Marked alterations in the distal gut microbiome linked to diet-induced obesity (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 20.
Published in final edited form as: Cell Host Microbe. 2008 Apr 17;3(4):213–223. doi: 10.1016/j.chom.2008.02.015
SUMMARY
We have investigated the inter-relationship between diet, gut microbial ecology and energy balance using a mouse model of obesity produced by consumption of a prototypic Western diet. Diet-induced obesity (DIO) produced a bloom in a single uncultured clade within the Mollicutes class of the Firmicutes, which became the dominant lineage within the distal gut microbiota. This bloom was diminished by subsequent dietary manipulations that limit weight gain and reduce adiposity. Transplantation of the microbiota from mice with DIO to lean germ-free recipients produced a significantly greater increase in adiposity than transplants from lean donors. Metagenomic sequencing of the gut microbiome, biochemical assays, plus sequencing and in silico metabolic reconstructions of a related human gut-associated Mollicute (E.dolichum), revealed features that may provide a competitive advantage for members of the bloom in the Western diet nutrient milieu, including genes involved in import and metabolism of simple sugars. Our study illustrates how combining comparative metagenomics with gnotobiotic mouse models and specific dietary manipulations can disclose the niches of previously uncharacterized members of the gut microbiota.
INTRODUCTION
Energy balance is an equilibrium between the amount of energy extracted from the diet and the amount expended. Studies in germ-free mice indicate that the structure and operations of the intestinal microbial community (the gut microbiota) should be factored into this equation. Colonization of adult germ-free animals with a distal gut microbiota harvested from the ceca of conventionally-raised donors fed a low-fat polysaccharide-rich diet produces a marked increase in body fat content within 10-14 days (Backhed et al., 2004; Turnbaugh et al., 2006). This increase in adiposity reflects the effect of the gut microbiota on both sides of the energy balance equation. The microbiota ferments complex dietary plant polysaccharides that cannot be digested by the host, which lacks the requisite glycoside hydrolases in its proteome (Sonnenburg et al., 2005; http://www.cazy.org/). The resulting monosaccharide and short-chain fatty acid (SCFA) products are absorbed by the host and delivered to the liver where they are converted to more complex lipids. The microbiota concomitantly regulates expression of host genes that promote the deposition of this absorbed energy in adipocytes (Backhed et al., 2004; 2007; Dumas et al., 2006; Martin et al., 2007).
As in humans, >90% of bacterial phylogenetic types (phylotypes) comprising the mouse distal gut microbiota are members of two bacterial divisions (phyla): the Bacteroidetes and the Firmicutes (Eckburg et al., 2005; Ley et al., 2005,2006b; Frank et al., 2007). Studies of genetically obese C57BL/6J mice homozygous for a mutation in the leptin gene (ob/ob), and their lean ob/+ and +/+ littermates, revealed that obesity in this model is associated with a division-wide increase in the relative abundance of the Firmicutes and a corresponding division-wide decrease in the relative abundance of the Bacteroidetes (Ley et al., 2005). Shotgun sequencing of DNA prepared from the cecal microbiota of ob/ob, ob/+, and +/+ littermates indicated that the obesity-associated gut ‘microbiome’ (genes present in the microbiota) had an increased capacity for fermenting polysaccharides relative to the lean-associated microbiome (Turnbaugh et al., 2006). These in silico predictions were supported by assays of intestinal contents. Furthermore, when adult wild-type germ-free mice fed a standard polysaccharide-rich chow diet were colonized with a microbiota harvested from ob/ob or lean (+/+) donors, adiposity in recipients of the obese microbiota increased to a significantly greater degree than in recipients of a lean microbiota, supporting the conclusion that the obese gut microbiota has an increased, and transmissible, capacity to promote fat deposition (Turnbaugh et al., 2006).
The linkage between adiposity and gut microbial ecology in mice appears to apply to humans. Culture-independent, 16S rRNA sequence-based methods have been used to survey the distal gut microbiota of 12 unrelated obese men and women before and after being randomly assigned to a fat-restricted (FAT-R) or a carbohydrate-restricted (CARB-R) low calorie diet (Ley et al., 2006b). Obese subjects had a lower relative abundance of the Bacteroidetes and higher relative abundance of the Firmicutes than lean controls. Over time, the relative abundance of the Bacteroidetes progressively increased, proportional to the degree of weight loss. These changes occurred in both the CARB-R and FAT-R groups and were division-wide: i.e., they were not due to blooms or extinctions of specific phylogenetic types (phylotypes). Correspondingly, overall levels of diversity in the microbiota of each individual remained constant as these changes occurred in their gut microbial ecology (Ley et al., 2006b).
The leptin deficient, ob/ob mouse model of obesity established a correlation between host adiposity, microbial community structure, and the efficiency of energy extraction from a standard, low-fat rodent chow diet that was rich in plant polysaccharides, but it did not allow us to investigate the effects of manipulating diet, or diminishing host adiposity on the gut microbiota and its microbiome. Furthermore, leptin deficiency is extremely rare in humans and is associated with a variety of other host phenotypes (Montague et al., 1997). There was also an important set of variables in the human study that could not be constrained. The subjects were unrelated and thus genetically distinct. Their gut microbial ecology also differed: the division-wide changes in Bacteroidetes and Firmicutes produced by weight loss were a shared feature but the collections of microbial lineages represented in each division in each individual were distinctive. In addition, despite general compliance to their assigned diets, the type of food consumed varied between individuals assigned to a given treatment group (FAT-R or CARB-R), and within individuals over time (Ley et al., 2006b). Therefore, in this report, we have turned to a mouse model of diet-induced obesity (DIO) produced by consumption of a prototypic high-fat/high-sugar Western diet, where all animals were genetically identical, ‘inherited’ a similar microbiota, and where once an obese state was achieved, specified diets could be imposed to reduce adiposity. Comparative metagenomic and functional analyses of the distal gut microbiota of these mice have allowed us to test the hypothesis that the relationship between diet and the representation of microbial lineages and genes in the microbiome, is dynamic and adjustable, and impacts the capacity of the microbiota to promote host adiposity. This study also illustrates how the marriage of gnotobiotic mouse models and metagenomics can reveal heretofore-unappreciated roles (niches/professions) played by previously uncharacterized members of the microbial community.
RESULTS
Diet-induced obesity alters gut microbial ecology
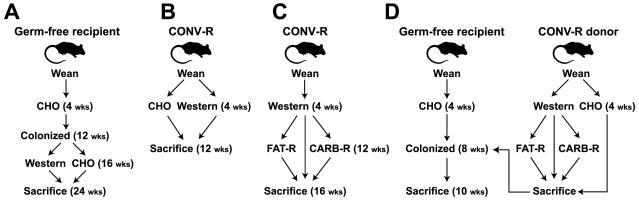
Ten germ-free male C57BL/6J mice were weaned onto a low-fat chow diet rich in structurally complex plant polysaccharides (‘CHO’ diet), and then gavaged at 12 weeks of age with a distal gut (cecal) microbiota harvested from a conventionally-raised donor (see Table S1 for the percentage of calories derived from protein, carbohydrate, and fat). This process of ‘conventionalization’ was designed to insure that all recipients inherited a similar microbiota. All recipients were subsequently maintained in gnotobiotic isolators. Four weeks later, five of the conventionalized mice were switched to ‘Western’ diet high in saturated and unsaturated fats (41% of total calories) and the types of carbohydrates commonly used as human food additives [sucrose (18% of chow weight), maltodextrin (12%), plus corn starch (16%); Tables S1 and S2]. The remaining five mice were continued on the CHO diet. All mice were sacrificed eight weeks later (24 weeks after birth) (Figure 1A). Mice on the Western diet gained significantly more weight than mice maintained on the CHO diet (5.3±0.8g versus 1.5±0.2g; p<0.05, Student’s t-test) and had significantly more epididymal fat (3.7±0.5% versus 1.7±0.1% of total body weight; p<0.01, Student’s t-test).
Figure 1. Experimental design.
(A) Diet-induced obesity (DIO) in germ-free mice colonized with a complex microbial community. (B) Conventionally-raised (CONV-R) wild-type mice fed a Western or CHO diet. (C) Specific dietary shifts after two months on the Western diet. (D) Microbiota transplantation experiments from donor mice on multiple diets to lean germ-free CHO-fed recipients. Numbers in parentheses refer to the age of mice at each step in the protocol. Mouse diets are labeled Western, FAT-R, CARB-R, and CHO (see Tables S1 and S2).
Cecal microbial community structure was defined in each mouse in each of the two groups by sequencing full-length 16S rRNA gene amplicons produced by PCR of community DNA (see Materials and Methods in Supplemental Data; Table S3). Communities were then compared using the UniFrac metric (Lozupone et al., 2006). The premise of UniFrac is that two microbial communities with a shared evolutionary history will share branches on a 16S rRNA phylogenetic tree, and that the fraction of branch length shared can be quantified and interpreted as the degree of community similarity.
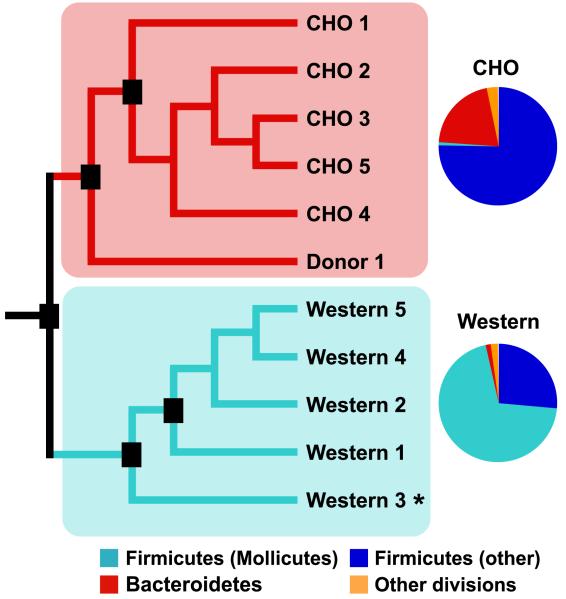
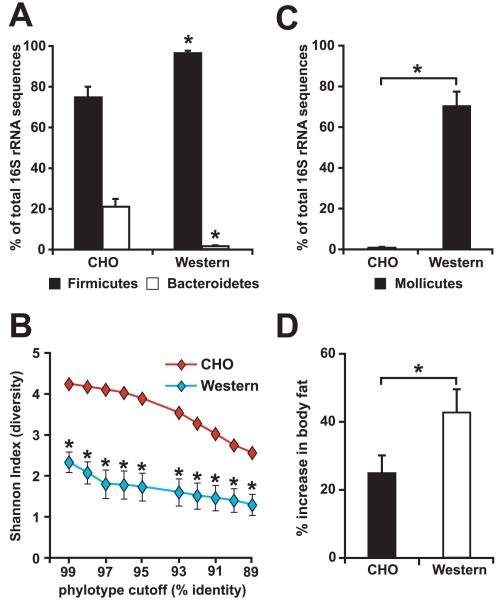
The results of UniFrac analysis revealed that the five Western diet-associated cecal communities were more similar to each other than to the five lean gut communities (Figure 2). As in the ob/ob model of obesity, the Western diet-associated cecal community had a significantly higher relative abundance of the Firmicutes and a significantly lower relative abundance of the Bacteroidetes (Figure 3A). Unlike the ob/ob microbiota, the observed shift in the Firmicutes was not division-wide: the overall diversity of the Western diet microbiota dropped dramatically, due to a bloom in a single class of the Firmicutes - the Mollicutes (Figure 2 and Figure 3B,C) (Note that the term Mollicutes, meaning ‘soft skin’, is often used to describe bacteria that lack a cell wall, such as the Mycoplasma. We use the term only in the phylogenetic sense, and in accordance with current taxonomy, to indicate a class of bacteria with shared evolutionary history based on their 16S rRNA gene sequences).
Figure 2. Diet-induced obesity alters gut microbial ecology in conventionalized mice.
Adult C57BL/6J conventionalized mice were fed a low-fat high-polysaccharide (CHO) or high-fat/high-sugar (Western) diet. 16S rRNA gene sequence-based surveys and UniFrac-based analyses were performed on the distal gut (cecal) contents of ten mice (n=5 mice/group) and the cecal contents from the donor mouse. Black boxes indicate nodes that were reproduced in >70% of all jackknife replications (n=96 sequences). Pie charts show the average relative abundance of bacterial lineages in the CHO diet versus Western diet cecal microbiota. The asterisk indicates that the sample was also analyzed based on whole community shotgun sequencing.
Figure 3. Diet-induced obesity (DIO) is linked to changes in gut microbial ecology, resulting in an increased capacity of the distal gut microbiota to promote host adiposity.
(A) The relative abundance (% of total 16S rRNA gene sequences) of the Firmicutes and Bacteroidetes divisions in the distal gut (cecal) microbiota of conventionalized, wild-type C57BL/6J mice fed a standard low-fat high-polysaccharide chow diet (CHO; n=5) or a high-fat/high-sugar Western diet (n=5). (B) DIO is associated with a marked reduction in the overall diversity of the cecal bacterial community. The Shannon index of diversity was calculated at multiple phylotype cutoffs (defined by % identity of 16S rRNA gene sequences) for each individual cecal dataset using DOTUR (Schloss and Handlesman, 2005). The average diversity at each cutoff is plotted for mice fed the CHO and Western diets. (C) DIO is linked to a bloom of the Mollicutes class of bacteria within the Firmicutes division. The relative abundance of the Mollicutes is shown for conventionalized mice fed the CHO or Western diet. (D) Microbiota transplantation experiments reveal that the DIO community has an increased capacity to promote host fat deposition. Total body fat was measured using dual-energy x-ray absorptiometry (DEXA) before and after a two-week colonization of adult germ-free CHO-fed C57BL/6J wild-type mice with a cecal microbiota harvested from mice maintained on CHO or Western diet (n=14 mice/treatment group). Mean values±SEM are shown. Asterisks in panels A-D indicate that the differences are statistically significant (Student’s t-test, p<0.05), after using the Bonferroni correction to limit false positives.
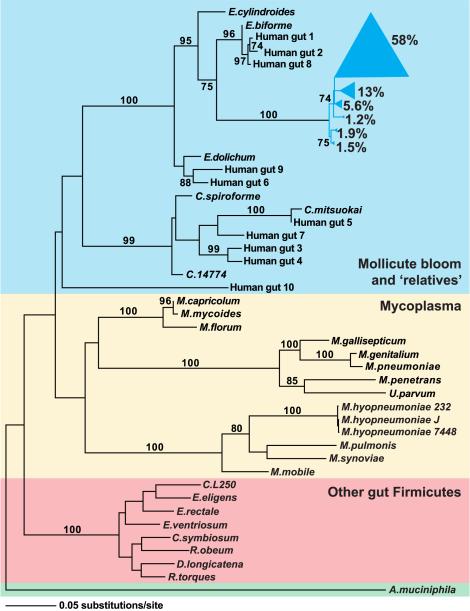
Using ≥99% sequence identity among 16S rRNA genes as a threshold cutoff, we identified 132 ‘strain’-level phylotypes represented within the Mollicute bloom: the bloom was dominated by six phylotypes that together comprised 81% of the Mollicute sequences (Figure 4; DOTUR; Schloss and Handelsman, 2005). Other Mollicutes phylogenetically related to this clade have been cultured from the human gut (e.g. Eubacterium dolichum, E.cylindroides, and E.biforme) and observed in 16S rRNA datasets generated from the fecal microbiota of obese humans (Ley et al., 2006b). However, there are no reported cultured representatives of the dominant phylotypes observed in the DIO mouse model (Figure 4).
Figure 4. Phylogeny of selected representatives from the Firmicutes division, including the Mollicute bloom and closely related human strains.
16S rRNA gene sequences for previously sequenced Firmicute genomes and Mollicute strains isolated from the human gut were identified in the RDP database (Cole et al., 2005). All Mollicute sequences obtained from conventionalized C57BL/6J mice fed a CHO or Western diet (n=801 sequences) and from our previous survey of obese humans (length>1250 nucleotides; n=571 sequences; Ley et al., 2006b) were separately binned into phylotypes using DOTUR (99% identity; Schloss and Handlesman, 2005). One representative of each of the six dominant mouse phylotypes was chosen (together comprising 81% of the mouse Mollicute sequences) in addition to one representative of each of the ten dominant human phylotypes. Likelihood parameters were determined using Modeltest (Posada and Crandall, 1998) and a maximum-likelihood tree was generated using PAUP (Swofford, 2003). Bootstrap values represent nodes found in >70 of 100 repetitions. Phylotypes from the Mollicute bloom are shown in blue; wedge size is proportional to the indicated relative abundance (% of Mollicute 16S rRNA gene sequences). The Mollicute bloom and relatives are shaded in blue, previously sequenced Mollicutes (including the obligate parasites, Mycoplasma, and Mesoplasma florum) are shaded in yellow, and recently sequenced Firmicutes found in the ‘normal’ distal human gut microbiota are shaded in red. Akkermansia muciniphila, a Verrucomicrobia, was used to root the tree (shaded in green).
To determine whether these diet-induced changes in gut microbial ecology also occur in mice exposed to microbes starting at birth, we conducted a follow-up study using a different experimental design. In this case, conventionally-raised C57BL/6J mice were weaned onto a Western or a CHO diet and then maintained, in separate cages, on those diets for 8-9 weeks (n=8 animals/group). All animals were sacrificed after 12 weeks of age (Figure 1B). Those on the Western diet gained significantly more weight (13.8±0.9g versus 10.9±0.9g; p<0.05, Student’s t-test) and had significantly greater adiposity (epididymal fat pad weight was 3.0±0.2% of total body weight in the Western diet group versus 1.6±0.1% in the CHO group; p<0.001, Student’s t-test). The cecal microbiota of these conventionally-raised mice fed the Western diet was dominated by the same Mollicute lineage that had been identified in the earlier conventionalization experiment involving germ-free animals (Figure S1).
The immune system is one of the host factors that influences gut microbial ecology (Suzuki et al., 2004; Ley et al., 2006a; Lupp et al., 2007; Peterson et al., 2007). However, this bloom occurred in all mice fed the Western diet and did not require a functional innate or adaptive immune system: i.e., the Mollicute bloom was present at a significantly higher abundance in the cecal microbiota of conventionally-raised Western diet-fed C57BL/6J mice that were wild-type, MyD88−/− or Rag1−/−, compared to their genotypically-matched CHO-fed siblings (Figure S1).
To directly test whether the DIO-associated gut microbial community possesses functional attributes that can increase host adiposity to a greater degree than a CHO-diet associated gut microbial community, we transplanted the cecal microbiota harvested from obese, conventionally-raised wild-type donors who had been on the Western diet for ≥8 weeks since weaning, or the cecal microbiota from lean CHO-fed controls, to 8-9 week-old germ-free CHO-fed recipients (n=1 donor and 4-5 recipients/treatment group/experiment; n=3 independent experiments, including one CHO-fed control group described in Turnbaugh et al., 2006). All recipients were maintained on a CHO diet (16% of kcal from fat, 61% from carbohydrates of which 2% are from fructose, glucose, lactose, maltose, and sucrose combined), and sacrificed 14d after receiving the microbiota transplant (Figure 1D). Mice colonized with a DIO-associated microbiota exhibited a significantly greater percentage increase in body fat, as defined by dual energy x-ray absorptiometry (DEXA), than mice who had been gavaged with a microbiota from CHO-fed donors (43.0±7.1 versus 24.8±4.9 percentage increase; p<0.05, Student’s t-test based on the combined data from all three experiments) (Figure 3D). Importantly, there were no statistically significant differences in chow consumption (14.5±0.3 versus 14.7±0.8 kcal/d) or initial weight (22.9±0.3 versus 23.8±0.7g) between recipients of the obese and lean cecal microbiotas.
To test the impact of defined shifts in diet on the body weight, adiposity, and distal gut microbial ecology of obese mice, we designed two custom chows that were modifications of the Western diet: one with reduced carbohydrates (CARB-R); the other with reduced fat (FAT-R) (see Table S1 and S2 for information about the composition and caloric density of these diets). Sixteen conventionally-raised C57BL/6J mice, representing two families derived from two mothers who were sisters to ensure that they all inherited a similar microbial community (Ley et al., 2005), were weaned onto the Western diet and maintained on it for two months. A subset of mice from each family was subsequently continued on the Western diet for four weeks (n=5; control group), while the remaining siblings were switched to the CARB-R (n=6), or FAT-R diets (n=5) for four weeks (Figure 1C).
Mice switched to the FAT-R or CARB-R diet consumed significantly fewer calories [12.5±0.1 kcal/d (FAT-R) and 12.0±0.2 kcal/d (CARB-R) versus 14.1±0.2 kcal/d (Western); p<0.0001, ANOVA], gained significantly less weight [0.6±0.3g (FAT-R) and 0.0±0.3g (CARB-R) versus 2.0±0.3g (Western); p<0.01, ANOVA], and had significantly less fat [epididymal fat pad weight: 1.9±0.3% of total body weight for FAT-R and 1.9±0.2% for CARB-R versus 2.8±0.2% (Western); p<0.05, ANOVA] than those maintained on the Western diet (Figure S2). This provided us with the animal model we had sought: diet-induced obesity followed by weight stabilization and reductions in adiposity, in genetically identical mice consuming defined diets who had inherited a similar microbiota from their mothers.
16S rRNA gene sequence-based surveys revealed that weight stabilization was accompanied by (i) a significant reduction in the relative abundance of the Mollicutes [31.9±11.6% of all bacterial sequences for FAT-R, and a significantly more pronounced decrease to 6.1±3.6% for CARB-R versus 50.3±6.1% for the Western diet; p<0.05, ANOVA], and (ii) a significant division-wide increase in the relative abundance of Bacteroidetes (2.8-fold on the FAT-R, and 2.2-fold on the CARB-R diets; p<0.05, ANOVA) (Figure S3).
To test if these alterations in gut microbial ecology had an effect on the ability of the microbiota to promote host adiposity, we colonized germ-free, CHO-fed recipients with a cecal microbiota harvested from conventionally-raised donors who had been on the Western diet since weaning (8 weeks) and then switched to a FAT-R or CARB-R diet (n=1 donor and 4-5 germ-free recipients/treatment group/experiment; n=2 independent experiments; Figure 1D). Unlike with recipients of the DIO-associated microbiota, there was no statistically significant difference in the amount of fat gained between mice colonized with the FAT-R or CARB-R communities, compared to mice colonized with a cecal microbiota from lean CHO-fed donors (33.6±8.7%, 37.4±10.6%, and 24.8±4.9% increases, respectively; p=0.2, ANOVA).
Combined, these results indicate that both the FAT-R and CARB-R diets repress multiple effects of Western diet-induced obesity: i.e. they decrease adipose tissue mass, diminish the bloom in a single uncultured Mollicute lineage, increase the relative abundance of Bacteroidetes, and reduce the ability of the microbiota to promote fat deposition.
The Western diet-associated gut microbiome
To further investigate the linkage between diet-induced obesity and the Mollicute bloom, we performed capillary sequencing of seven cecal samples obtained from seven mice: (i) three samples were from animals fed the Western diet (one that had been conventionalized, two that were conventionally-raised), (ii) two were from conventionally-raised mice that had been switched from the Western to FAT-R diet for 4 weeks, and (iii) two were from conventionally-raised mice that had been switched to the CARB-R diet for 4 weeks (one mouse/family/diet; as noted above, the conventionally-raised mice were from two mothers who were sisters; Table S4). A total of 48 Mb of high-quality sequence data was generated (average of 7 Mb/cecal DNA sample; Table S5).
Taxonomic assignments
All seven datasets were dominated by sequences homologous to known bacterial genomes (49.97±2.52%), followed by sequences with no significant homology to any entries in the non-redundant (NR) database (34.82±1.89%) or that could not be confidently assigned (10.28±0.45%), followed by sequences homologous to eukarya (4.56±1.02%), archaea (0.27±0.05%), and viruses (0.10±0.01%) [BLASTX assignments performed with MEGAN (Huson et al., 2007); for further details see methods described in Supplemental Data] (Figure S4A). The sequences homologous to eukarya could be assigned to two principal groups: metazoa (largely derived from host cells) and apicomplexa.
Consistent with the PCR-based 16S rRNA data, the largest group of sequences in all seven cecal microbiomes was homologous to the Firmicutes division of Bacteria. Analysis of 16S rRNA gene fragments culled from the metagenomic datasets confirmed the presence of the Mollicute bloom in the Western diet-associated cecal microbiome (Figure S4D). However, all of the datasets, including those from mice on the Western diet, had a low relative abundance of sequences homologous to previously sequenced Mollicute genomes (Figure S4C). These results support the conclusion that the genetic make-up of the DIO-associated Mollicute bloom is distinct from that of previously sequenced Mollicutes.
Analysis of 16S rRNA gene fragments and NR-based taxonomic assignments confirmed that both the FAT-R and the CARB-R diets resulted in an increased relative abundance of sequences homologous to the Bacteroidetes (Figure S4B,D). To focus on the microbiomes’ bacterial and archaeal gene content, all sequences that could be confidently assigned to eukarya were removed before conducting the analyses described below.
Functional predictions
Metagenomic sequencing reads were subsequently assigned to orthologous groups from the STRING-extended COG database (von Mering et al., 2007) and the Kyoto Encyclopedia for Genes and Genomes (KEGG; Kanehisa et al., 2004). KEGG pathway-based metabolic reconstructions of cecal microbiomes harvested from mice fed the Western, CARB-R, or FAT-R diets revealed a variety of differences associated with the various diets (Table 1). Notably, the Western diet microbiome is significantly enriched for KEGG pathways involved in the import and fermentation of simple sugars and host glycans, including ‘fructose and mannose metabolism’ and ‘phosphotransferase system’ (p<0.05 based on bootstrap analysis of pathways in the Western diet-versus CARB-R microbiomes).
Table 1.
Metabolic pathways from KEGG (Kyoto Encylopedia of Genes and Genomes) found at significantly higher relative abundance (enriched) or lower relative abundance (depleted) in the Western diet microbiome relative to the CARB-R microbiome*
| Metabolic pathway | |
|---|---|
| Enriched | Phosphotransferase system (PTS) |
| Fructose and mannose metabolism | |
| Glycolysis / Gluconeogenesis | |
| Glutamate metabolism | |
| Carbon fixation** | |
| Unclassified (non-enzyme) | |
| Pyrimidine metabolism | |
| Protein export | |
| Phenylalanine, tyrosine and tryptophan biosynthesis | |
| Oxidative phosphorylation | |
| Depleted | ABC transporters |
| Bacterial chemotaxis | |
| Bacterial motility proteins | |
| Flagellar assembly | |
| Protein kinases | |
| Two-component system | |
| Pentose and glucuronate interconversions | |
| Other amino acid metabolism | |
| Starch and sucrose metabolism | |
| Ribosome |
Phosphotransferase systems (PTS) are a class of transport systems involved in the uptake and phosphorylation of a variety of carbohydrates (Deutscher et al., 2006). Each transporter involves three linked enzymes that act as phosphoryl group recipients and donors: two are cytoplasmic enzymes that act on all imported PTS carbohydrates (HPr and EI); the other is a carbohydrate-specific complex (EII) comprising one or two hydrophobic integral membrane domains (EIIC/D) and two hydrophilic domains (EIIA/B) (Deutscher et al., 2006). Phosphoenolpyruvate, produced though glycolysis, can be used to generate ATP (via pyruvate kinase), or used to drive the import of additional sugars through transfer of a phosphoryl group to EI of the PTS (Figure 5). PTS genes are found in multiple divisions of bacteria, including Proteobacteria such as E.coli, as well as multiple sequenced Firmicutes (e.g., the Mollicutes Mycoplasma genitalium, M.pneumoniae, M.pulmonis, M.penetrans, M.gallisepticum, M.mycoides, M.mobile, M.hyopneumoniae, M.synoviae, and M.capricolum; KEGG version 40; Kanehisa et al., 2004). The PTS also plays a role in regulating microbial gene expression through catabolite repression, allowing the cell to preferentially import simple sugars over other carbohydrates (Deutscher et al., 2006).
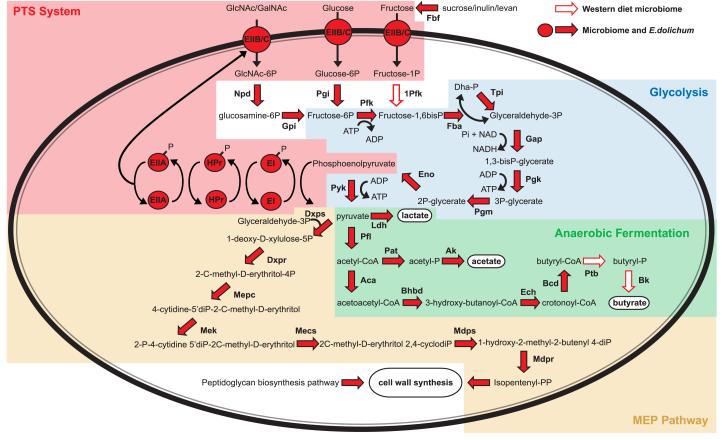
Figure 5. Metabolic reconstructions of the Eubacterium dolichum genome and the Western diet microbiome.
Predicted gene presence calls for the Western diet microbiome and/or the E.dolichum genome are displayed in the upper right. Fermentation end-products and cellular biomass are highlighted in white ellipses. Note that culture-based studies of E.dolichum have demonstrated its ability to produce lactate, acetate, and butyrate (Moore et al., 1976), suggesting that the apparent gap in the pathway for generating butyrate reflects the draft nature of the genome assembly or the possibility that this organism uses novel enzymes to generate this end-product of anaerobic fermentation. Abbreviations for enzymes (in boldface): Pgi, phosphoglucose isomerase; Pfk, phosphofructokinase; Fba, fructose-1,6-bisphosphate aldolase; Tpi, triose-phosphate isomerase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, enolase; Pyk, pyruvate kinase; EI, PTS enzyme I; HPr, PTS protein HPr; EIIA/B/C, PTS proteins; DXPS, 1-deoxy-D-xylulose-5-phosphate synthase; DXPR, DXP-reductoisomerase; MEPC, MEP cytidylyltransferase; MEK, CDP-ME kinase; MECS, MECDP-synthase; MDPS, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; MDPR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; Ldh, L-lactate dehydrogenase; Pfl, pyruvate formate-lyase; Pat, phosphate acetyltransferase; Ak, acetate kinase; Aca, acetyl-CoA C-acetyltransferase; Bhbd, 3-hydroxybutyryl-CoA dehydrogenase; Ech, enoyl-CoA hydratase; Bcd, butyryl-CoA dehydrogenase; Ptb, phosphotransbutyrylase; Bk, butyrate kinase; 1-Pfk, 1-phosphofructokinase; Npd, N-acetylglucosamine-6-phosphate deacetylase; Gpi, phosphoglucosamine isomerase; Fbf, fructan beta-fructosidase.
Multiple components of the PTS are present in the Western diet microbiome (EI and HPr plus EII), which could allow the import of simple sugars (e.g., glucose and fructose that together comprise sucrose, an abundant component of the Western diet), as well as sugars associated with the host gut mucosa (N-acetyl-galactosamine) (Figure 5). The Western diet microbiome also contains genes that support metabolism of these phosphorylated sugars to various end-products of anaerobic fermentation (e.g. lactate and the short-chain fatty acids butyrate and acetate; Figure 5). In addition, the Western diet microbiome is enriched for genes encoding beta-fructosidase, a glycoside hydrolase capable of fermenting beta-fructosidases such as sucrose, inulin, or levan (p<0.05 based on a χ2 test of Western versus CARB-R microbiome).
The Western diet-associated cecal microbiome contains genes for cell wall biosynthesis and cell division: (i) orthologous groups COG0707, COG0766, and COG0768-COG0773 (together, found at a slightly higher relative abundance in the Western versus CARB-R microbiome; p=0.3 based on a χ2 test); (ii) multiple components of the KEGG pathway for peptidoglycan biosynthesis; and (iii) all enzymes in the 2-methyl-D-erythritol 4-phosphate (MEP) pathway that converts pyruvate to isopentyl-pyrophosphate (IPP; Figure 5). IPP provides, among other things, a precursor for peptidoglycan biosynthesis [with the aid of genes for farnesyl diphosphate synthase (K00795) and undecaprenyl diphosphate synthetase (K00806) that were also identified in the microbiome]. Together, these findings indicate that unlike other Mollicutes (e.g., the mycoplasmas), members of the bloom have the capacity to construct a cell wall.
Additionally, unlike the more diverse Firmicutes-enriched ob/ob and CARB-R microbiomes, the Western diet-associated microbiome is depleted for genes assigned to KEGG pathways involved in motility, including (i) ‘bacterial chemotaxis’, (ii) ‘bacterial motility proteins’, and (iii) ‘flagellar assembly’ (Table 1). This observation suggests that the Mollicute bloom is either non-motile or utilizes a mechanism for gliding motility, such as that found recently in other Mollicutes, that is independent of the known pathways for bacterial chemotaxis and flagellar biosynthesis (Jaffe et al., 2004; Hasselbring and Krause, 2007). .
Assembly and analysis of contigs
All seven microbiome datasets were assembled individually and as one pooled dataset using the program ARACHNE (Batzoglou et al., 2002). As expected, the reduced diversity of the Western diet microbiome produced the largest contiguous ‘genome fragments’ (Table S6). Manual inspection of genome fragments from the combined assembly (N50 contig length = 1738 bases; Figure S5), revealed multiple contigs containing genes that were enriched in the Western diet microbiome, including those involved in the degradation of beta-fructosides such as sucrose, inulin, and levan (fructan beta-fructosidase) and the import of simple sugars (PTS genes for fructose and glucose transport). A large contig was also found that contained multiple genes involved in the import of amino acids (ABC transporters) (Figure S5). Interestingly, the two genome fragments containing PTS genes were each flanked by another gene involved in carbohydrate metabolism: in one case, an alpha-amylase (starch degradation) and in the other fragment, fructose-bisphosphate aldolase (glycolysis). These genome fragments are likely derived from the expanded uncultured Mollicute clade: they are composed of reads from microbiomes with a high relative abundance of the bloom and share the highest degree of homology with Bacillus and Mollicute genomes (Table S7).
Validation of PTS expression
We constructed a cDNA library from mRNA-enriched total community RNA that had been isolated from the cecum of an obese mouse fed the Western diet (see Materials and Methods in Supplemental Data for information about the mRNA enrichment procedure). Sequence analysis of the inserts in this library confirmed that a gene encoding EII of the fructose, mannose, and N-acetyl-galactosamine specific PTS transporter (COG3716) was expressed. The low representation of mRNA-derived sequences in our library precluded further (cost-effective) characterization of the DIO cecal microbiome’s transcriptome. However, sequencing of 16S rRNA-derived inserts in the library provided further support of the high abundance of the Mollicute bloom: 80.6% of expressed 16S rRNAs had a best-BLAST-hit to Mollicute gene sequences (BLASTN comparisons with the NCBI nucleotide database, e-value < 10−25).
Validation of enhanced fermentation in the DIO microbiota
To verify our in silico predictions concerning metabolic activities that are enriched in the Western-diet associated gut microbiome, we performed gas-chromatography-mass spectrometric and microanalytic assays of the concentrations of short chain fatty acids and lactate in aliquots of the same cecal samples that had been used for 16S rRNA surveys and metagenomic sequencing of community DNA (See Methods in Supplemental Data). As predicted from our metabolic reconstructions, the cecal contents of mice fed the Western diet (on average, 50% Mollicutes) had a significantly higher concentration of multiple end-products of bacterial fermentation, including lactate, acetate, and butyrate compared to the cecal contents of CARB-R mice (on average, 6% Mollicutes) (Figure S6).
Whole genome sequencing and analysis of a human gut-associated Mollicute
We have yet to successfully culture representatives of the Mollicute clade that blooms in the distal gut microbiota of mice fed a Western diet. Therefore, to obtain additional insights about genomic and metabolic features that may allow this lineage to bloom in the cecal habitat of mice fed a Western diet, and to validate our comparative metagenomic predictions, we sequenced the genome of Eubacterium dolichum strain ATCC29143, a related Mollicute (Figure 4) isolated from the human gut microbiota (Table S8). A deep draft assembly of its genome was produced, based on 49-fold coverage with reads from a 454 FLX pyrosequencer (106 Mb), and 9-fold coverage with reads from a traditional ABI 3730xl capillary sequencer (GenBank accession ABAW00000000; http://genome.wustl.edu/pub/organism/).
We first compared this deep draft assembly of the E.dolichum genome to eight other deep-draft assemblies of human gut-associated Firmicutes and to fourteen finished Mollicute genomes (Figures 6 and S7). The program MetaGene (Noguchi et al., 2006) was used to predict the protein products of these diverse Firmicute/Mollicute genomes and the proteins assigned to the STRING-extended COG database and the KEGG database using BLASTP homology searches (e-value < 10−5; von Mering et al., 2007; Kanehisa et al., 2004).
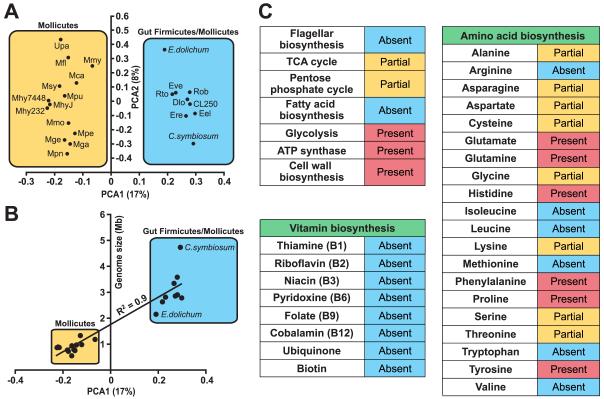
Figure 6. Metabolic and principal component analysis (PCA) of sequenced Firmicute genomes.
(A) PCA analysis of 14 previously sequenced Mollicute genomes (mostly Mycoplasma) and draft genome assemblies of nine human gut-associated Firmicutes (http://genome.wustl.edu/pub/). MetaGene was used to predict proteins from each genome (Noguchi et al., 2006). Proteins were then assigned to KEGG orthologous groups based on homology (BLASTP e-value<10−5; KEGG version 40; Kanehisa et al., 2004). Genomes were clustered based on the relative abundance of KEGG metabolic pathways (number of assignments to a given pathway divided by total number of pathway assignments). Only pathways found at >0.6% relative abundance in at least two genomes were included. The first two components are shown, representing 17% and 8% of the variance respectively. Abbreviations: Mca, Mycoplasma capricolum; Mfl_, Mesoplasma florum L1;_ Mga_, Mycoplasma gallisepticum R,_ Mge_, Mycoplasma genitalium G37;_ Mhy232_, Mycoplasma hyopneumoniae 232;_ Mhy7448_, Mycoplasma hyopneumoniae 7448;_ MhyJ_, Mycoplasma hyopneumoniae J;_ Mmo_, Mycoplasma mobile 163K;_ Mmy_, Mycoplasma mycoides subsp. mycoides SC str. PG1;_ Mpe_, Mycoplasma penetrans HF-2;_ Mpn_, Mycoplasma pneumoniae M129;_ Mpu_, Mycoplasma pulmonis UAB CTIP;_ Msy_, Mycoplasma synoviae 53;_ Upa, Ureaplasma parvum; E.dolichum, Eubacterium dolichum; CL250, Clostridium sp. L2-50; C.symbiosum, Clostridium symbiosum; Dlo_, Dorea longicatena;_ Eel_, Eubacterium eligens;_ Ere_, Eubacterium rectale;_ Eve_, Eubacterium ventriosum;_ Rob_, Ruminococcus obeum;_ and Rto_, Ruminococcus torques._ (B) KEGG pathway relative abundance has a significant correlation with genome size. A linear regression was performed comparing PCA1 to genome size (or draft assembly size). PCA1 has a significant correlation to genome size (R2=0.9, p<0.05). (C) Metabolic pathways in E.dolichum. Pathways are marked partial if most genes are present and absent if ≤2 genes are present.
Principal component analysis (PCA) of KEGG pathway representation in all 23 genomes revealed a clear clustering of the previously sequenced Mollicute genomes and the recently sequenced commensal gut Firmicutes, including E. dolichum (Figure 6A). The total size of the E.dolichum assembly is over twice the average Mollicute genome (2.2 versus 0.91 Mb), and two-thirds the average size of the recently sequenced gut Firmicute genomes (3.2 Mb). Our analyses revealed that the genome size reduction and corresponding gene loss that has occurred during Mollicute evolution has produced small genomes that are largely restricted to encoding components of metabolic pathways essential for life (Figure S8). Accordingly, bacterial genome size significantly correlates with the clustering results (Figure 6B; R2=0.9, p<0.05). As expected from its relatively restricted genome size, E. dolichum is enriched for many KEGG pathways involved in essential cellular functions such as “Cell division”, “Replication, Recombination, and Repair”, “Ribosome”, and others (Figure S7) but is missing a number of metabolic pathways similar to other ‘streamlined’ genomes (e.g. the mycoplasma, and oceanic α-proteobacteria; Jaffe et al., 2004; Giovannoni et al., 2005). Its genome lacks predicted proteins involved in bacterial chemotaxis and flagellar biosynthesis, the tricarboxylic acid cycle, the pentose phosphate cycle, and fatty acid biosynthesis (Figure 6C). It is also significantly depleted for ABC transporters relative to the other gut Firmicutes (Figure S7), and a variety of metabolic pathways for the de novo synthesis of vitamins and amino acids are incomplete or undetectable (Figure 6C).
E. dolichum has a number of genomic features that could promote fitness in the cecal nutrient metabolic milieu created by the host’s consumption of the Western diet. As in the metagenomic dataset generated from the Western diet-associated cecal microbiome, its genome is enriched for predicted PTS proteins involved in the import of simple sugars including glucose, fructose, and N-acetyl-galactosamine (Figures 5 and S7). STRING-based protein networks constructed from the E.dolichum genome revealed that many of these PTS orthologous groups are found in the Western diet microbiome, but not in all nine recently sequenced gut Firmicutes (Figure S8). In addition, the E.dolichum genome encodes a beta-fructosidase capable of degrading fructose-containing carbohydrates such as sucrose, genes for the metabolism of PTS-imported sugars to lactate, butyrate, and acetate, plus a complete 2-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis - all genetic features of the Western-diet-associated cecal microbiome (Figures 5 and S8).
DISCUSSION
The findings presented in this study emphasize the importance of viewing our metabolome in a ‘supra-organismal’ context, where both microbial and host contributions must be considered in order to achieve a fuller understanding of the factors that regulate energy balance. This study also demonstrates the power of combining gnotobiotic mouse models with comparative metagenomics to define relationships between diet, gut microbial ecology, microbial gene content, and the functional attributes associated with a (gut) microbial community.
In a recent study of germ-free and conventionalized mice fed low-fat high-polysaccharide versus Western diets, we examined the effects of the gut microbiota on host metabolism, without examining the effects of host adiposity on the structure or metabolic activities of the microbial community (Backhed et al., 2007). This study revealed that germ-free C57BL/6J mice were resistant to obesity caused by consumption of the same high-fat/high-sugar Western diet used in the current report. This resistance was associated with increased host metabolism of fatty acids: (i) without a gut microbiota, levels of the active, phosphorylated form of AMP-activated kinase (AMPK) were increased in skeletal muscle and liver, as were the activities of AMPK’s downstream targets, acetylCoA carboxylase and carnitine palmitoyltransferase (Cpt1); (ii) the absence of a microbiota also increased intestinal epithelial expression of angiopoietin-like 4 (also known as fasting induced adipocyte protein), a secreted, circulating inhibitor of lipoprotein lipase in mice and humans that induces peroxisomal proliferator activated receptor co-activator (Pgc-1α) and hence increases mitochondrial fatty acid oxidation (Backhed et al., 2007).
We can now add another component to the complex and dynamic interplay between diet, the microbiota and the energy balance equation: namely that with administration of a Western diet, there is a restructuring of the distal gut microbial community so that a Mollicute lineage in the Firmicutes, normally present at low abundance in the mouse colon, expands dramatically to dominate this body habitat. The Mollicute bloom is accompanied by a division-wide suppression of Bacteroidetes, indicating that this Mollicute lineage has an increased fitness, not only relative to other Firmicutes, but also relative to all other Bacteroidetes identified in the community. By comparing the Western, FAT-R, and CARB-R distal gut microbiomes with a deep draft assembly of a human gut associated Mollicute closely related to the phylotype that dominates the DIO-associated microbiome, we find that members of this bloom have evolved the capacity to import the type of carbohydrates found in the model Western diet administered to mice, as well as in foods commonly consumed by humans living in highly Westernized societies (e.g. glucose, fructose, and sucrose), and to metabolize these imported sugars to short chain fatty acids that are readily absorbed by the host. While these data are consistent with the hypothesis that the bloom is highly efficient at competing for oligo- and monosaccharide components of the Western diet, other alternative hypotheses for its success remain to be explored, including changes in the gut environment (e.g. bile acids, motility, pH) and physiology due to alterations in host adiposity or components of the diet.
Gut microbiota transplantation experiments from donors consuming the various diets to lean germ-free recipients, indicate that the significant association between the proportional representation of the Mollicute bloom in the microbiota and host adiposity may be causal rather than casual. The increased adiposity produced by transplantation of a Western versus CHO diet-associated microbiome occurs in germ-free recipients consuming a low fat, polysaccharide-rich CHO diet that has more modest levels of the sugars that are abundantly represented in the Western diet. This raises the possibility that the Mollicute-enriched community not only facilitates transfer of calories from the diet to the host, but also has effects on host metabolism of absorbed calories.
Mining the 16S rRNA datasets we obtained from the fecal microbiota of the 12 unrelated obese humans who were followed for a year while they consumed low calorie FAT-R or CARB-R diets (see Introduction) revealed a decrease in the relative abundance of the Mollicutes class of bacteria as the individuals lost weight (from 4.6±1.3% of 16S rRNA gene sequences prior to the diet to 2.9±1.0% one year later, p=0.1). However, given the small sample size and the varied diets of the subjects, additional studies of lean and obese humans consuming high-fat/high-sugar Western-type diets will be necessary to determine whether or not the abundance of various Mollicute lineages is significantly linked to diet and/or adiposity.
A recent study explored the relationship between the representation of selected members of the Firmicutes, and consumption of high and low carbohydrate diets in 19 obese volunteers over a 4-week period (Duncan et al., 2007). They found that the abundance of a specific group within the Firmicutes that includes Roseburia spp. and Eubacterium rectale, decreased with decreased carbohydrate intake, as did fecal butyrate concentrations. These types of studies, combined with our results, suggest that gut microbes are highly niche-partitioned and that a grand challenge is to match microbes to their preferred substrates.
Our report describes an initial effort to establish a pipeline that combines gnotobiotic mice, specific dietary manipulations, and comparative metagenomics in order to identify and characterize organisms or groups of organisms that play important roles in nutrient and energy harvest. Some of these organisms may become therapeutic targets for manipulation. This effort is predicated on the concept that the energy and nutrient content of food is a relative term, based in part on the gut microbial ecology of the consumer and is motivated by the fact that obesity (and malnutrition) represent extremely serious global problems.
EXPERIMENTAL PROCEDURES
Animals
All experiments involving mice were performed using protocols approved by the Washington University Animal Studies Committee.
Conventionalization
Germ-free male 8-9 week old C57BL/6J mice were maintained in plastic gnotobiotic isolators, under a strict 12-h light cycle and fed an autoclaved low-fat, polysaccharide-rich chow diet (CHO) ad libitum (Hooper et al., 2002; Backhed et al., 2004). Conventionalization was performed by harvesting cecal contents from conventionally-raised animals, and introducing them, by gavage, into germ-free recipients, as described in Backhed et al. (2007).
Conventionally-raised mice
Once C57BL/6J littermates were weaned, they were housed individually in microisolator cages where they were maintained in a specified pathogen-free state, under a 12-h light cycle, and fed a CHO diet (PicoLab, Purina), a high-fat/high-sugar Western diet (Harlan-Teklad TD96132), a fat-restricted (FAT-R) diet (Harlan-Teklad TD05633), or a carbohydrate-restricted (CARB-R) diet (Harlan-Teklad TD05634) ad libitum.
Microbiota transplantation experiments
Adult germ-free C57BL/6J mice ≥8 weeks old were colonized with a cecal microbiota obtained from wild-type (+/+) C57BL/6J donor mice fed CHO, Western, FAT-R, or CARB-R diets. Recipient mice, maintained on a CHO diet, were anesthetized at 0.5 and 14 days post colonization with an intraperitoneal injection of ketamine (10 mg/kg body weight) and xylazine (10mg/kg) and total body fat content was measured by dual-energy x-ray absorptiometry (DEXA; Lunar PIXImus Mouse, GE Medical Systems; Bernal-Mizrachi et al., 2002). Recipient mice were housed individually in microisolator cages within gnotobiotic isolators throughout the experiment to avoid exposure to the microbiota of the other mice, and to allow the direct monitoring of the chow consumed by each mouse. Animals were sacrificed immediately after the final DEXA on day 14.
Shotgun sequencing and assembly of cecal microbiomes
DNA samples were used to construct pOTw13-based libraries (GC10 cells, Gene Choice) for capillary-based sequencing with an ABI 3730xl instrument. Unidirectional (forward) sequencing reads were generated from each library (an average of 10,600 reads/library). Reverse reads were also generated to improve assembly (768-1536 per library; total of 7,680 reads;). Sequences were trimmed based on quality score and vector sequences were removed prior to analysis (Applied Biosystems; KB Basecaller). Each dataset was assembled individually, in addition to a combined assembly of all seven datasets, using ARACHNE (Batzoglou et al., 2002; parameters: maxcliq1=500; maxcliq2=500; genome size = 1 Gb). ARACHNE was chosen because it has been shown to generate reliable contigs from complex simulated metagenomic datasets (Mavromatis et al., 2007). Genes were predicted from individual sequencing reads and contigs using MetaGene (Noguchi et al., 2006).
Microbiome functional analysis
NCBI BLAST was used to query the STRING-extended COG database (version 7; von Mering, 2007) and the KEGG database (version 40; Kanehisa et al., 2004). COG and KEGG comparisons were performed by using NCBI BLASTX employing default parameters. A cutoff of e-value < 10−5 was used for environmental gene tag (EGT) assignments and sequence comparisons. Predicted proteins were searched for conserved domains and assigned functional identifiers with InterProScan (version 4.3; Mulder et al., 2005). Predicted glycoside hydrolases were confirmed based on criteria used for the Carbohydrate Active Enzymes (CAZy) database (http://www.cazy.org/; Bernard Henrissat, personal communication).
Other methods
See Supplemental Data for descriptions of protocols used for (i) preparing DNA from the cecal microbiota, (ii) performing 16S rRNA sequence-based surveys of the cecal microbiota, (iii) making taxonomic assignments of shotgun sequencing reads of the cecal microbiome, (iv) transcriptional profiling of the cecal microbiome, (v) sequencing and assembling the E.dolichum genome, (vi) assaying metabolic end-products of fermentation, and (vii) statistical analyses.
This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the project accession XXX-XXX. The version described in this paper is the first version, XXX-XXX. All reads have been deposited in the NCBI Trace Archive. PCR-derived 16S rRNA gene sequences are deposited in GenBank under accession numbers XXX-XXX.
Supplementary Material
01
Figure S1 – The Mollicute bloom occurs in conventionally-raised wild-type C57BL/6J mice as well as in mice without an intact innate or adaptive immune system. Wild-type (+/+), MyD88 −/−, or Rag1 −/− C57BL/6J mice were weaned onto a standard low-fat polysaccharide-rich (CHO) or high-fat/high-sugar (Western) diet. 16S rRNA gene sequence-based surveys were performed; sequences were aligned (NAST; DeSantis et al., 2006), and inserted into an ARB neighbor-joining tree (Ludwig et al., 2004). Asterisks indicate significant differences (Student’s t-test p<0.001).
Figure S2 – Mice with diet-induced obesity that are switched to a FAT-R or CARB-R diet exhibit stabilization of weight, decreased caloric intake and reduced adiposity. (A) Weight gain (g) and (B) percentage epidydymal fat-pad weight to body weight in wild type C57BL/6J mice that were initially weaned onto a Western diet for 8 weeks, and then maintained on the Western diet, or switched to a FAT-R or CARB-R diet for four weeks (n=5-6 mice/treatment group). Weight was monitored during the four-week period. (C) Chow consumption (kcal/d) is decreased in mice switched to a FAT-R or CARB-R diet. Data are represented as mean±SEM. Asterisks indicate significant differences (ANOVA of FAT-R or CARB-R versus Western, *p<0.05, **p<0.01, ***p<0.0001).
Figure S3 – Switching from a Western to FAT-R or CARB-R diet results in a division-wide increase in the relative abundance of Bacteroidetes, and a decrease in the relative abundance of Mollicutes. UniFrac-based analysis of bacterial community membership shows an impact of diet on gut microbial ecology: cecal communities analyzed from two families of C57BL/6J wild-type mice (Table S3) generally cluster based on host diet (Western, FAT-R, and CARB-R). The average relative abundance (% of total 16S rRNA gene sequences) of bacterial lineages within the cecal microbiota of all mice fed a Western, FAT-R, or CARB-R diet is displayed as pie charts. Black boxes indicate nodes that were reproduced in >50% of all jackknife replications (n=126 sequences were randomly re-sampled). Asterisks indicate cecal samples that were analyzed by whole community shotgun sequencing.
Figure S4 – Taxonomic assignments of metagenomic sequencing reads from seven cecal microbiome datasets based on BLAST homology searches, and by alignment of 16S rRNA gene fragments. (A) The cecal microbiome is dominated by sequences homologous to Bacteria. Sequencing reads were trimmed based on quality and vector sequence and the resulting datasets were used as queries against the NCBI non-redundant database (e-value<10−5). Sequences were assigned to the lowest taxonomic group that would include all significant hits, using MEGAN (Huson et al., 2007). Pie charts are shown for each individual dataset and for the average of all datasets. Colors indicate assignments to bacteria (red), archaea (green), eukarya (yellow), viruses (blue), sequences that could not be confidently assigned to a group (purple), and sequences with no significant BLASTX matches (orange). **(B)** Relative abundance of microbiome sequences homologous to genomes from four bacterial divisions: Bacteroidetes (red), Proteobacteria (yellow), Actinobacteria (orange), and Firmicutes (blue). All divisions observed at >1% relative abundance are shown. (C) Relative abundance of microbiome sequences homologous to genomes from bacterial classes within the Firmicutes division: Bacilli (dark blue), Clostridia (yellow), and Mollicutes (light blue). (D) Taxonomic assignments of 16S rRNA gene fragments obtained from cecal microbiome datasets. 16S rRNA gene fragments were identified by querying the Ribosomal Database Project (RDP) database (version 9.33; BLASTN e-value < 10−5; Cole et al., 2005). 16S rRNA gene fragments were aligned with NAST (DeSantis et al., 2006) and added to an ARB neighbor-joining tree (Ludwig et al., 2004). 16S rRNA gene fragments from the Bacteroidetes (red), Proteobacteria (yellow), Verrucomicrobia (green), Mollicutes (light blue), and other Firmicutes (dark blue) are shown.
Figure S5 - Assembly of metagenomic sequence data reveals physical linkage between the Mollicute phosphotransferase system (PTS) and other genes involved in carbohydrate metabolism. The pooled mouse gut microbiome dataset was assembled using ARACHNE (n=7 combined datasets; Batzoglou et al., 2002; see Tables S6 and S7 for assembly statistics). The contig length is shown as a solid black bar. Arrows indicate predicted proteins. Functional assignments were derived from the NCBI annotations and verified by BLASTP comparisons of each predicted protein with the STRING-extended COG database (von Mering et al., 2007) and the KEGG database (Kanehisa et al., 2004), in addition to Hidden Markov Model (HMM)-based protein domain searching with InterProScan (Mulder et al., 2005). Contigs 23 and 73 are >98% identical over the region in pink (234/238 nucleotides): they are likely different ends of the same gene that were not joined due to the relatively stringent assembly parameters employed.
Figure S6 – Concentration of bacterial fermentation end-products in the ceca of Western, FAT-R, and CARB-R mice. Acetate and butyrate levels (μmol per g wet weight cecal contents) were measured by gas chromatography mass spectrometry. Lactate levels (mM per kg protein) were measured using established microanalytic methods (see Methods above). Data are represented as mean±SEM. Asterisks indicate significant differences (Student’s t-test of Western versus CARB-R, *p<0.05, **p<0.01).
Figure S7 – KEGG metabolic pathways significantly enriched in the human gut-derived Eubacterium dolichum strain DSM 3991 genome relative to eight human gut-associated Firmicutes. Pathways whose relative representation is significantly different between the E. dolichum genome and the pooled gut Firmicute genomes (n=8) were identified using a bootstrap comparison of the abundance of sequences assigned to all KEGG pathways (xipe version 2.4; confidence level = 0.98, sample size = 10,000; Rodriguez-Brito et al., 2006). The relative abundance of all KEGG pathways with significantly different representation found at a relative abundance >0.6% in at least two microbiome datasets was transformed into a z-score and clustered by genome and pathway using a Euclidean distance metric (de Hoon et al., 2004). Enrichment (yellow) and depletion (blue) are defined as a relative abundance greater or less than the mean for all datasets (i.e. a z-score greater or less than zero, respectively). For full strain names see Figure 6.
Figure S8 – STRING-based protein network analysis of the predicted E. dolichum proteome. MetaGene (Noguchi et al., 2006) was used to predict proteins from the E. dolichum deep draft assembly. Proteins were subsequently assigned to COGs based on homology (BLASTP e-value<10−5; STRING database version 7, von Mering et al., 2007). Annotated COG interactions were used to organize the protein network, including interactions based on neighborhood, gene fusion, co-occurrence, homology, co-expression, experiments, databases, and text mining (Medusa Java appet; Hooper and Bork, 2005). Nodes, each representing a different orthologous group, are colored as follows: green, present in all analyzed Firmicute genomes (including the mycoplasma); blue, present in all recently sequenced gut Firmicute genomes; red, present in the Western diet-associated cecal microbiome (based on BLAST homology searches, e-value<10−5 and the deposited annotations in the STRING database, version 7). 89% of the COGs found in the E.dolichum genome were also found in the Western diet microbiome. Most of the COGs in green are involved in essential cellular functions such as transcription and translation (56% of the COG category assignments are to ‘Information storage and processing’). Some clusters of interest are highlighted, including the phosphotransferase system (PTS), the 2-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis (MEP), cell wall biosynthesis, ABC transporters, and V-type ATPases for H+ import.
Supplementary Table 1: Protein, carbohydrate, and fat composition of various mouse chow diets
Supplementary Table 2: Percent weight of chow ingredients
Supplementary Table 3: 16S rRNA gene-sequence libraries
Supplementary Table 4: Nomenclature used to designate microbiome datasets obtained from the cecal microbiota of C57BL/6J mice
Supplementary Table 5: Microbiome sequencing statistics
Supplementary Table 6: Microbiome assembly statistics
Supplementary Table 7: Read placements in contigs and BLAST results
Supplementary Table 8: E.dolichum draft genome sequencing statistics
ACKNOWLEDGEMENTS
We thank David O’Donnell and Maria Karlsson for husbandry of gnotobiotic mice, Jan Crowley, Sabrina Wagoner, and Jill Manchester for outstanding technical support, Ruth Ley, Michael Mahowald, Buck Samuel, Justin Sonnenburg, Marios Giannakis, Andrew Goodman, Priya Psudarsanam, and Rob Knight for their many helpful suggestions during the course of these studies, our colleagues Bob Fulton, Jennifer Godfrey, William Courtney, Jian Xu, and Sandy Clifton in the Genome Sequencing Center for their assistance with metagenomic sequencing and with the deep draft assembly of the Eubacterium dolicum genome, Clay Semenkovich and Trey Coleman for assistance with dual energy x-ray absorptiometry, Edward DeLong (MIT) for generously sharing his protocols for enriching mRNA from total microbial community RNA prior to cDNA synthesis and cloning, and Bernard Henrissat [Universités Aix-Marseille I and II and the Carbohydrate Active Enzymes (CAZy) Database] for confirming the annotation of cecal microbiome- and _E.dolichum-_associated glycoside hydrolases cited in this report. This work was supported in part by grants from the NIH (DK70977 and DK30292), and the W.M. Keck Foundation.
REFERENCES
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S, Jaffe DB, Stanley K, Butler J, Gnerre S, Maucell E, Berger B, Mesirov JP, Lander ES. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Mizrachi C, Weng S, Li B, Nolte LA, Feng C, Coleman T, Holloszy JO, Semenkovich CF. Respiratory uncoupling lowers blood pressure through a leptin-dependent mechanism in genetically obese mice. Arterioscler. Thromb. Vasc. Biol. 2002;22:961–968. doi: 10.1161/01.atv.0000019404.65403.71. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Env. Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe JD, et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Hasselbring BM, Krause DC. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol. Microbiol. 2007;63:44–53. doi: 10.1111/j.1365-2958.2006.05507.x. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Mills JC, Roth KA, Stappenbeck TS, Wong MH, Gordon JI. Combining gnotobiotic mouse models with functional genomics to define the impact of the microflora on host physiology. In: Sansonetti P, Zychlinsky A, editors. Methods in Microbiology, Molecular Cellular Microbiology. Vol. 31. Academic Press; London: 2002. pp. 559–589. [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd CM, McClelland GH. Data analysis: a model-comparison approach. Harcourt Brace Jovanovich; San Diego: 1989. [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006a;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006b;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Martin FJ, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- Montague CT, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- Moore WEC, Johnson JL, Holdeman LV. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int. J. Syst. Bacteriol. 1976;26:238–252. [Google Scholar]
- Mulder NJ, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 33:D201–205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007 doi: 10.1016/j.chom.2007.09.013. in press. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4 Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Figure S1 – The Mollicute bloom occurs in conventionally-raised wild-type C57BL/6J mice as well as in mice without an intact innate or adaptive immune system. Wild-type (+/+), MyD88 −/−, or Rag1 −/− C57BL/6J mice were weaned onto a standard low-fat polysaccharide-rich (CHO) or high-fat/high-sugar (Western) diet. 16S rRNA gene sequence-based surveys were performed; sequences were aligned (NAST; DeSantis et al., 2006), and inserted into an ARB neighbor-joining tree (Ludwig et al., 2004). Asterisks indicate significant differences (Student’s t-test p<0.001).
Figure S2 – Mice with diet-induced obesity that are switched to a FAT-R or CARB-R diet exhibit stabilization of weight, decreased caloric intake and reduced adiposity. (A) Weight gain (g) and (B) percentage epidydymal fat-pad weight to body weight in wild type C57BL/6J mice that were initially weaned onto a Western diet for 8 weeks, and then maintained on the Western diet, or switched to a FAT-R or CARB-R diet for four weeks (n=5-6 mice/treatment group). Weight was monitored during the four-week period. (C) Chow consumption (kcal/d) is decreased in mice switched to a FAT-R or CARB-R diet. Data are represented as mean±SEM. Asterisks indicate significant differences (ANOVA of FAT-R or CARB-R versus Western, *p<0.05, **p<0.01, ***p<0.0001).
Figure S3 – Switching from a Western to FAT-R or CARB-R diet results in a division-wide increase in the relative abundance of Bacteroidetes, and a decrease in the relative abundance of Mollicutes. UniFrac-based analysis of bacterial community membership shows an impact of diet on gut microbial ecology: cecal communities analyzed from two families of C57BL/6J wild-type mice (Table S3) generally cluster based on host diet (Western, FAT-R, and CARB-R). The average relative abundance (% of total 16S rRNA gene sequences) of bacterial lineages within the cecal microbiota of all mice fed a Western, FAT-R, or CARB-R diet is displayed as pie charts. Black boxes indicate nodes that were reproduced in >50% of all jackknife replications (n=126 sequences were randomly re-sampled). Asterisks indicate cecal samples that were analyzed by whole community shotgun sequencing.
Figure S4 – Taxonomic assignments of metagenomic sequencing reads from seven cecal microbiome datasets based on BLAST homology searches, and by alignment of 16S rRNA gene fragments. (A) The cecal microbiome is dominated by sequences homologous to Bacteria. Sequencing reads were trimmed based on quality and vector sequence and the resulting datasets were used as queries against the NCBI non-redundant database (e-value<10−5). Sequences were assigned to the lowest taxonomic group that would include all significant hits, using MEGAN (Huson et al., 2007). Pie charts are shown for each individual dataset and for the average of all datasets. Colors indicate assignments to bacteria (red), archaea (green), eukarya (yellow), viruses (blue), sequences that could not be confidently assigned to a group (purple), and sequences with no significant BLASTX matches (orange). **(B)** Relative abundance of microbiome sequences homologous to genomes from four bacterial divisions: Bacteroidetes (red), Proteobacteria (yellow), Actinobacteria (orange), and Firmicutes (blue). All divisions observed at >1% relative abundance are shown. (C) Relative abundance of microbiome sequences homologous to genomes from bacterial classes within the Firmicutes division: Bacilli (dark blue), Clostridia (yellow), and Mollicutes (light blue). (D) Taxonomic assignments of 16S rRNA gene fragments obtained from cecal microbiome datasets. 16S rRNA gene fragments were identified by querying the Ribosomal Database Project (RDP) database (version 9.33; BLASTN e-value < 10−5; Cole et al., 2005). 16S rRNA gene fragments were aligned with NAST (DeSantis et al., 2006) and added to an ARB neighbor-joining tree (Ludwig et al., 2004). 16S rRNA gene fragments from the Bacteroidetes (red), Proteobacteria (yellow), Verrucomicrobia (green), Mollicutes (light blue), and other Firmicutes (dark blue) are shown.
Figure S5 - Assembly of metagenomic sequence data reveals physical linkage between the Mollicute phosphotransferase system (PTS) and other genes involved in carbohydrate metabolism. The pooled mouse gut microbiome dataset was assembled using ARACHNE (n=7 combined datasets; Batzoglou et al., 2002; see Tables S6 and S7 for assembly statistics). The contig length is shown as a solid black bar. Arrows indicate predicted proteins. Functional assignments were derived from the NCBI annotations and verified by BLASTP comparisons of each predicted protein with the STRING-extended COG database (von Mering et al., 2007) and the KEGG database (Kanehisa et al., 2004), in addition to Hidden Markov Model (HMM)-based protein domain searching with InterProScan (Mulder et al., 2005). Contigs 23 and 73 are >98% identical over the region in pink (234/238 nucleotides): they are likely different ends of the same gene that were not joined due to the relatively stringent assembly parameters employed.
Figure S6 – Concentration of bacterial fermentation end-products in the ceca of Western, FAT-R, and CARB-R mice. Acetate and butyrate levels (μmol per g wet weight cecal contents) were measured by gas chromatography mass spectrometry. Lactate levels (mM per kg protein) were measured using established microanalytic methods (see Methods above). Data are represented as mean±SEM. Asterisks indicate significant differences (Student’s t-test of Western versus CARB-R, *p<0.05, **p<0.01).
Figure S7 – KEGG metabolic pathways significantly enriched in the human gut-derived Eubacterium dolichum strain DSM 3991 genome relative to eight human gut-associated Firmicutes. Pathways whose relative representation is significantly different between the E. dolichum genome and the pooled gut Firmicute genomes (n=8) were identified using a bootstrap comparison of the abundance of sequences assigned to all KEGG pathways (xipe version 2.4; confidence level = 0.98, sample size = 10,000; Rodriguez-Brito et al., 2006). The relative abundance of all KEGG pathways with significantly different representation found at a relative abundance >0.6% in at least two microbiome datasets was transformed into a z-score and clustered by genome and pathway using a Euclidean distance metric (de Hoon et al., 2004). Enrichment (yellow) and depletion (blue) are defined as a relative abundance greater or less than the mean for all datasets (i.e. a z-score greater or less than zero, respectively). For full strain names see Figure 6.
Figure S8 – STRING-based protein network analysis of the predicted E. dolichum proteome. MetaGene (Noguchi et al., 2006) was used to predict proteins from the E. dolichum deep draft assembly. Proteins were subsequently assigned to COGs based on homology (BLASTP e-value<10−5; STRING database version 7, von Mering et al., 2007). Annotated COG interactions were used to organize the protein network, including interactions based on neighborhood, gene fusion, co-occurrence, homology, co-expression, experiments, databases, and text mining (Medusa Java appet; Hooper and Bork, 2005). Nodes, each representing a different orthologous group, are colored as follows: green, present in all analyzed Firmicute genomes (including the mycoplasma); blue, present in all recently sequenced gut Firmicute genomes; red, present in the Western diet-associated cecal microbiome (based on BLAST homology searches, e-value<10−5 and the deposited annotations in the STRING database, version 7). 89% of the COGs found in the E.dolichum genome were also found in the Western diet microbiome. Most of the COGs in green are involved in essential cellular functions such as transcription and translation (56% of the COG category assignments are to ‘Information storage and processing’). Some clusters of interest are highlighted, including the phosphotransferase system (PTS), the 2-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis (MEP), cell wall biosynthesis, ABC transporters, and V-type ATPases for H+ import.
Supplementary Table 1: Protein, carbohydrate, and fat composition of various mouse chow diets
Supplementary Table 2: Percent weight of chow ingredients
Supplementary Table 3: 16S rRNA gene-sequence libraries
Supplementary Table 4: Nomenclature used to designate microbiome datasets obtained from the cecal microbiota of C57BL/6J mice
Supplementary Table 5: Microbiome sequencing statistics
Supplementary Table 6: Microbiome assembly statistics
Supplementary Table 7: Read placements in contigs and BLAST results
Supplementary Table 8: E.dolichum draft genome sequencing statistics