellular level. The ability to manipulate particular nanoparticle features, such as their physical, chemical, and biological properties, opens up a host of possibilities for researchers interested in rationally designing these nanoparticles for use in drug delivery, as image contrast agents, and for diagnostic purposes.
Nanoparticles for such medical applications are frequently given via parenteral administration. As with any foreign material, the body mounts a biological response to an administered nanoparticle. This response is the result of a complex interplay of factors, not just the intrinsic characteristics of the nanoparticle. In particular, most materials, upon contact with biological matrices, are immediately coated by proteins, leading to a protein “corona” [2–4]. Protein coronas are complex and variable. The complete plasma proteome is expected to contain as many as 3700 proteins [5,6], of which approximately 50 have been identified in association with various nanoparticles [7–10].
Certain components of the nanoparticle corona, called opsonins, may enhance uptake of the coated material by cells of the reticuloendothelial system (RES) [11,12]. The presence of opsonins on the particle surface creates a “molecular signature” which is recognized by immune cells and determines the route of particle internalization [13,14]. The route of internalization, then, may affect the eventual fate of the nanoparticle in the body (i.e. its rate of clearance from the bloodstream, volume of distribution, organ disposition, and rate and route of clearance from the body) [15,16].
The effect of this protein corona may have special significance for nanoparticles, due to the increased importance of surface effects for particles of this size. Several studies have shown that biological responses to nanoparticles tend to scale with surface area rather than mass [17–21]. As things become smaller, their surface areas shrink much more slowly than their volumes, causing nanoscale materials to have far greater surface-to-volume ratios than larger particles. A larger surface-to-volume ratio also implies more proteins will bind a nanoparticle (relative to its mass) than a particle of larger size.
Even for larger particles, protein binding is established as one of the most important factors influencing biodistribution [22–25]. In current preclinical testing of pharmaceutical molecules, evaluation of plasma protein binding is recognized as an important element in an assessment of drug efficacy, safety, and disposition [26,27]. It has been shown to be important for understanding the pharmacokinetics and pharmacodynamics of the drug inside the body [26,28–30]. It is also used to extrapolate preclinical data to models that predict potential drug efficacy and/or toxicity in humans [26]. For biomaterials used as medical implants, it has long been understood that the nature of the deposited protein layer onto these medical devices is responsible for the early immunological response in patients [6]. In fact, even late stage biological responses are influenced and dictated by the surface composition of the material and how this surface interacts with surrounding tissue.
Of course, there are other factors that play a role in determining how nanoparticles distribute within the body. These include particle properties such as size, shape, surface charge (zeta potential), solubility, surface modifications (including targeting), and route of administration. There have been numerous review articles dwelling on the importance of these factors and how each one can influence biodistribution [11,31–38]. What is missing, however, is an understanding of how these factors influence plasma protein binding to nanoparticles, and the importance of nanoparticle–protein interactions to biodistribution, biocompatibility, and therapeutic efficacy of nanoparticles. The mechanism of protein binding is not well understood, nor is it currently known how more or less protein binding influences the biological response to a given nanoparticle (e.g. uptake by phagocytic cells of the RES and clearance). However, it is clear that both the amount and identities of proteins on the particle surface play a part in affecting the biodistribution of nanoparticles. To fully understand the protein corona, one must understand not only which proteins are attached to the particle, but the kinetics, affinities, and stoichiometries of protein association and dissociation with the nanoparticle [2].
This review article will cover these aspects of protein binding to nanoparticles. We will discuss methods used to isolate and identify proteins bound to nanoparticles, what factors influence protein binding, how these factors can be manipulated to enhance or decrease protein binding for drug development, the kinetics of protein binding, and the importance of protein binding to drug delivery and distribution throughout the body.
2. Methods for nanoparticle separation from plasma
Many methods are available and have been used to identify particle-bound proteins. One common challenge is the isolation of the nanoparticle–protein complex from excess proteins without disrupting the complex or inducing additional protein binding [2,39]. For most protein identification studies, nanoparticles are incubated with plasma so that the plasma levels are in excess of the available particle surface area. This nanoparticle/protein ratio is more representative of the true biological situation [40] and is the most appropriate way to mimic the conditions in the bloodstream. The complete composition of the protein corona at any given time is determined by the concentrations of the plasma proteome, which consists of over 3700 proteins, as well as their kinetic properties (equilibrium constants, on/off rates/binding affinity) for the particular nanoparticle [5,6].
To date, the preferred method for isolating nanoparticle–protein complexes is centrifugation. Relative to other techniques, centrifugation is easy and requires little material. Separation by centrifugation has been used to identify major plasma proteins such as human serum albumin, immunoglobulins, and fibrinogen bound to the nanoparticles [2,40]. These are some of the most prevalent proteins found in human plasma, so it is not surprising to find these proteins bound to injected nanoparticles.
While centrifugation is the conventional method for separation of the nanoparticle–protein complex from excess proteins, it has many limitations. The outcome can be affected by the duration of washing as well as the solution volumes used during the washing steps in the centrifugation protocol. High abundance proteins may be identified due to insufficient washing, while large proteins and protein aggregates that sediment to the bottom of the centrifugation tube may also be falsely identified. While centrifugation can provide enough protein for identification, these assays should be performed with utmost care and accompanied by other methods to exclude false positive results [5,39]. Such methods include, but are not limited to, gel filtration using size-exclusion chromatography, magnetic separation, and microfiltration [10,41].
The importance of using multiple techniques is illustrated by a study in which researchers compared techniques to isolate proteins bound to polysaccharide-stabilized iron oxide nanoparticles [10]. These techniques included centrifugation, gel filtration, magnetic separation, and membrane-based static microfiltration. The analysis of the proteins from all four techniques showed generally similar adsorption patterns. However, albumin binding appeared to depend on the method of separation. Negligible amounts of albumin were detected when magnetic separation was utilized compared to the other separation techniques. The study authors posited that this could be the result of albumin sticking to the filtration system, or somehow avoiding detection with this separation method. This finding emphasizes that while many techniques are available to separate proteins from nanoparticles, each individual method will have its nuances and specific limitations. As in the case of centrifugation separation, care must be taken when interpreting the results of each method to assure no false negative (or positive) results are obtained.
The most commonly used methods for determining total protein binding to small molecules and macromolecules are equilibrium dialysis and ultracentrifugation [42]. Identification of drug-bound proteins is routinely performed through techniques such as solid phase extraction, affinity chromatography, electrospray ionization mass spectrometry, equilibrium dialysis, ultrafiltration, ultracentrifugation, gel filtration, HPLC, capillary electrophoresis, and protein microarrays [26,43–45]. However, many of these methods are not applicable to some nanoparticles, depending on their size and unique chemical properties. For example, certain nanoparticles have inherent absorbance or fluorescence properties which can interfere with protein microarrays. These types of interferences must be identified and understood.
3. Protein separation and identification
Once the nanoparticle–protein complex has been separated from excess plasma proteins, the proteins bound to the nanoparticle surface need to be individually separated and identified. A common technique for the separation of the proteins is two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) [23,46,47] (Fig. 1). To identify individual proteins, it is common practice to compare the 2D protein gels to a 2D master map of human plasma proteins [8,9,15,23,25,47–52]. However, differences in donor plasma and anticoagulants used during the blood collection process (EDTA, sodium citrate, lithium heparin), can result in variations in plasma protein maps, and hence may contribute to a misinterpretation of the 2D data when compared to a specific protein master map.
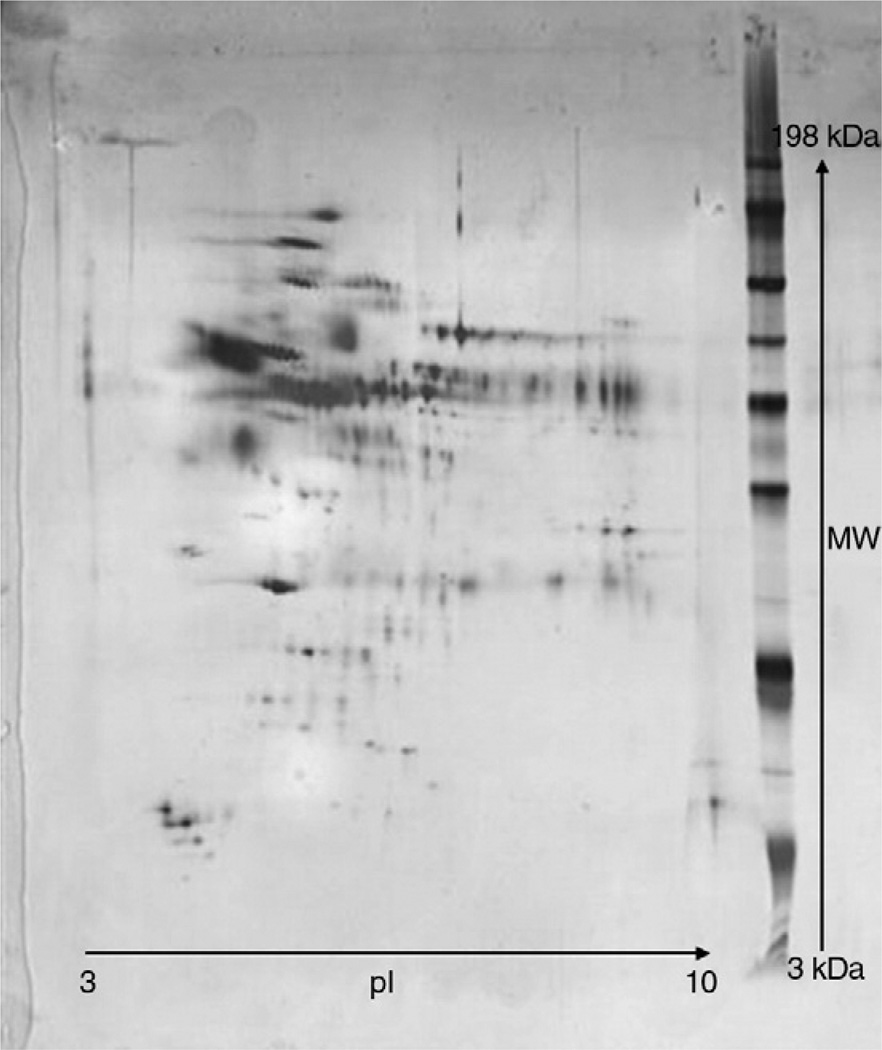
Fig. 1.
Example of a 2D protein gel used for protein identification. The protein adsorption pattern was analyzed for a sample of 30 nm colloidal gold. The colloidal gold sample was incubated with human plasma and then recovered by centrifugation. The proteins bound to the nanoparticles were separated by 2D gel electrophoresis.
To overcome this problem, researchers have begun to use N-terminal sequencing on individual spots to properly identify proteins [49–51], rather than simply relying on 2D separation patterns. Mass spectrometry, followed by peptide sequencing, is also conducted on individual excised protein spots from the 2D gel and compared to a known database of proteins [5,9,22,53,54]. For the identification of particular proteins of interest, immunoblotting and Western blotting have also been applied [55–58].
Aside from 2D-PAGE, other separation methods involving gel filtration, such as size-exclusion chromatography or affinity chromatography, are also used to separate proteins from plasma as well as identify individual proteins [10,40]. In some situations, the protein fractions collected from a chromatography run contain individual proteins, and no further separation is required. These proteins can then be identified through N-terminal sequencing or mass spectrometry and peptide sequencing.
Differences among the many types of nanoparticles can result in different protein binding profiles. Table 1 provides a general overview of the major proteins identified in the literature bound to an assortment of nanoparticles of different size, chemical make-up, and surface properties. Table 1 also describes the methods used for separating the proteins from plasma, as well as determining the protein binding profile. Though some proteins were specific to certain types of nanoparticles, seemingly dependent on the nanoparticle structure, the proteins that were identified on virtually all nanoparticles were those mentioned before, mainly albumin, immunoglobulin G (IgG), fibrinogen, and apolipoproteins. Cedervall et al. suggested that these proteins are found bound to nanoparticles due to their large abundance in blood [5]. What is unknown, however, is the affinity of the proteins' binding the nanoparticles. These proteins may dominate the surface of the nanoparticles initially, but then be displaced by proteins of lower abundance, higher affinity, and slower kinetics. This may lead to the presence of other nanoparticle-bound proteins at later times, such as other immunoglobulins, apolipoproteins, and components of the complement system [40]. These proteins, along with albumin and fibrinogen, are important in the clearance process of the body [59,60].
Table 1.
Summary of various nanoparticles and protein binding.
| Nanoparticles | Identified proteins | Method of protein isolation | Method of protein separation andidentification | Reference |
|---|---|---|---|---|
| Polystyrene with poloxamer 184,188, 407 | Factor B, transferrin, albumin, fibrinogen, IgG, apolipoproteins | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [46] |
| Liposomes | Albumin, fibrinogen, apolipoproteins, IgG, α1-antitrypsin, α2-macroglobulin, IgM | Gel filtration | 2D-PAGE; comparison to known 2D gel of plasma | [48] |
| Single-walled carbon nanotubes | Albumin | Centrifugation | Western Blot | [22] |
| Solid lipid nanoparticles with Tween 80 | Fibrinogen, IgG, IgM, apolipoproteins (including ApoE), transthyretin | Centrifugation, gel filtration | 2D-PAGE; comparison to known 2D gel of plasma | [74] |
| Solid lipid nanoparticles with poloxamer 188 | Fibrinogen, IgG, IgM, apolipoproteins (excluding ApoE), transthyretin, albumin | Centrifugation, gel filtration | 2D-PAGE; comparison to known 2D gel of plasma | [74] |
| Poly(lactic acid) nanoparticles with PEG | Albumin, fibrinogen, apolipoproteins, IgG | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma, and N-terminal sequencing | [49] |
| Polyhexadecylcyanoacrylate nanoparticles | Albumin, apolipoproteins, IgG, transferrin | Centrifugation | 2D-PAGE; comparison to known 2D gel of serum and mass spectroscopy | [9,55] |
| Poly(ε-caprolacton) nanoparticles | IgG, apolipoproteins | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma, and N-terminal sequencing | [50] |
| Polycyanoacrylate nanoparticles | Albumin, IgG, IgM, fibrinogen, apolipoproteins | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [52] |
| Iron oxide nanoparticles | Albumin, IgG, IgM, fibrinogen | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [10] |
| Albumin, IgM, fibrinogen, C3b, apolipoprotein A-1 | Gel filtration | |||
| Albumin, IgG, IgM, fibrinogen, C3b | Static filtration | |||
| IgG, IgM, fibrinogen | Magnetic separation | |||
| Various polymer/copolymer composition nanoparticles | Albumin, IgG, fibrinogen, apolipoproteins | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [129] |
| Poly(d,l-lactic acid) nanoparticles | Albumin, IgG, fibrinogen, IgM, apolipoproteins, antithrombin III | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [130] |
| Polystyrene with Rhodamine B | Albumin, IgG, fibrinogen, apolipoproteins, PLS:6 | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [131] |
| Various polymer/copolymer composition nanoparticles | Albumin, IgG, fibrinogen, IgM, apolipoproteins, PLS:6, U2 | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma, and N-terminal sequencing | [25] |
| Poly(isobutylcyanoacrylate) with Dextran | Albumin, IgG, fibrinogen, apolipoproteins, serotransferrine, transthyretine | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [75] |
| Single- and double-walled carbon nanotubes | Albumin, fibrinogen, apolipoproteins, C1q | “Affinity chromatography,” centrifugation of beads | SDS-PAGE with N-terminal sequencing, mass spectroscopy, or western blotting | [132] |
| Polybutylcyanoacrylate nanoparticles with polysorbate 80 | Albumin, IgG, fibrinogen, IgM, apolipoproteins | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [15] |
| Solid lipid nanoparticles with poloxamer or poloxamine coating | Albumin, fibrinogen, apolipoproteins | Centrifugation | 2D-PAGE; comparison to known 2D gel of plasma | [8] |
4. Kinetics of protein binding
The composition of the protein corona on a given nanoparticle, at a given time, depends on the concentrations and kinetic properties of the proteins found in plasma. Therefore, it is important to not only determine which proteins are adsorbed onto the surface of the nanoparticle, but also understand the binding affinities and stoichiometries. Protein affinities for nanoparticle surfaces likely differ from their affinities to analogous bulk materials due to a variety of effects including size, surface area, and the curvature of the nanoparticle surface. The composition of the nanoparticle–protein complex is in constant flux for the duration of the nanoparticle's interaction with the body. It is highly probable that proteins at high concentrations in plasma and with high association rates will initially occupy the surface of the nanoparticle. However, over time, these proteins may dissociate and be replaced by proteins of lower concentrations, slower exchange rates, and/or higher affinities [2,6]. The whole process of competitive adsorption of proteins onto a limited surface based on abundance, affinities, and incubation time is collectively known as the “Vroman Effect” [24,61,62]. This effect is important to consider in regards to particle distribution throughout the body. As the particles distribute from the blood to various locations, the differences in protein levels, as well as their affinities for binding, may play a part in determining how the protein corona evolves as the nanoparticle is shuttled from compartment to compartment.
Previous kinetic studies on solid lipid nanoparticles (SLN) showed that initial protein binding was predominated by albumin, replaced over time by fibrinogen, which was then replaced by IHRP (inter-α-trypsin inhibitor family heavy chain-related protein) and apolipoproteins [24,46]. Also shown was that with increasing plasma concentrations, the amount of fibrinogen on the solid lipid nanoparticle surface decreased while the amount of apolipoproteins steadily increased [24,63]. It was also shown that at high concentrations of plasma, fibrinogen can be displaced within seconds, if not fractions of a second, by apolipoproteins. Even though the concentrations of the several apolipoproteins found in human plasma are substantially lower than that of fibrinogen, the affinity (_K_d) of apolipoproteins, especially to hydrophobic surfaces, is much higher [24].
The kinetics of protein adsorption can be monitored in many ways. A study on 50 nm lecithin-coated polystyrene nanospheres used a combination of SDS-PAGE and Western blotting to analyze samples at various time points [58]. This study revealed a quantitative and qualitative profile of serum proteins adsorbed on the surface of the nanoparticles over periods from 5 min to 360 min. The quantitative profile showed an initial spike in the total amount of adsorbed plasma proteins from 60 min to 180 min, followed by a continual increase in adsorbed plasma proteins throughout the rest of the 360 minute experiment. According to the study, the qualitative profile showed a change in the types of proteins adsorbed, with some individual proteins increasing in quantity over time while others decreased. Of those increasing over time, the two main proteins identified were complement C3 and IgG. Other serum proteins, such as apolipoprotein E (ApoE) and immunoglobulin A (IgA), also increased with time. Conversely, albumin concentrations appeared to be constant over the course of the 360 min. No specific proteins were mentioned whose concentrations decreased with time.
While SDS-PAGE and Western blotting can be very useful in obtaining general information about the proteins adsorbed onto the nanoparticles, limitations are applicable. It is very difficult to obtain quantitative results from these gel methods as they are predominantly used for comparison purposes. Mobility on the gel can also be affected by variations in the proteins of interest, causing inaccurate results for molecular weight determination. Sensitivity limitations are also an issue. SDS-PAGE can detect anywhere between 1 and 50 ng of a single protein band, depending on the stain used, while Western blot limitations are dependent on the antibodies used as well as the conjugated substrate of choice. All of these conditions would have to be optimized for the particular protein of interest.
Another study evaluated the kinetics of protein binding to poly (ethylene glycol)-polyhexadecylcyanoacrylate (PEG-PHDCA) nanoparticles using a system that separates fluorescent dye labeled protein–SDS complexes electrokinetically in a gel media, and separates the proteins based on molecular weight [9]. Fluorescence measurement provides an estimation of the protein concentration with time. In this study, the two main proteins that were identified and monitored were albumin and ApoE. Initially, the concentration of albumin on the nanoparticles rapidly increased, then leveled off after 5 min. However, a steady increase in ApoE concentrations over the 60 minute time course was observed.
While no formal description of sensitivity limitations of this system have been described, it is expected to match or exceed the sensitivity of SDS-PAGE analysis. This system is currently being used with lab-on-chip technology, and this affords to the limitation of the chip, possibly requiring multiple chips to analyze various molecular weight range samples.
A few established techniques have recently been used to analyze the kinetic properties of protein binding to nanoparticle surfaces in a less potentially disruptive manner than centrifugation. These techniques include size-exclusion chromatography (SEC), isothermal titration calorimetry (ITC), and surface plasmon resonance (SPR) [2,40]. For various _N-iso_propylacrylamide/_N-tert_-butylacrylamide (NIPAM/BAM) copolymer nanoparticles, ITC was used to determine the stoichiometry and affinity of human serum albumin (HSA) to the nanoparticle surface. The results showed a similar association constant (_K_a) of ~2×106 M−1 for all particles irrespective of composition or size. On the other hand, the number of proteins bound to the nanoparticles increased with increasing hydrophobicity of the nanoparticle surface, as well as with increasing size. Hydrophobicity was controlled by varying the NIPAM/ BAM copolymer ratio from the more hydrophilic ratio of 85:15 NIPAM/ BAM to the more hydrophobic ratio of 50:50 NIPAM/BAM. For 70 nm particles, the number of bound proteins was 60+/−20 and 350+/− 70 for the hydrophilic and hydrophobic particles, respectively. For 200 nm particles, the number of bound proteins was 980+/−780 and 5400+/−1700 for the hydrophilic and hydrophobic particles, respectively [2].
With those same copolymer nanoparticles, SEC was used to determine protein exchange rates. A clear difference in exchange rates was observed, and this difference was attributed to the differences in particle hydrophobicity and protein identity [2]. SPR on thiol-conjugated nanoparticles bound to gold surfaces was used in an attempt to study the kinetics of association and dissociation of the NIPAM/BAM copolymers with plasma. While rate constants could not be fitted for plasma due to the vast excess of proteins, a fit of two rate constants revealed different kinetics, suggesting that the nanoparticles bind more than one type of protein with different rates [2].
All three of the above mentioned techniques also have their limitations in terms of sensitivity and resolution. Another point of limitation for these techniques is the nanoparticles themselves. Since the proteins are not disassociated from the nanoparticles, the nanoparticles are introduced into the system of analysis. This causes complexity when looking at various types of nanoparticles, particularly metallic (e.g. colloidal gold) nanoparticles, which can interfere with the analysis of techniques such as SPR. Care must be taken to avoid this interference, and confirmation by complimentary techniques is usually required to garner all the information from the experiment.
While some light has been shed on the kinetics of the nanoparticle–protein complex, the time-dependence of nanoparticle–protein binding seems highly specific to the type of nanoparticle and its size and surface properties. Unfortunately, no overall, distinct trends have emerged from these studies. One important area that is particularly poorly understood is the mechanism of the association and interaction of proteins with the particle surface. While hydrophobic/hydrophilic and electrostatic interactions (including Van Der Walls) most likely play a role, no one has explored these interactions thoroughly [64–68]. In-depth knowledge of these interactions may well lead to a better understanding of what influences the kinetics of protein binding and what trends, if any, may be applicable to this understanding. New developments in analytical ultracentrifugation for characterization of nanoparticles may be useful in understanding various particle–protein interactions, but no definitive uses of this technique for that purpose have been described yet [69–71]. Greater analytical improvements and techniques are greatly needed to fully understand the complexity involved in protein binding.
5. Properties that influence protein binding
The majority of studies examining the influence of protein binding on uptake have been conducted by either preincubating particles with bulk serum/plasma, or by preincubating particles with individual proteins or attaching individual proteins to the surface of the particles, and evaluating uptake by macrophages. This research has showed that neutrally charged particles have a distinctively slower opsonization rate than charged particles, demonstrating a direct correlation between surface charge and protein binding [60,72]. A study on the influence of surface charge density (i.e. zeta potential) of negatively charged polymeric nanoparticles (keeping their size and surface hydrophobicity approximately constant), showed an increase in plasma protein absorption with surface charge density, but showed virtually no difference in the profile of detected protein species [23]. On the other hand, studies on polystyrene nanoparticles have shown that positively charged particles (bearing basic functional groups) preferentially adsorbed proteins with isoelectric points less than 5.5 (such as albumin), while negatively charged particles (and particles with surfaces bearing acidic functional groups) predominantly bound proteins with isoelectric points greater than 5.5 (such as IgG) [73]. This study also showed that distinct proteins such as albumin and IgG preferentially bind particles bearing strong basic or weak acid groups on their surface.
The hydrophobicity of a nanoparticle surface has previously been shown to influence not only the amounts of protein bound to the particle, but also the identities of the bound proteins [5,8,15,25,74,75]. Generally, hydrophobic particles are opsonized more quickly than hydrophilic particles, due to the enhanced absorbability of plasma proteins onto the surface of hydrophobic particles [60,76–78]. In one example, a less hydrophobic 85:15 NIPAM/BAM copolymer particle was compared to its more hydrophobic counterpart, a 50:50 copolymer particle. The less hydrophobic copolymer particle-bound virtually no proteins, except for small amounts of HSA, while the more hydrophobic copolymer particles preferentially bound apolipoproteins AI, AII, AIV, and E as well as HSA, fibrinogen, and various other proteins [5]. A study on 200 nm NTPAM/BAM hydrophobic and hydrophilic copolymers showed that while the protein pattern was the same for the copolymers, the more hydrophobic particles bound ~ 50-fold more ApoA-I than the hydrophilic copolymer. On the other hand, an unidentified protein band at 75 kDa was similar for both particles. Caution must be taken when analyzing polymers in terms of their hydrophobicity; changing the hydrophobicity of polymers is very difficult without inadvertently affecting the size, charge, and/or composition of the particles. Since all these properties are important influences on the protein binding profile of a nanoparticle, these factors must also be considered.
The roles of particle size, surface curvature, and particle surface area in protein binding have also been investigated. Investigators have shown that for 50:50 NIPAM/BAM copolymer particles varying in diameter from 70 to 700 nm, the amount of bound protein varied with size and surface curvature. However, the protein pattern was the same for all sizes considered [5].
Though particle composition (base material type, shape, and size) clearly influences protein binding, the surface properties (charge and hydrophobicity) are likely to be more important. Whether a nanoparticle binds proteins at all seems to depend on surface chemistry (charge and hydrophobicity), but the majority of nanoparticles which bind proteins tend to bind the same types of proteins irrespective of their composition (only the amounts of bound protein change). This is clearly illustrated in Table 1 and Table 2. While there are exceptions, this rule seems to hold true for the majority of nanoparticles that have been investigated.
Table 2.
Table Nanoparticle properties that influence protein binding.
| Nanoparticle propertiesthat influence proteinbinding | Observation/effect | Reference |
|---|---|---|
| Surface charge | Neutral particles have slower opsonization rates than charged particles | [60,72] |
| Hydrophobicity | Hydrophobicity influences both the amount of opsonization and the identities of bound proteins. | [5,8,15,25,74,75] [60,76–78] |
| Hydrophobic nanoparticles are opsonized more quickly than hydrophilic particles. | ||
| Size/morphology/shape/ surface curvature | Influences the amount of bound protein but not identities of bound proteins. | [5] |
An interesting set of studies have shed some light on the protein–nanoparticle interface and how protein structure, activity/function, and stability can change upon binding to a nanoparticle surface [79–81], and how this may be influenced by properties such as curvature and surface chemistry [82]. Studies have shown that for specific proteins (lysozyme and human carbonic anhydrase), smaller nanoparticles, possibly due to their higher surface curvature, had a higher retention of native-like protein structure and function than their larger counterparts [82–84]. Other studies showed single-walled carbon nanotubes have the ability to stabilize proteins under harsh conditions better than flat supports [82,85,86], and that certain nanoparticles may induce protein aggregation and fibrillation [87–89]. Several studies on the interactions of 4th and 5th generation polyamidoamine dendrimers with various proteins (including HSA, BSA, and acetyl cholinesterase to name a few) have shown changes in the enzyme activities, binding properties, and protein confirmation of these proteins [90–93]. Finally, in terms of surface chemistry, a study comparing hydrophobic and hydrophilic silica spheres showed a greater change in the secondary structure of fibrinogen and BSA for the hydrophobic particles [82,94]. No hypothesis was made as to how the changes in protein structure might influence macrophage uptake. Such a study would be interesting and helpful to understand the role of bound protein structure in nanoparticle uptake.
6. Effect of protein binding on nanoparticle biodistribution
Just as the properties of a nanoparticle influence the protein binding profile, the binding profile influences biodistribution. Protein binding can cause a change in nanoparticle size and surface charge [22,31,34,58]. These changes affect the internalization process of these nanoparticles into macrophages and the overall distribution throughout the body. While many studies have considered the importance of physical characteristics (i.e. size, surface charge, etc.) for biodistribution and how they can be manipulated to induce a desired effect, this section will focus primarily on the changes in biodistribution caused by protein binding. Protein binding has been posited to be one of the key factors that influence biodistribution [22–25], and (as discussed above) many properties, such as size and surface chemistry of the nanoparticle, influence protein binding. Understanding this “cause and effect” relationship can help in understanding the biodistribution of nanoparticles.
Specific bound proteins can have a direct effect on particle internalization and biodistribution [34,60,95]. Certain bound proteins allow macrophages of the reticuloendothelial system (RES) to more easily recognize nanoparticles [60]. Binding of opsonins such as IgG, complement factors, and fibrinogen are said to promote phagocytosis and the eventual removal of the particles from systemic circulation via cells of the RES [24,96,97]. These particles tend to sequester in the RES organs very rapidly and concentrate in the liver and spleen [60,98–100]. On the other hand, dysopsonins such as albumin are said to promote prolonged circulation times in the blood [24,101,102].
For polystyrene nanospheres, fetuin was shown to mediate uptake of these particles by liver macrophages (Kupffer cells) via the scavenger receptor [103]. Other serum proteins were also shown to aid in this uptake, but were not specifically identified. Another study involving polystyrene nanospheres showed the formation of a complex with fibronectin, allowing these particles to be primarily taken up by Kupffer cells [22,101]. In bulk plasma, a study showed that over time, an increase in the amount of C3 and IgG on the surface of lecithin-coated polystyrene nanospheres was directly correlated with an increase in hepatic uptake by Kupffer cells [58]. The concentrations of ApoE and IgA were also shown to increase, but a correlation between this increase and potential influence of uptake by hepatocytes is still under investigation. Covalent attachment of various apolipoproteins, mainly ApoE, ApoA-I, and ApoB- 100, to either HSA or poly(butylcya-noacrylate) nanoparticles, was shown to enable drug transport into the brain by enabling the interaction of these particles with brain endothelial cells [104–106].
There is still some debate on the benefits or disadvantages of protein binding [67]. In some cases, it is useful to have these proteins bind as they can direct or target the nanoparticle to a particular area of the body. For example, the binding of certain apolipoproteins to the surface of the particle can be useful for distribution of the particle across the blood brain barrier (BBB) into the brain [15,74,107,108]. However, binding of these proteins has also been shown to correlate with rapid uptake into the liver and the spleen, and clearance of the particles by the RES, a negative effect if one is attempting to increase circulation and retention time of the nanoparticles in the body [60].
The attachment of polymers has widely been used to prevent protein binding, or at least decrease the amount, and/or change the overall profile of proteins bound [109]. In one case, the attachment of a poloxamine 908 coating to polystyrene nanospheres reduced fibronectin adsorption considerably when compared to the uncoated nanospheres, and also reduced liver accumulation [22,101]. In another study, the addition of a Pluronic F127 coating to both single-walled carbon nanotubes and amorphous silica particles enhanced dispersion of the nanoparticles, while greatly reducing adsorption of serum proteins, and resulted in reduced toxicity to RAW 264.7 cells [22]. Table 3 describes the effects of various coatings on nanoparticles in terms of protein binding and biodistribution. On a separate note, impurities and products of oxidative degradation of polymers were associated with certain pharmacological and immunological effects [reviewed in 110]. This emphasizes the importance of nanoparticle physicochemical characterization prior to assigning observed effects to nanoparticles.
Table 3.
Table Protein binding compared to uncoated nanoparticles.
| Coating | Proteinbindingcomparedtouncoated | Targeting tospecificphagocytic uptake(binding of C3, IgG) | Prevention ofbiodistributionto RES(in vivo) | References |
|---|---|---|---|---|
| Poly (ethylene glycol) (PEG) | Less | Yes | Yes | [52,111] |
| Less | Yes | N/A | [49] | |
| Less | N/A | Yes | [133] | |
| N/A | N/A | Yes | [113] | |
| Poloxamer | N/A | N/A | Yes | [57] |
| Poloxamine | N/A | N/A | Yes | [101] |
| Dextran | Less | Yes | N/A | [50] |
| Pluronic F127 | Less | N/A | N/A | [22] |
| Polysorbate | N/A | Yes | N/A | [15] |
| Poly(oxyethylene) triblocks | Less | No | N/A | [51] |
The addition of poly(ethylene glycol) (PEG), or “PEGylation,” is the most common and preferred method of “masking” nanoparticles from immune recognition [49,52,60]. It has been shown to decrease interactions of various nanoparticles with blood proteins and help avoiding recognition by the RES, in essence prolonging blood circulation [9,49,50,111–113]. PEGylation can be performed by covalently linking, entrapping, or adsorbing PEG chains onto the surface of the particle [60]. The theory behind PEGylation is that the addition of PEG can add protein/opsonization resistance properties by preventing interactions between the particle surface and the plasma proteins [60,114]. In one study comparing PHDCA and PEG-PHDCA, roughly twice as much protein was adsorbed onto the non-PEGylated nanoparticle [9]. The composition of adsorbed proteins was also shown to be slightly different.
Various structure activity relationship studies have been conducted to determine how a change in the thickness and density of a PEG coating affects opsonization and biodistribution. In one study, poly (lactic acid) was coated with varying molecular weights (MW) of PEG (from 2000 to 20,000 Da) as well as varying PEG contents to change surface density of PEG [49]. Protein adsorption decreased with increasing MW, and was much less than uncoated particles. In terms of surface density of PEG, again, protein adsorption decreased with increasing PEG coating density, and was much less than uncoated particles. In all cases, protein adsorption was not completely avoided (i.e. albumin, fibrinogen, IgG, and apolipoproteins were detected), but was lessened with PEGylation. The PEG coatings were also shown to decrease uptake by both polymorphonuclear cells and THP-1 cells, proportional to the surface density of the PEG [49]. Numerous other studies have also shown that increasing the molecular weight of a PEG coating can decrease protein binding and cellular uptake and increase the particle's blood circulation half-life [52,60,115]. A study on fluorescien-labeled poly(d,l-lactide) nanoparticles compared covalently linked PEG nanoparticles versus multiblock copolymer PEG nanoparticles. Results showed that the covalently linked PEG nanoparticles prevented protein adsorption to a greater degree than did copolymer nanoparticles, but even the copolymer nanoparticles bound less protein than uncoated nanoparticles [67]. In this case, the results were attributed to more surface PEG on the covalently linked nanoparticles as opposed to the copolymer nanoparticles. While none of the particles were cytotoxic, RAW264.7 cells showed a reduced uptake for the covalently linked PEG nanoparticles compared to the copolymer PEG nanoparticles, in accordance with the protein binding data. In a study on cytotoxic PHDCA nanoparticles, PEGylated particles were shown to have decreased protein binding, increased blood circulation time, and decreased cytotoxicity [111].
While changes due to protein binding might be difficult to predict, understanding their effects can help researchers develop particles that make use of this phenomenon, and help in the rational design of nanoparticles for use in drug delivery, as image contrast agents, and for diagnostic purposes. The notion of cloaking nanoparticles to prevent protein binding and avoid uptake into macrophages, rendering longer circulation times in the body, can be very important for a researcher attempting to develop a nanoparticle-based therapy with limited side effects caused by non-specific uptake. On the other hand, specific bound proteins can also help target or direct the nanoparticle towards a particular pathway or area of the body [24]. Therefore, engineering a particle that can specifically bind certain proteins of interest for targeting purposes, or engineering particles with these proteins already bound to the particle, can greatly enhance the ability to develop directed nanoparticles as drug candidates (Fig. 2).
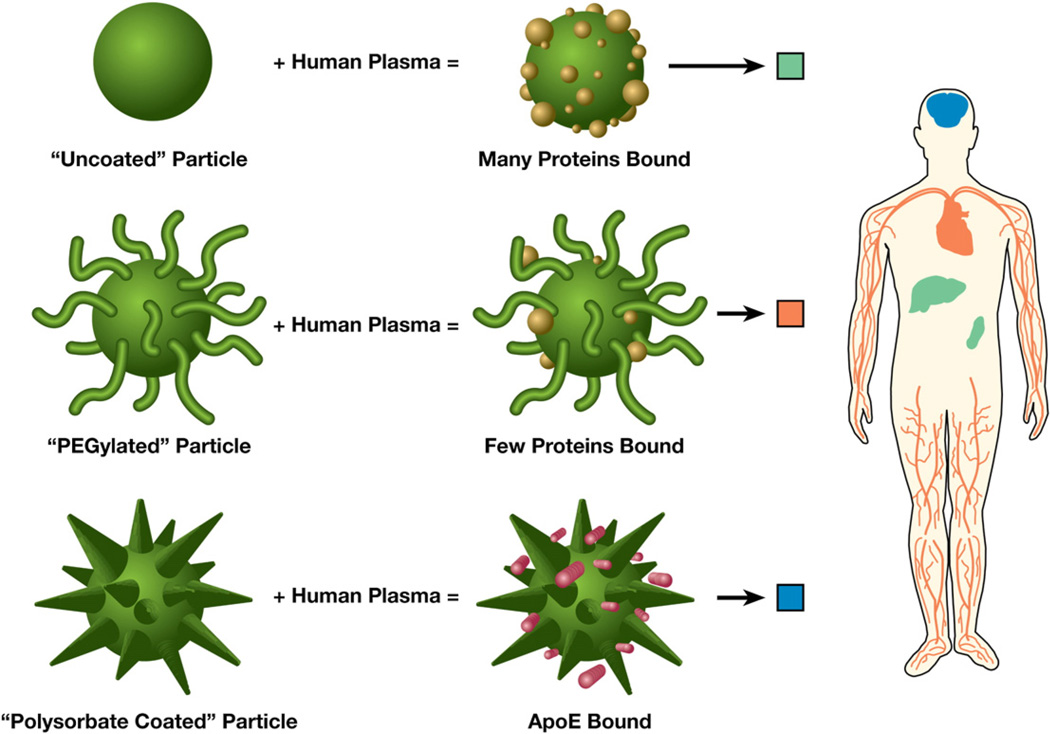
Fig. 2.
Biodistribution of nanoparticles with varying coatings and bound proteins. Uncoated particles bind proteins and are taken up by the RES into the liver and spleen. “PEGylated” particles bind very few proteins, avoid uptake by the RES, and are longer circulating in the blood. “Polysorbate-coated” particles can specifically bind ApoE and selectively target to the brain across the blood brain barrier.
One study showed that injection of 30 nm colloidal gold with bound tumor necrosis factor resulted in a rapid accumulation of gold in the liver and spleen, and that the gold did not dissipate out of these organs even after one month's time [113]. However, upon PEGylation, this nanoparticle avoided detection and clearance by the RES, most likely due to the lack of protein binding on the surface of the nanoparticle [113]. These particles were also shown to be targeted to, and accumulated in, the tumor in a tumor-burden mouse model. It is important to keep in mind that while PEGylation can help in avoidance of rapid recognition by the RES, complete avoidance is rarely achieved as nanoparticles may still get recognized and taken up by the RES system, as evident by the results of the above mentioned study [113]. The reason for this is currently unknown and has not been thoroughly investigated. A study with radiolabeled poloxamer 407 and poloxamine 908, however, showed the potential for desorption of the coating or displacement by serum proteins [116]. This may be a possible mechanism for PEG as well, resulting in opsonization and RES uptake of certain PEGylated nanoparticles.
PEG is not the only polysaccharide that can be attached to a nanoparticle to avoid immune recognition and/or influence the proteins that bind to the surface of the nanoparticle. This is clearly illustrated in Table 2. Various other polymers and polysaccharides have been utilized in place of PEG.
In terms of specific ApoE binding to nanoparticles for delivery across the BBB into the brain, the adsorption of apolipoproteins has been shown to be important for the transportation of drugs across the BBB and into the brain [15], though there is still some debate over the mechanism of transport [108]. Researchers observed a correlation between the amounts of ApoE bound to solid lipid nanoparticles versus the chain length of the poly(ethylene oxide) (PEO) monomer on the poloxamer coating attached to the solid lipid nanoparticle. Shorter PEO chain lengths resulted in greater ApoE adsorption [8]. Another study also showed a similar correlation between PEO chain length on poloxamer coatings of polystyrene nanoparticles and amount of ApoE adsorbed [46]. For different coatings, it was shown that a polysorbate 80 coating onto poly(butylcyanoacrylate) (PBCA) nanoparticles resulted in the preferential adsorption of ApoE [15]. Researchers showed an enhanced binding of ApoE and ApoB-100 to PEG-PHDCA nanoparticles compared to the non-PEGylated nanoparticles [55]. Even a difference in a coating of the same nanoparticle can make a difference. Gessner et al. coated solid lipid nanoparticles with either Tween 80 or poloxamer 188 [74]. The particles coated with Tween 80 showed an abundance of binding of various apolipoproteins, especially ApoE. Poloxamer 188-coated nanoparticles bound various apolipoproteins as well, but did not bind ApoE. It did, however, bind albumin, which was not abundant on the Tween 80-coated particles. Goppert et al. also showed varying binding of ApoE to solid lipid nanoparticles based on the different types of polysorbate coatings [15].
One can expect that the direct attachment of apolipoproteins to a particle surface would aid in drug delivery to brain tissue. Indeed, a study showed that the covalent attachment of either ApoE3, ApoA-I, or ApoB-100 to albumin nanoparticles significantly facilitated transport of a bound study drug, loperamide, across the BBB as opposed to controls of loperamide alone or nanoparticles with no apolipoproteins attached [104]. Other drugs such as doxorubicin, tubocurarine, and dalargin, which normally have poor brain diffusion, were also able to be delivered to the brain using polysorbate 80-coated PBCA nanoparticles, which preferentially adsorbed ApoE compared to uncoated nanoparticles [108,117–121]. The knowledge of the potential targeting properties of apolipoproteins, along with anti-toxicity and targeting effects of albumin, make for a potent combination for drug delivery vehicles.
7. Use in medical applications
Knowledge of specific proteins that are adsorbed onto particle surfaces and their biological effects, including biodistribution and targeting, can be very useful when attempting to design drug candidates. Besides the ability to design particles to interact with particular proteins, one can attach these proteins specifically to a nanoparticle to obtain the desired targeting effect. A good example of this technology at work is the FDA approved drug Abraxane™. Abraxane is a nanoparticle albumin-bound (nab) form of paclitaxel [122]. It has been shown to target tumors, enhance tumor penetration, and minimize toxicity compared to other forms of paclitaxel [123]. Albumin trans-cytosis and uptake into endothelial cells is meditated by binding of albumin to a cell surface 60-kDa glycoprotein (gp60), which then signals the rest of the cascade to internalize albumin and albumin-bound particles [124–128]. Albumin is a logical choice for binding to nanoparticles as it has been shown to be one of the prominent plasma proteins that bind to nanoparticles when they come in contact with plasma (Table 1). Pre-binding nanoparticles with albumin can help overcome some of the toxicities associated with solvent-based formulations, and its targeting properties make it a promising choice for nanoparticle formulations.
The ability to use the nab technology, or related technology, in the future can greatly enhance the targeting ability of drugs while reducing systemic toxicity. The nab technology is currently being applied to two applications currently in early clinical trials. These are _nab_-Docetaxel for targeting solid tumors and _nab_-Rapamycin for intravenous administration of rapamycin as an anti-cancer agent [125] (as opposed to the oral administration of rapamycin for use as an antibiotic or immunosuppressant). Future applications of the nab technology are also under development.
8. Conclusion
A first step in fully utilizing and developing nanoparticles as successful drug candidates is understanding the nature of the nanoparticle–protein complex and realizing that the nature of the proteins adsorbed onto the particles (encompassing the protein corona) influence nanoparticle uptake and traffic throughout the body. The ability to manipulate nanoparticles to achieve particular medical functions depends on this understanding. Designing nanoparticles to specifically adsorb certain proteins, with trafficking proteins, or to prevent adsorption of proteins, is a potentially powerful tool of nanomedicine. As researchers recognize additional factors influencing protein binding, this kind of knowledge could allow nanoparticle delivery to specific locations in the body by manipulation of the factors influencing the protein corona.
Many challenges remain before this can be put into practice, though much important work in this area has been conducted. A wealth of information has come from studies in which particles were incubated with bulk serum, plasma, or with solutions of individual proteins. Here, we have discussed these and other techniques for studying protein binding and the particular technical challenges facing each technique. The results of protein binding studies have clearly shown that nanoparticle physical properties such as composition, size, charge, and surface coating dramatically influence protein binding. There is also much information in the literature on protein kinetics, though this data primarily illustrates that the exception is the rule; protein binding kinetics are highly specific to the type of nanoparticle and its particular size and surface properties. Finally, we reviewed data showing the importance of protein binding to cellular uptake and how it links to the biodistribution of nanoparticles.
Further study of the nanoparticle–protein complex is still required. Greater knowledge of how proteins bind to nanoparticles, factors that determine affinity, and better ways to manipulate specificity, are all necessary. More information is required to determine which specific proteins are required to direct nanoparticles towards specific parts of the body (e.g. ApoE for brain delivery) and what physical properties of nanoparticles can be even more finely tuned to help achieve desired biodistribution patterns (e.g. PEGylation to change surface charge). The effects of nanoparticle binding on protein function is also an area of ongoing study. Ultimately, all this information will help in the rational design of nanoparticle-based drugs and allow full realization of this promising area of technology.
Acknowledgements
We are grateful to Allen Kane for assistance with illustrations. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
☆
This review is part of the Advanced Drug Delivery Reviews theme issue on “Identifying and Assessing Biomaterial Nanotoxicity in Translational Research for Preclinical Drug Development”
References
- 1.National Nanotechnology Initiative. The Initiative and Its Implementation Plan. 2000 [Google Scholar]
- 2.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahoo B, Goswami M, Nag S, Maiti S. Spontaneous formation of a protein corona prevents the loss of quantum dot fluorescence in physiological buffers. Chem. Phys. Lett. 2007;445:217–220. [Google Scholar]
- 4.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–47. [Google Scholar]
- 5.Cedervall T, Lynch I, Foy M, Berggard T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem., Int Ed. Engl. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 6.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv. Colloid Interface Sci. 2007;134–135:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW, Hall JB, Mcneil SE. Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomedicine. doi: 10.1016/j.nano.2008.08.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goppert TM, Muller RH. Protein adsorption patterns on poloxamer- and poloxamine-stabilized solid lipid nanoparticles (SLN) Eur. J. Pharm. Biopharm. 2005;60:361–372. doi: 10.1016/j.ejpb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Kim HR, Andrieux K, Delomenie C, Chacun H, Appel M, Desmaele D, Taran F, Georgin D, Couvreur P, Taverna M. Analysis of plasma protein adsorption onto PEGylated nanoparticles by complementary methods: 2-DE, CE and Protein Lab-on-chip system. Electrophoresis. 2007;28:2252–2261. doi: 10.1002/elps.200600694. [DOI] [PubMed] [Google Scholar]
- 10.Thode K, Luck M, Semmler W, Muller RH, Kresse M. Determination of plasma protein adsorption on magnetic iron oxides: sample preparation. Pharm. Res. 1997;14:905–910. doi: 10.1023/a:1012104017761. [DOI] [PubMed] [Google Scholar]
- 11.Patel HM. Serum opsonins and liposomes: their interaction and opsonophago-cytosis. Crit. Rev. Ther. Drug Cam Syst. 1992;9:39–90. [PubMed] [Google Scholar]
- 12.Chonn A, Semple SC, Cullis PR. Association of blood proteins with large unilamellar liposomes in vivo. J. Biol. Chem. 1992;267:18,759–18,765. [PubMed] [Google Scholar]
- 13.Kiwada H, Miyajima T, Kato Y. Studies on the uptake of mechanism of liposomes by perfused rat liver. II. An indispensable factor for liver uptake in serum. Chem. Pharm. Bull. 1987;35:1189–1195. doi: 10.1248/cpb.35.1189. [DOI] [PubMed] [Google Scholar]
- 14.Tyrrell DA, Richardson VJ, Ryman BE. The effect of serum protein fractions on liposome-cell interactions in cultured cells and the perfused rat liver. Biochim. Biophys. Acta. 1977;497:469–480. doi: 10.1016/0304-4165(77)90204-5. [DOI] [PubMed] [Google Scholar]
- 15.Goppert TM, Muller RH. Polysorbate-stabilized solid lipid nanoparticles as colloidal carriers for intravenous targeting of drugs to the brain: comparison of plasma protein adsorption patterns. J. Drug Target. 2005;13:179–187. doi: 10.1080/10611860500071292. [DOI] [PubMed] [Google Scholar]
- 16.Muller RH, Heinemann S. Surface modelling of microparticles as parenteral systems with high tissue affinity. In: Gurny R, Junginger HE, editors. Bioadhesion-Possibilities and Future Trends. Wissenschaftliche Verlagsgesellschaft, Stuttgart; 1989. pp. 202–213. [Google Scholar]
- 17.Brown DM, Wilson MR, MacNee W, Stone V, Donaldson K. Size-dependent proinflammatory effects of ultrafine polystyrene particles: a role for surface area and oxidative stress in the enhanced activity of ultrafines. Toxicol. Appl. Pharmacol. 2001;175:191–199. doi: 10.1006/taap.2001.9240. [DOI] [PubMed] [Google Scholar]
- 18.Donaldson K, Brown D, Clouter A, Duffin R, MacNee W, Renwick L, Tran L, Stone V. The pulmonary toxicology of ultrafine particles. J. Aerosol Med. 2002;15:213–220. doi: 10.1089/089426802320282338. [DOI] [PubMed] [Google Scholar]
- 19.Donaldson K, Li XY, Macnee W. Ultrafine (nanometre) particle mediated lung injury. J. Aerosol Sci. 1998;29:553–560. [Google Scholar]
- 20.Oberdorster G, Ferin J, Gelein R, Soderholm SC, Finkelstein J. Role of the alveolar macrophage in lung injury: studies with ultrafine particles. Environ. Health Perspect. 1992;97:193–199. doi: 10.1289/ehp.97-1519541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran CL, Buchanan D, Cullen RT, Searl A, Jones AD, Donaldson K. Inhalation of poorly soluble particles. II. Influence of particle surface area on inflammation and clearance. Inhal. Toxicol. 2000;12:1113–1126. doi: 10.1080/08958370050166796. [DOI] [PubMed] [Google Scholar]
- 22.Dutta D, Sundaram SK, Teeguarden JG, Riley BJ, Fifield LS, Jacobs JM, Addleman SR, Kaysen GA, Moudgil BM, Weber TJ. Adsorbed proteins influence the biological activity and molecular targeting of nanomaterials. Toxicol. Sci. 2007;100:303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- 23.Gessner A, Lieske A, Paulke B, Muller R. Influence of surface charge density on protein adsorption on polymeric nanoparticles: analysis by two-dimensional electrophoresis. Eur. J. Pharm. Biopharm. 2002;54:165–170. doi: 10.1016/s0939-6411(02)00081-4. [DOI] [PubMed] [Google Scholar]
- 24.Goppert TM, Muller RH. Adsorption kinetics of plasma proteins on solid lipid nanoparticles for drug targeting. Int. J. Pharm. 2005;302:172–186. doi: 10.1016/j.ijpharm.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 25.Luck M, Paulke BR, Schroder W, Blunk T, Muller RH. Analysis of plasma protein adsorption on polymeric nanoparticles with different surface characteristics. J. Biomed. Mater. Res. 1998;39:478–485. doi: 10.1002/(sici)1097-4636(19980305)39:3<478::aid-jbm19>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Musteata FM, Pawliszyn J, Qjan MG, Wu JT, Miwa GT. Determination of drug plasma protein binding by solid phase microextraction. J. Pharm. Sci. 2006;95:1712–1722. doi: 10.1002/jps.20558. [DOI] [PubMed] [Google Scholar]
- 27.Olson RE, Christ DD. Plasma protein binding of drugs. Annu. Rep. Med. Chem. 1996;31:327–336. [Google Scholar]
- 28.Banker MJ, Clark J-A, Williams TH. Development and validation of a 96-well equilibrium dialysis apparatus for measuring plasma protein binding. J. Pharm. Sci. 2003;92:967–974. doi: 10.1002/jps.10332. [DOI] [PubMed] [Google Scholar]
- 29.Kariv I, Cao H, Oldenburg KR. Development of a high throughput equilibrium dialysis method. J. Pharm. Sci. 2001;90:580–587. doi: 10.1002/1520-6017(200105)90:5<580::aid-jps1014>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Sarre S, Van Belle K, Smolders I, Krieken G, Michotte Y. The use of microdialysis for the determination of plasma protein binding of drugs. J. Pharm. Biomed. Anal. 1992;10:735–739. doi: 10.1016/0731-7085(91)80073-i. [DOI] [PubMed] [Google Scholar]
- 31.Chithrani BD, Ghazani AA, Chan CW. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 32.McNeil SE. Nanotechnology for the biologist. J. Leukoc. Biol. 2005;78:585–594. doi: 10.1189/jlb.0205074. [DOI] [PubMed] [Google Scholar]
- 33.Moghimi SM, Davis SS. Innovations in avoiding particle clearance from blood by Kupffer cells: cause for reflection. Crit Rev. Then Drug Cam Syst. 1994;11:31–59. [PubMed] [Google Scholar]
- 34.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol. Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 35.Poznansky MJ, Juliano RL. Biological approaches to the controlled delivery of drugs: a critical review. Pharmacol. Rev. 1984;36:277–336. [PubMed] [Google Scholar]
- 36.Dejong WH, Hagens WI, Krystek P, Burger MC, Sips AJ, Geertsma RE. Particle size-dependent organ distribution of gold nanoparticles after intravenous administration. Biomaterials. 2008;29:1912–1919. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 37.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharmacol. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Biochem. Soc. Trans. 2007;035:61–67. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]
- 39.Patri AK, Dobrovolskaia MA, Stern ST, Mcneil SE. Chapter 7: preclinical characterization of engineered nanoparticles intended for cancer therapeutics. In: Amiji MM, editor. Nanotechnology for Cancer Therapy. Boca Raton: CRC/Taylor & Francis; 2007. pp. 105–137. [Google Scholar]
- 40.Klein J. Probing the interactions of proteins and nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2029–2030. doi: 10.1073/pnas.0611610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahley RW, Innerarity LT, Rail SC, Jr, Weisgraber KH. Plasma lipoproteins: apolipoprotein structure and function. J. Lipid Res. 1984;25:1277–1294. [PubMed] [Google Scholar]
- 42.Eriksson MA, Gabrielsson J, Nilsson LB. Studies of drug binding to plasma proteins using a variant of equilibrium dialysis. J. Pharm. Biomed. Anal. 2005;38:381–389. doi: 10.1016/j.jpba.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Kurz H, Trunk H, Weitz B. Evaluation of methods to determine protein-binding of drugs. Equilibrium dialysis, ultrafiltration, ultracentrifugation, gel filtration. Arzneimittelforschung. 1977;27:1373–1380. [PubMed] [Google Scholar]
- 44.Salcius M, Michaud GA, Schweitzer B, Predki PF. Identification of small molecule targets on functional protein microarrays. Methods Mol. Biol. 2007;382:239–248. doi: 10.1007/978-1-59745-304-2_15. [DOI] [PubMed] [Google Scholar]
- 45.Zsila F, Visy J, Mady G, Fitos I. Selective plasma protein binding of antimalarial drugs to alphal-acid glycoprotein. Bioorg. Med. Chem. 2008;16:3759–3772. doi: 10.1016/j.bmc.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 46.Blunk T, Hochstrasser DF, Sanchez JC, Muller BW, Muller RH. Colloidal carriers for intravenous drug targeting: plasma protein adsorption patterns on surface-modified latex particles evaluated by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 1993;14:1382–1387. doi: 10.1002/elps.11501401214. [DOI] [PubMed] [Google Scholar]
- 47.Seehof K, Kresse M, Mader K, Muller RH. Interactions of nanoparticles with body proteins-improvement of 2D-PAGE-analysis by internal standard. Int J. Pharm. 2000;196:231–234. doi: 10.1016/s0378-5173(99)00429-9. [DOI] [PubMed] [Google Scholar]
- 48.Diederichs JE. Plasma protein adsorption patterns on liposomes: establishment of analytical procedure. Electrophoresis. 1996;17:607–611. doi: 10.1002/elps.1150170332. [DOI] [PubMed] [Google Scholar]
- 49.Gref R, Luck M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Muller RH. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf., B Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 50.Lemarchand C, Gref R, Passirani C, Garcion E, Petri B, Muller R, Costantini D, Couvreur P. Influence of polysaccharide coating on the interactions of nanoparticles with biological systems. Biomaterials. 2006;27:108–118. doi: 10.1016/j.biomaterials.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 51.Luck M, Pistel KF, Li YX, Blunk T, Muller RH, Kissel T. Plasma protein adsorption on biodegradable microspheres consisting of poly(d,l-lactide-coglycolide), poly(l-lactide) or ABA triblock copolymers containing poly(oxyethylene). Influence of production method and polymer composition. J. Control. Release. 1998;55:107–120. doi: 10.1016/s0168-3659(98)00030-3. [DOI] [PubMed] [Google Scholar]
- 52.Peracchia MT, Harnisch S, Pinto-Alphandary H, Gulik A, Dedieu JC, Desmaele D, d'Angelo J, Muller RH, Couvreur P. Visualization of in vitro protein-rejecting properties of PEGylated stealth polycyanoacrylate nanoparticles. Biomaterials. 1999;20:1269–1275. doi: 10.1016/s0142-9612(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 53.Berggard T, Arrigoni G, Olsson O, Fex M, Linse S, James P. 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J. Proteome. Res. 2006;5:669–687. doi: 10.1021/pr050421l. [DOI] [PubMed] [Google Scholar]
- 54.Ehrenberg M, McGrath JL. Binding between particles and proteins in extracts: implications for microrheology and toxicity. Acta. Biomater. 2005;1:305–315. doi: 10.1016/j.actbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Kim HR, Andrieux K, Gil S, Taverna M, Chacun H, Desmaele D, Taran F, Georgin D, Couvreur P. Translocation of poly(ethylene glycol-co-hexadecyl)cyanoacrylate nanoparticles into rat brain endothelial cells: role of apolipoproteins in receptor-mediated endocytosis. Biomacromolecules. 2007;8:793–799. doi: 10.1021/bm060711a. [DOI] [PubMed] [Google Scholar]
- 56.Norman ME, Williams P, Ilium L. Influence of block copolymers on the adsorption of plasma proteins to microspheres. Biomaterials. 1993;14:193–202. doi: 10.1016/0142-9612(93)90023-u. [DOI] [PubMed] [Google Scholar]
- 57.Stolnik S, Daudali B, Aden A, Whetstone J, Heald CR, Garnett MC, Davis SS, Ilium L. The effect of surface coverage and conformation of poly(ethylene oxide) (PEO) chains of poloxamer407 on the biological fate of model colloidal drug carriers. Biochim. Biophys. Acta. 2001;1514:261–279. doi: 10.1016/s0005-2736(01)00376-5. [DOI] [PubMed] [Google Scholar]
- 58.Nagayama S, Ogawara K, Fukuoka Y, Higaki K, Kimura T. Time-dependent changes in opsonin amount associated on nanoparticles alter their hepatic uptake characteristics. Int J. Pharm. 2007;342:215–221. doi: 10.1016/j.ijpharm.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 59.Frank MM, Fries LF. The role of complement in inflammation and phagocytosis. Immunol. Today. 1991;12:322–326. doi: 10.1016/0167-5699(91)90009-I. [DOI] [PubMed] [Google Scholar]
- 60.Owens DE, Peppas NP. Opsonization, biodistribution and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 61.Moghimi SM, Szebeni J. Stealth liposomes long circulating nanoparticles: criticale issues in pharmacokinetics, opsonization and protein-binding properties. Prog. Lipid Res. 2003;42:463–478. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 62.Vroman L, Adams AL, Fischer GC, Munoz PC. Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces. Blood. 1980;55:156–159. [PubMed] [Google Scholar]
- 63.Harnisch S, Muller RH. Adsorption kinetics of plasma proteins on oil-in-water emulsions for parenteral nutrition. Eur. J. Pharm. Biopharm. 2000;49:41–46. doi: 10.1016/s0939-6411(99)00064-8. [DOI] [PubMed] [Google Scholar]
- 64.Bousquet Y, Swart PJ, Schmitt-Colin N, Velge-Roussel F, Kuipers ME, Meijer DK, Bru N, Hoebeke J, Breton P. Molecular mechanisms of the adsorption of a model protein (human serum albumin) on poly(methylidene malonate 2.1.2) nanoparticles. Pharm. Res. 1999;16:141–147. doi: 10.1023/a:1018843401077. [DOI] [PubMed] [Google Scholar]
- 65.Liu HS, Wang YC. The sorption of lysozyme and ribonuclease onto ferromagnetic nickel powder. 2. Desorption and competitive adsorption. Colloids Surf., B Biointerfaces. 1995;5:35–42. [Google Scholar]
- 66.Patil S, Sandberg A, Heckert E, Self W, Seal S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials. 2007;28:4600–4607. doi: 10.1016/j.biomaterials.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sant S, Poulin S, Hildgen P. Effect of polymer architecture on surface properties, plasma protein adsorption, and cellular interactions of pegylated nanoparticles. J. Biomed. Mater. Res. A. 2008;87:885–895. doi: 10.1002/jbm.a.31800. [DOI] [PubMed] [Google Scholar]
- 68.Yin G, Liu Z, Zhan J, Ding EX, Yuan NJ. Impacts of the surface charge property on protein adsorption on hydroxyapatite. Chem. Eng.J. 2002;87:181–186. [Google Scholar]
- 69.Colfen H, Machtle W, Borger L, editors. Analytical Ultracentrifugation of Polymers and Nanoparticles. Anal. Bioanal. Chem. 2006 [Google Scholar]
- 70.Laue T. Analytical centrifugation: equilibrium approach. In: Coligan JE, editor. Current Protocols in Protein Science. Hoboken: Wiley Interscience; 2001. p. Unit 20.23. [DOI] [PubMed] [Google Scholar]
- 71.Laue TM. Analytical ultracentrifugation. In: Coligan JE, editor. Current Protocols in Protein Science. Hoboken: Wiley Interscience; 2001. p. Unit 7.5. [DOI] [PubMed] [Google Scholar]
- 72.Roser M, Fischer D, Kissel T. Surface-modified biodegradable albumin nano- and microspheres. II: Effect of surface charges on in vitro phagocytosis and biodistribution in rats. Eur. J. Pharm. Biopharm. 1998;46:255–263. doi: 10.1016/s0939-6411(98)00038-1. [DOI] [PubMed] [Google Scholar]
- 73.Gessner A, Lieske A, Paulke BR, Muller RH. Functional groups on polystyrene model nanoparticles: influence on protein adsorption. J. Biomed. Mater. Res. A. 2003;65:319–326. doi: 10.1002/jbm.a.10371. [DOI] [PubMed] [Google Scholar]
- 74.Goppert TM, Muller RH. Plasma protein adsorption of Tween 80- and poloxamer 188-stabilized solid lipid nanoparticles. J. Drug Target. 2003;11:225–231. doi: 10.1080/10611860310001615956. [DOI] [PubMed] [Google Scholar]
- 75.Labarre D, Vauthier C, Chauvierre C, Petri B, Muller R, Chehimi MM. Interactions of blood proteins with poly(isoburylcyanoacrylate) nanoparticles decorated with a polysaccharidic brush. Biomaterials. 2005;26:5075–5084. doi: 10.1016/j.biomaterials.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 76.Carstensen H, Muller RH, Muller BW. Particle-size surface hydrophobicity and interaction with serum of parenteral fat emulsions and model-drug carriers as parameters related to res uptake. Clin. Nutr. 1992;11:289–297. doi: 10.1016/0261-5614(92)90006-c. [DOI] [PubMed] [Google Scholar]
- 77.Muller RH, Wallis KH, Troster SD, Kreuter J. In vitro characterization of poly (methyl-methacrylate) nanoparticles and correlation to their in vivo fate. J. Control. Release. 1992;20:237–246. [Google Scholar]
- 78.Norman ME, Williams P, Illum L. Human serum-albumin as a probe for surface conditioning (opsonization) of block copolymer-coated microspheres. Biomaterials. 1992;13:841–849. doi: 10.1016/0142-9612(92)90177-p. [DOI] [PubMed] [Google Scholar]
- 79.Karajanagi SS, Vertegel AA, Kane RS, Dordick JS. Structure and function of enzymes adsorbed onto single-walled carbon nanotubes. Langmuir. 2004;20:11,594–11,599. doi: 10.1021/la047994h. [DOI] [PubMed] [Google Scholar]
- 80.Teichroeb J, Forrest J, Ngai V, Jones L. Anomalous thermal denaturing of proteins adsorbed to nanoparticles. Eur. Phys. J., E Soft Matter. 2006;21:19–24. doi: 10.1140/epje/i2006-10040-2. [DOI] [PubMed] [Google Scholar]
- 81.Kaufman ED, Belyea J, Johnson MC, Nicholson ZM, Ricks JL, Shah PK, Bayless M, Pettersson T, Feldoto Z, Blomberg E, Claesson P, Franzen S. Probing protein adsorption onto mercaptoundecanoic acid stabilized gold nanoparticles and surfaces by quartz crystal microbalance and zeta-potential measurements. Langmuir. 2007;23:6053–6062. doi: 10.1021/la063725a. [DOI] [PubMed] [Google Scholar]
- 82.Asuri P, Bale SS, Karajanagi SS, Kane RS. The protein–nanomaterial interface. Curr. Opin. Biotechnol. 2006;17:562–568. doi: 10.1016/j.copbio.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Lundqvist M, Sethson I, Jonsson BH. Protein adsorption onto silica nanoparticles: conformational changes depend on the particles' curvature and the protein stability. Langmuir. 2004;20:10,639–10,647. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- 84.Vertegel AA, Siegel RW, Dordick JS. Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir. 2004;20:6800–6807. doi: 10.1021/la0497200. [DOI] [PubMed] [Google Scholar]
- 85.Asuri P, Karajanagi SS, Dordick JS, Kane RS. Directed assembly of carbon nanotubes at liquid–liquid interfaces: nanoscale conveyors for interfacial biocatalysis. J. Am. Chem. Soc. 2006;128:1046–1047. doi: 10.1021/ja0573965. [DOI] [PubMed] [Google Scholar]
- 86.Asuri P, Karajanagi SS, Yang H, Yim TJ, Kane RS, Dordick JS. Increasing protein stability through control of the nanoscale environment. Langmuir. 2006;22:5833–5836. doi: 10.1021/la0528450. [DOI] [PubMed] [Google Scholar]
- 87.Linse S, Cabaleiro-Lago C, Xue WF, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kogan MJ, Bastus NG, Amigo R, Grillo-Bosch D, Araya E, Turiel A, Labarta A, Giralt E, Puntes VF. Nanoparticle-mediated local and remote manipulation of protein aggregation. Nano Lett. 2006;6:110–115. doi: 10.1021/nl0516862. [DOI] [PubMed] [Google Scholar]
- 89.Colvin VLK, M K. Nanoparticles as catalysts for protein fibrillation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8679–8680. doi: 10.1073/pnas.0703194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gabellieri E, Strambini GB, Shcharbin D, Klajnert B, Bryszewska M. Dendrimer–protein interactions studied by tryptophan room temperature phosphorescence. Biochim. Biophys. Acta. 2006;1764:1750–1756. doi: 10.1016/j.bbapap.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 91.Klajnert B, Sadowska M, Bryszewska M. The effect of polyamidoamine dendrimers on human erythrocyte membrane acetylcholinesterase activity. Bioelectrochemistry. 2004;65:23–26. doi: 10.1016/j.bioelechem.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 92.Shcharbin D, Jokiel M, Klajnert B, Bryszewska M. Effect of dendrimers on pure acetylcholinesterase activity and structure. Bioelectrochemistry. 2006;68:56–59. doi: 10.1016/j.bioelechem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 93.Shcharbin D, Klajnert B, Mazhul V, Bryszewska M. Dendrimer interactions with hydrophobic fluorescent probes and human serum albumin. J. Fluoresc. 2005;15:21–28. doi: 10.1007/s10895-005-0209-7. [DOI] [PubMed] [Google Scholar]
- 94.Roach P, Farrar D, Perry CC. Surface tailoring for controlled protein adsorption: effect of topography at the nanometer scale and chemistry. J. Am. Chem. Soc. 2006;128:3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- 95.Patel HM, Moghimi SM. Serum-mediated recognition of liposomes by phagocytic cells of the reticuloendothelial system – the concept of tissue specificity. Adv. Drug Deliv. Rev. 1998;32:45–60. doi: 10.1016/s0169-409x(97)00131-2. [DOI] [PubMed] [Google Scholar]
- 96.Camner P, Lundborg M, Lastbom L, Gerde P, Gross N, Jarstrand C. Experimental and calculated parameters on particle phagocytosis by alveolar macrophages. J. Appl. Physiol. 2002;92:2608–2616. doi: 10.1152/japplphysiol.01067.2001. [DOI] [PubMed] [Google Scholar]
- 97.Leroux JC, De Jaeghere F, Anner B, Doelker E, Gurny R. An investigation on the role of plasma and serum opsonins on the internalization of biodegradable poly (d,l-lactic acid) nanoparticles by human monocytes. Life Sci. 1995;57:695–703. doi: 10.1016/0024-3205(95)00321-v. [DOI] [PubMed] [Google Scholar]
- 98.Gref R, Domb A, Quellec P, Blunk T, Muller RH, Verbavatz JM, Langer R. The controlled intravenous delivery of drugs using peg-coated sterically stabilized nanospheres. Adv. Drug Deliv. Rev. 1995;16:215–233. doi: 10.1016/0169-409X(95)00026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Illum L, Davis SS, Muller RH, Mak E, West P. The organ distribution and circulation time of intravenously injected colloidal carriers sterically stabilized with a block copolymer-poloxamine 908. Life Sci. 1987;40:367–374. doi: 10.1016/0024-3205(87)90138-x. [DOI] [PubMed] [Google Scholar]
- 100.Panagi Z, Beletsi A, Evangelatos G, Livaniou E, Ithakissios DS, Avgoustakis K. Effect of dose on the biodistribution and pharmacokinetics of PLGA and PLGA-mPEG nanoparticles. Int. J. Pharm. 2001;221:143–152. doi: 10.1016/s0378-5173(01)00676-7. [DOI] [PubMed] [Google Scholar]
- 101.Moghimi SM, Muir IS, Illum L, Davis SS, Kolb-Bachofen V. Coating particles with a block co-polymer (poloxamine-908) suppresses opsonization but permits the activity of dysopsonins in the serum. Biochim. Biophys. Acta. 1993;1179:157–165. doi: 10.1016/0167-4889(93)90137-e. [DOI] [PubMed] [Google Scholar]
- 102.Ogawara K, Furumoto K, Nagayama S, Minato K, Higaki K, Kai T, Kimura T. Pre-coating with serum albumin reduces receptor-mediated hepatic disposition of polystyrene nanosphere: implications for rational design of nanoparticles. J. Control Release. 2004;100:451–455. doi: 10.1016/j.jconrel.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 103.Nagayama S, Ogawara K, Minato K, Fukuoka Y, Takakura Y, Hashida M, Higaki K, Kimura T. Fetuin mediates hepatic uptake of negatively charged nanoparticles via scavenger receptor. Int. J. Pharm. 2007;329:192–198. doi: 10.1016/j.ijpharm.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 104.Kreuter J, Hekmatara T, Dreis S, Vogel T, Gelperina S, Langer K. Covalent attachment of apolipoprotein A-I and apolipoprotein B-100 to albumin nanoparticles enables drug transport into the brain. J. Control. Release. 2007;118:54–58. doi: 10.1016/j.jconrel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 105.Kreuter J, Shamenkov D, Petrov V, Ramge P, Cychutek K, Koch-Brandt C, Alyautdin R. Apolipoprotein-mediated transport of nanoparticle-bound drugs across the blood-brain barrier. J. Drug Target. 2002;10:317–325. doi: 10.1080/10611860290031877. [DOI] [PubMed] [Google Scholar]
- 106.Michaelis K, Hoffmann MM, Dreis S, Herbert E, Alyautdin RN, Michaelis M, Kreuter J, Langer K. Covalent linkage of apolipoprotein e to albumin nanoparticles strongly enhances drug transport into the brain. J. Pharmacol. Exp. Ther. 2006;317:1246–1253. doi: 10.1124/jpet.105.097139. [DOI] [PubMed] [Google Scholar]
- 107.Kreuter J. Nanoparticulate systems for brain delivery of drugs. Adv. Drug Deliv. Rev. 2001;47:65–81. doi: 10.1016/s0169-409x(00)00122-8. [DOI] [PubMed] [Google Scholar]
- 108.Olivier JC. Drug transport to brain with targeted nanoparticles. NeuroRx. 2005;2:108–119. doi: 10.1602/neurorx.2.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moghimi SM, Hunter AC. Capture of stealth nanoparticles by the body's defences. Crit. Rev. Ther. Drug Carr. Syst. 2001;18:527–550. [PubMed] [Google Scholar]
- 110.Moghimi SM, Hunter AC. Poloxamers and poloxamines in nanoparticle engineering and experimental medicine. Trends Biotechnol. 2000;18:412–420. doi: 10.1016/s0167-7799(00)01485-2. [DOI] [PubMed] [Google Scholar]
- 111.Peracchia MT, Fattal E, Desmaele D, Besnard M, Noel JP, Gomis JM, Appel M, d'Angelo J, Couvreur P. Stealth PEGylated polycyanoacrylate nanoparticles for intravenous administration and splenic targeting. J. Control. Release. 1999;60:121–128. doi: 10.1016/s0168-3659(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 112.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263:160–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 113.Paciotti GF, Myer L, Weinreich D, Goia D, Pavel N, McLaughlin RE, Tamarkin L. Colloidal gold: a novel nanoparticle vector for tumor directed drug delivery. Drug Deliv. 2004;11:169–183. doi: 10.1080/10717540490433895. [DOI] [PubMed] [Google Scholar]
- 114.Jeon SI, Lee JH, Andrade JD, Degennes PG. Protein surface interactions in the presence of polyethylene oxide. 1. Simplified theory. J. Colloid Interface Sci. 1991;142:149–158. [Google Scholar]
- 115.Zahr AS, Davis CA, Pishko MV. Macrophage uptake of core-shell nanoparticles surface modified with poly(ethylene glycol) Langmuir. 2006;22:8178–8185. doi: 10.1021/la060951b. [DOI] [PubMed] [Google Scholar]
- 116.Neal JC, Stolnik S, Schacht E, Kenawy ER, Garnett MC, Davis SS, Illum L. In vitro displacement by rat serum of adsorbed radiolabeled poloxamer and poloxamine copolymers from model and biodegradable nanospheres. J. Pharm. Sci. 1998;87:1242–1248. doi: 10.1021/js970462j. [DOI] [PubMed] [Google Scholar]
- 117.Kreuter J, Alyautdin RN, Kharkevich DA, Ivanov AA. Passage of peptides through the blood-brain barrier with colloidal polymer particles (nanoparticles) Brain Res. 1995;674:171–174. doi: 10.1016/0006-8993(95)00023-j. [DOI] [PubMed] [Google Scholar]
- 118.Gulyaev AE, Gelperina SE, Skidan IN, Antropov AS, Kivman GY, Kreuter J. Significant transport of doxorubicin into the brain with polysorbate 80-coated nanoparticles. Pharm. Res. 1999;16:1564–1569. doi: 10.1023/a:1018983904537. [DOI] [PubMed] [Google Scholar]
- 119.Steiniger SC, Kreuter J, Khalansky AS, Skidan IN, Bobruskin AI, Smirnova ZS, Severin SE, Uhl R, Kock M, Geiger KD, Gelperina SE. Chemotherapy of glioblastoma in rats using doxorubicin-loaded nanoparticles. Int. J. Cancer. 2004;109:759–767. doi: 10.1002/ijc.20048. [DOI] [PubMed] [Google Scholar]
- 120.Alyautdin R, Gothier D, Petrov V, Kharkevich D, Kreuter J. Analgesic activity of the hexapeptide dalargin adsorbed on the surface of polysorbate 80-coated poly (butyl cyanoacrylate) nanoparticles. Eur. J. Pharm. Sci. 1995;41:44–48. [Google Scholar]
- 121.Alyautdin RN, Tezikov EB, Ramge P, Kharkevich DA, Begley DJ, Kreuter J. Significant entry of tubocurarine into the brain of rats by adsorption to polysorbate 80-coated polybutylcyanoacrylate nanoparticles: an in situ brain perfusion study. J. Microencapsul. 1998;15:67–74. doi: 10.3109/02652049809006836. [DOI] [PubMed] [Google Scholar]
- 122.Abraxane Data Sheet. 2005 [Google Scholar]
- 123.Foote M. Using nanotechnology to improve the characteristics of antineoplastic drugs: improved characteristics of nab-paclitaxel compared with solvent-based paclitaxel. Biotechnol. Annu. Rev. 2007;13:345–357. doi: 10.1016/S1387-2656(07)13012-X. [DOI] [PubMed] [Google Scholar]
- 124.Desai N, Trieu V, Yao Z, Louie L, Ci S, Yang A, Tao C, De T, Beals B, Dykes D, Noker P, Yao R, Labao E, Hawkins M, Soon-Shiong P. Increased antitumor activity intratumor paclitaxel concentrations endothelial cell transport of cremophor-free albumin-bound paclitaxel ABI-007 compared with cremophor-based paclitaxel. Clin. Cancer Res. 2006;12:1317–1324. doi: 10.1158/1078-0432.CCR-05-1634. [DOI] [PubMed] [Google Scholar]
- 125.Hawkins MJ, Soon-Shiong P, Desai N. Protein nanoparticles as drug carriers in clinical medicine. Adv. Drug Deliv. Rev. 2008;60:876–885. doi: 10.1016/j.addr.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 126.John TA, Vogel SM, Tiruppathi C, Malik AB, Minshall RD. Quantitative analysis of albumin uptake transport in the rat microvessel endothelial monolayer. Am. J. PhysiolLung Cell. Mol. Physiol. 2003;284:L187–L196. doi: 10.1152/ajplung.00152.2002. [DOI] [PubMed] [Google Scholar]
- 127.Minshall RD, Tiruppathi C, Vogel SM, Malik AB. Vesicle formation and trafficking in endothelial cells and regulation of endothelial barrier function. Histochem. Cell Biol. 2002;117:105–112. doi: 10.1007/s00418-001-0367-x. [DOI] [PubMed] [Google Scholar]
- 128.Vogel SM, Minshall RD, Pilipovic M, Tiruppathi C, Malik AB. Albumin uptake transcytosis in endothelial cells in vivo induced by albumin-binding protein. Am. J. Physiol., Lung Cell. Mol. Physiol. 2001;281:L1512–L1522. doi: 10.1152/ajplung.2001.281.6.L1512. [DOI] [PubMed] [Google Scholar]
- 129.Gessner A, Waicz R, Lieske A, Paulke B, Mader K, Muller RH. Nanoparticles with decreasing surface hydrophobicities: influence on plasma protein adsorption. Int. J. Pharm. 2000;196:245–249. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 130.Allemann E, Gravel P, Leroux JC, Balant L, Gurny R. Kinetics of blood component adsorption on poly(d,l-lactic acid) nanoparticles: evidence of complement C3 component involvement. J. Biomed. Mater. Res. 1997;37:229–234. doi: 10.1002/(sici)1097-4636(199711)37:2<229::aid-jbm12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 131.Muller RH, Ruhl D, Luck M, Paulke BR. Influence fluorescent labelling of polystyrene particles on phagocytic uptake hydrophobicity and plasma protein adsorption. Pharm. Res. 1997;14:18–24. doi: 10.1023/a:1012043131081. [DOI] [PubMed] [Google Scholar]
- 132.Salvador-Morales C, Flahaut E, Sim E, Sloan J, Green ML, Sim RB. Complement activation and protein adsorption by carbon nanotubes. Mol. Immunol. 2006;43:193–201. doi: 10.1016/j.molimm.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 133.Fang C, Shi B, Pei YY, Hong MH, Wu J, Chen HZ. In vivo tumor targeting of tumor necrosis factor-alpha-loaded stealth nanoparticles: effect of MePEG molecular weight and particle size. Eur. J. Pharm. Sci. 2006;27:27–36. doi: 10.1016/j.ejps.2005.08.002. [DOI] [PubMed] [Google Scholar]