Role of TNF alpha and PLF in bone remodeling in a rat model of repetitive reaching and grasping (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 20.
Published in final edited form as: J Cell Physiol. 2010 Oct;225(1):152–167. doi: 10.1002/jcp.22208
Abstract
We have previously developed a voluntary rat model of highly repetitive reaching that provides an opportunity to study effects of non-weight bearing muscular loads on bone and mechanisms of naturally occurring inflammation on upper limb tissues in vivo. In this study, we investigated the relationship between inflammatory cytokines and matricellular proteins (Periostin-like-factor, PLF, and connective tissue growth factor, CTGF) using our model. We also examined the relationship between inflammatory cytokines, PLF and bone formation processes. Rats underwent initial training for 5 weeks, and then performed a high repetition high force (HRHF) task (12 reaches/min, 60% maximum grip force, 2 hr/day, 3 days/week) for 6 weeks. We then examined the effect of training or task performance with or without treatment with a rat specific TNFα antibody on inflammatory cytokines, osteocalcin (a bone formation marker), PLF, CTGF, and behavioral indicators of pain or discomfort. The HRHF task decreased grip strength and induced forepaw mechanical hypersensitivity in both trained control and 6-week HRHF animals. Two weeks of anti-TNFα treatment improved grip strength in both groups, but did not ameliorate forepaw hypersensitivity. Moreover, anti-TNFα treatment attenuated task-induced increases in inflammatory cytokines (TNFα, IL-1α, and MIP2 in serum; TNFα in forelimb bone and muscles) and serum osteocalcin in 6-week HRHF animals. PLF levels in forelimb bones and flexor digitorum muscles increased significantly in 6-week HRHF animals, increases attenuated by anti-TNFα treatment. CTGF levels were unaffected by task performance or anti-TNFα treatment in 6-week HRHF muscles. In primary osteoblast cultures, TNFα, MIP2 and MIP3a treatment increased PLF levels in a dose dependent manner. Also in primary osteoblast cultures, increased PLF promoted proliferation and differentiation, the latter assessed by measuring Runx2, alkaline phosphatase (ALP) and osteocalcin mRNA levels; ALP activity; as well as calcium deposition and mineralization. Increased PLF also promoted cell adhesion in MC3T3-E1 osteoblast-like cell cultures. Thus, tissue loading in vivo resulted in increased TNFα, which increased PLF, which then induced anabolic bone formation, the latter results confirmed in vitro.
Keywords: repetitive strain injury, musculoskeletal disorder, cytokines, chemokines, Periostin-like-factor
Introduction
We have developed a voluntary rat model of repetitive reaching and grasping that permits the examination of both tissue and behavioural responses to non-weight bearing muscular loads on bone and mechanisms of naturally occurring inflammation in upper limb tissues(Barbe et al., 2003; Barbe et al., 2008; Barr et al., 2003; Elliott et al., 2009b; Elliott et al., 2008). Thus far, we have reported evidence of early local tissue and systemic inflammatory responses and then fibrosis with performance of high repetition low force tasks using this model (Al-Shatti et al., 2005; Barbe et al., 2003; Barr et al., 2003; Clark et al., 2003; Elliott et al., 2009b; Elliott et al., 2008) and that performance of higher demand tasks (e.g. high repetition high force tasks) leads to further inflammation and fibrosis(Clark et al., 2004; Fedorczyk et al., 2009). With performance of high repetition tasks, with or without high force, early bone and muscle adaptive remodeling was observed, which was then followed by pathological remodeling with continued task performance (Barr et al., 2003; Rani et al., 2009a; Rani et al., 2009b), although we have yet to examine possible mechanisms underlying these changes. In addition, we also found increased fibroblast production of collagen type I and Connective Tissue Growth factor (CTGF), the latter a matricellular protein, in the fibrotic repair process in nerves and tendons (Clark et al., 2004; Clark et al., 2003; Fedorczyk et al., 2009), but have yet to examine its increase in other tissues in this model. Finally, we have reported an early but transient increase in Periostin-like-factor (PLF), also a matricellular protein, in loaded forelimb bones in this model, an increase that was associated temporally with a transient increase in bone formation, although both, PLF and bone formation decreased as inflammatory processes continued to increase with continued task performance (Rani et al., 2009b). These latter findings are suggestive of a reactive response of musculoskeletal tissues to repetitive movement that may involve PLF. Therefore in this study, we explored the early adaptive remodeling response in bone and some of the underlying mechanisms, specifically, the relationship between inflammatory cytokines, two matricellular proteins, PLFD and CTGF, and bone formation.
The term ‘matricellular’ was coined in 1995 (Bornstein, 1995; Bornstein, 2009), to denote a subset of proteins that are extracellular matrix (ECM) proteins with properties that can be distinguished from structural macromolecules and bioactive proteins, such as growth factors, cytokines, and proteases, which are also part of the ECM. Matricellular proteins function mainly as adapters and modulators of cell–matrix interactions (Bornstein and Sage, 2002). They regulate cell function by interacting with cell-surface receptors, cytokines, growth factors, proteases, and extracellular matrix structural proteins. Some of the processes that they mediate are adhesion, differentiation, inflammation and fibrosis (Bornstein, 2009; Chen and Lau, 2009; Jugdutt et al., 2009; Rhee et al., 2009). Two matricellular proteins PLF and CTGF, relevant to our studies are briefly described below.
Periostin is a member of the fasciclin family of proteins, classified together due to the presence of multiple 150 amino acid long fasciclin or repeat domains. These domains are involved in adhesion, migration and other cellular processes. Other family members include Fasciclin I, PLF, βig-H3 and Stabilin I and II (Litvin et al., 2005). Periostin and Periostin-like-factor are alternatively spliced forms of the same gene that differ only in one exon at the carboxy terminus (Litvin et al., 2005). Periostin is expressed in multiple tissues like heart, bone and periodontal ligament during development (Kuhn et al., 2007; Lie-Venema et al., 2008; Norris et al., 2008), and is re-expressed in adults after bone fracture (Nakazawa et al., 2004) and myocardial, vascular, and skeletal muscle injuries (Goetsch et al., 2003; Kuhn et al., 2007; Lindner et al., 2005; Stanton et al., 2000). In addition, Periostin is highly expressed in various types of cancers (Bao et al., 2004; Kanno et al., 2008; Kudo et al., 2006; Kudo et al., 2007; Puglisi et al., 2008; Sasaki et al., 2003; Takanami et al., 2008; Tsunoda et al., 2009; Yoshioka et al., 2002). We have reported that Periostin is highly up regulated in forelimb bones, muscles and tendons in our animal model of repetitive strain injury with performance of high repetition high force tasks (Rani et al., 2009a; Rani et al., 2009b), although, once elevated early during the initial training period, the levels do not change with continued task performance.
Similar to Periostin, PLF plays an important role in development of tissues like bone, cartilage, heart, brain and nerves during embryogenesis (Zhu et al., 2008). It is also expressed at very low levels in musculoskeletal and nerve tissues in normal adults, but increases in these tissues after incidences of cellular or tissue stress, and/or injury (Litvin et al., 2006; Zhu et al., 2009; Rani et al, 2008a,b). In contrast to Periostin, in animals performing a HRHF task for 12 weeks, levels of PLF progressively increase during training and early task performance in musculoskeletal tissues, peaking at week 6 in forelimb bones (Rani et al., 2009b). In our model we also observed increased pro-inflammatory cytokines, including TNFα, ILα and macrophage inflammatory proteins (MIPs) in serum, and TNFα in musculoskeletal tissues with performance of either a high repetition negligible force task or a moderate repetition high force task, both moderate demand tasks (Barbe et al., 2008; Elliott et al., 2009a), although we have yet to examine the effects of performing a high demand task involving high repetition as well as high force on inflammatory cytokine levels in serum, bone or muscle. Since we have observed (1) a temporal association between inflammatory responses, and bone remodeling (Barr et al., 2003), (2) that PLF increased early in our model during a period of early adaptive bone formation, and (3) that PLF is anabolic in bone (Zhu et al., 2009), we focused here on finding a mechanistic link between inflammation, PLF, and anabolic bone changes occurring with performance of repetitive tasks.
CTGF is a secreted, extracellular matrix protein that belongs to the CCN family of immediate-early genes needed for differentiation and tissue repair (Arnott et al., 2007; Brigstock, 2009; de Winter et al., 2008; Xu et al., 2000). CTGF mRNA and protein is expressed in fibroblasts, endothelial cells, hypertrophic chondrocytes, osteoblasts, vascular smooth muscle cells, Schwann cells, and epithelial cells, with varying functions by cell type (Clark et al., 2004; Clark et al., 2003; Igarashi et al., 1996; Igarashi et al., 1993). It is a mediator of profibrotic activity of transforming growth factor beta 1 (TGFβ1), a known regulator of fibrosis, but also is abundant in fibrotic responses in the absence of noticeably elevated levels of TGFβ1 (Brigstock, 2009; Chen and Lau, 2009; Shi-Wen et al., 2008). Another regulator of CTGF is TNFα (Barrientos et al., 2008; Brigstock, 2009; Chen and Lau, 2009), a cytokine that increases in musculoskeletal and nerve tissues as well as serum in our repetitive strain injury model (Al-Shatti et al., 2005; Barbe et al., 2008; Elliott et al., 2009b). We have previously found increased production of CTGF in nerve and tendon after 12 weeks of performance of either high repetition low force or high repetition high force tasks either following or temporally associated with increased TNFα (Clark et al., 2004; Clark et al., 2003; Fedorczyk et al., 2009). These findings further suggest possible links between inflammatory responses and expression of matricellular proteins.
TNFα is a pro-inflammatory cytokine that regulates proliferation of many cell types including osteoclasts and fibroblasts (Ochi et al., 2009; Yoshimatsu et al., 2009). The differentiation and function of osteoblasts is also affected by TNFα, which in turn alters the balance between bone formation and bone resorption (Minamitani et al., 2009; Ochi et al., 2009; Yamazaki et al., 2009). TNFα directly promotes osteoclastogenesis via engagement of TNFR1 on osteoclast precursor cells (Huang et al., 2003; Ma et al., 2004; Ritchlin et al., 2003; Zhang et al., 2001) or indirectly via induction of M-CSF and RANKL expression on mesenchymal cells (Hsu et al., 1999). Moreover, TNFα is known to stimulate bone resorption and inhibit bone formation, with inflammatory bone loss partly due to its effect on osteoblasts, such as the induction of Smurf-mediated degradation of Runx2, which leads to the inhibition of osteoblasts (Kaneki et al., 2006). We hypothesize that the early adaptive remodeling of bone tissue undergoing repetitive loading in our model would be modulated by the superimposed inflammatory response. Therefore, we examined the effect of a rat specific TNFα antibody, which binds both the membrane bound and soluble form of TNFα, on the expression of key inflammatory cytokines and chemokines, matricellular proteins, osteocalcin (a bone formation marker), and behavioral indicators of inflammatory pain (grip strength, an indicator of movement-related hyperalgesia, and forepaw cutaneous sensation). We hypothesize that inflammatory cytokine/chemokine increases occurring early in our in vivo model are associated with increased PLF, and therefore, anabolic bone remodeling, and that reducing inflammatory cytokine levels during this remodeling phase would reduce both PLF and bone formation responses. We further sought to investigate how these responses might be mediated by examining the effect of increased PLF in vitro on proliferation, adhesion and differentiation of cultured primary osteoblasts and MC3T3E-1 cells. Additionally, since we have observed increased CTGF, another matricellular protein, by 12 weeks of task performance in past studies, we also examined whether reducing inflammatory cytokine levels in tissues would affect CTGF.
Materials and methods
Source of animals
For this study, the Temple University Institutional Animal Care and Use Committee approved the animal protocols in compliance with NIH guidelines for the humane care and use of laboratory animals. Studies were conducted on female Sprague-Dawley rats (Ace, PA), housed in a central animal facility in separate cages on a 12-hour (hr) light: dark cycle with free access to water.
Primary osteoblast cultures were derived from a colony of Sprague-Dawley rats maintained at Temple University School of Medicine (Philadelphia, PA). All animals were maintained and used according to the principles in the NIH Guide for the Care and Use of Laboratory Animals (U.S. Department of Health and Human Services, Publ. No. 86-23, 1985) and guidelines established by the Institutional Animal Care and Use Committee at Temple University.
Behavioral apparatus and repetitive motion task performance
Rats were trained and performed a HRHF task in a force apparatus, which was custom-designed (by Dr. Ann Barr and Custom Medical Research Equipment, Glendora, NJ) and integrated into an existing commercially available operant training system (Med Associates, Georgia, VT). The testing chamber and force apparatus have been described in detail previously (Clark et al., 2004; Rani et al., 2009b). Experimental rats performed a high repetition high force (HRHF) task regimen of 12 reaches/min at 60 ± 5% maximum pulling force (MPF).
Fifty-three 3 month old, Sprague-Dawley rats were used for these experiments. Thirty-eight rats were trained to perform a repetitive handle-pulling task with food reward using standard operant conditioning procedures during a 5-week training period. During this training period, rats learned the HRHF task in the operant test chamber until they could reach into the tube dispenser with no specified reach rate at 60 ± 5% maximum pulling force for 10 min/day. Once the animals were able to perform the task consistently (usually after 5 weeks), rats were randomly divided into a trained only control group (TC) and a HRHF group. Then the HRHF experimental animals (n =19) began the task regimen at the rate of 12 reaches/min at 60 ± 5% maximum pulling force (MPF), for 2 h/day, 3 days/week (Monday, Wednesday, Friday) for 6 weeks. The daily task was divided into four, 0.5-hour training sessions separated by 1.5 h. The TC rats (n =19) were age and weight-matched to HRHF task rats, but did not perform beyond this initial training period. Fifteen age-matched, normal control (NC) rats were also included that were not exposed to task training or performance.
Administration of anti-TNFα
An anti-rat TNFα antibody (CNTO 1081; generously provided by Centocor R&D Inc.) was injected intraperitoneally (i.p., 15 mg/kg body wt) into HRHF rats (n=6), first at the beginning of task week 4. A 2nd injection was administered at the end of task week 4, and a 3rd injection was administered at the end of task week 5. All HRHF rats were then euthanized at the end of task week 6 (6-week HRHF without anti-TNFα (6HRHF), n =_13; 6_-week HRHF with anti-TNFα (6HRHF/αTNF), n=6). Six of the trained rats (TC) received anti-TNFα treatments on this same schedule. All TC rats were euthanized at matched time points to HRHF rats, i.e. they were trained for 5 weeks, and then rested for 6 weeks before euthanasia (TC without anti-TNFα (TC), n =13; TC with anti-TNFα (TC/αTNF); n=6).
Sensorimotor behavioral testing
Maximum forearm grip strength and cutaneous sensation was determined in all rats on the day of euthanasia. Briefly, for assessment of grip strength, rats were lifted gently by the tail and allowed to grasp a rigid bar attached to a force transducer and digital display unit (Stoelting, Wood Dale, IL), as described previously (Barbe et al., 2003; Clark et al., 2004; Clark et al., 2003; Rani et al., 2009b). When the first signs of active grasp were observed, the rats are pulled upward slowly by the tail with increasing firmness until their grasp was overcome. The peak force was recorded as maximum grip strength. The test was repeated 3–5 times/limb, and maximum grip strength per trial included in the statistical analysis. Mechanical cutaneous sensation in the forepaws of the preferred reach limb was assessed using the up-down method of Von Frey monofilament testing as described previously (Clark et al., 2004).
Serum biomarker analyses
Serum was collected from all rats following euthanasia (Nembutal, 120 mg/kg body weight), 18–36 h after completion of the final task session, blood was collected by cardiac puncture using a 23-gauge needle. The blood was centrifuged immediately at 1000 g for 20 min at 4°C. Serum was collected, separated into 200 μl aliquots, flash-frozen, and stored at −80 °C until analyzed. Levels of Cytokine-Induced Neutrophil Chemoattractant 2a (CINC2a), IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-10, Interferon Gamma (IFNβ), granulocyte-macrophage-colony-stimulating factor (GMCSF), MIP-2, MIP3a, monocyte chemoattractant protein-1((MCP1), Regulated on Activation, Normal T-cell Expressed and Secreted (RANTES), and TNFα were analyzed in serum by Aushon Biosystems Searchlight multiplex ELISA according to manufacturer’s protocols, with pg/ml reported, as previously described in Barbe et al., 2009. Serum levels of osteocalcin were analyzed using a commercially available ELISA kit (Nordic Bioscience).
ELISA and Western blot analysis of musculoskeletal tissues
The distal end of forearm bones (radius and ulna) and flexor digitorum muscles were collected from 32 rats: 8 NC, 8 TC, 3 TC/αTNF, 10 6HRHF, and 3 6HRHF/αTNF. The collected tissues were homogenized in RIPA buffer [25 mM Tris HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail (Sigma)], incubated overnight at 4 °C, supernatant collected and stored at −80 °C. A bone powder was prepared from the radius and ulna bones by rapidly pulverizing frozen samples in a Bessman tissue pulverizer (Fisher Scientific) pre-cooled in a bath of dry ice/ethanol, and then homogenized in RIPA buffer. For ELISA of muscles and bones, total protein was determined using BCA-200 protein assays (Bicin Choninic Acid, Pierce). Tissue lysates were analyzed for TNFα using a commercially available ELISA kit according to manufacturer’s protocols (BioSource International). Each sample was run in duplicate. ELISA assay data (pg cytokine protein) were normalized to μg total protein.
For western blot analysis (n=3/group), 25 μg of protein sample was mixed with 5X Laemmli sample buffer (10% SDS, 50% Glycerol, 25% β-mercaptoethanol, 300 mM Tris HCl pH 6.8, and 0.04% Bromophenol blue), boiled for 5 min, and resolved by 10% SDS-PAGE. Protein samples were electroblotted to a nitrocellulose membrane at 90 V for 1 h at 4 °C, membrane was blocked in 5% non-fat dry milk in phosphate-buffered saline (PBS) with 0.1% Tween-20 (PBST) for 1 h and incubated in 0.3 μg/ml PLF (custom made) or 0.2μg/ml CTGF specific primary antibody (Santa Cruz) overnight at 4 °C. Specificity of the PLF primary antibody has been previously described (Zhu et al., 2008). The membrane was washed with PBST, incubated with 0.2 μg/ml of HRP-conjugated goat anti-rabbit IgG secondary antibody (Pierce, Rockford, IL) for 1 h at room temperature, and the chemiluminescent signal was detected using the ECL kit (Pierce, Rockford, IL). Blots were stripped, washed and reprobed with 0.2 μg/ml GAPDH-specific primary antibodies overnight at 4 °C, using the method described above. Cell lysates were also analyzed for Periostin as described above.
Immunohistochemical analyses
In accordance with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association, animals were euthanized (Nembutal, 120 mg/kg body weight) and perfused transcardially with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). Tissues from limbs (n=3/group) were collected and postfixed “en bloc” by immersion overnight. Post fixation, flexor digitorum muscle and tendon, with the median nerve were separated en bloc as a flexor mass from the bones, cryoprotected in 30% sucrose in phosphate buffered saline (PBS) and then frozen-sectioned on a cryostat into 15 μm longitudinal sections, as previously described (Barbe et al., 2003). Forearm bones (radius and ulna) were collected; decalcified, paraffin embedded, and sectioned as described previously (Barbe et al., 2003; Rani et al., 2009b). Longitudinal sections from the forelimb bone or flexor digitorum muscles were treated with 3% H2O2 in methanol for 30 min, washed, and then blocked with 4% goat serum in phosphate buffered saline and 0.1% Triton-X100 (PBST) for 30 min at room temperature. Sections were then incubated overnight at 4°C with primary antibody diluted in 5% goat serum/PBS. Anti-PLF (custom made) was used at a 1:1000 dilution. Specificity of the anti-PLF primary antibody has been previously described (Zhu et al., 2008). After washing, sections were incubated with secondary antibody, diluted 1:500 for 30 min at room temperature. The sections were then washed and treated for 30 min with signal amplification kit, (ABC kit, Vector laboratories), signal was visualized using diaminobenzidine (DAB). DAB-treated sections were counterstained with hematoxylin and examined using bright field microscopy. Negative control staining was performed by omitting the primary antibody.
Recombinant Periostin-Like-Factor (rPLF)
The complete mouse PLF cDNA insert from the pgem3zf- vector (Litvin et al., 2004) was amplified using PLF specific primers, with EcoRI and SalI restriction enzyme sequences added to the 3′ and 5′ end of the primers, respectively to achieve a directionally placed insert in the expression vector, pGEX6P-1 (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). Protein was expressed as directed by the manufacturer and the quality and specificity of the bacterially expressed protein was determined by western blot analysis. The effect of rPLF on proliferation and differentiation of osteoblasts was determined as described below.
Reagents to over express or knock down PLF: Generation of adenovirus
Small interfering RNA was directed at a region in exon 17, specific to PLF (AGTCATTCAAGGCAGTCTTCAG), was inserted into the IMG-1200 adenoviral vector to obtain better infectivity. The adenovirus (AdsiPLF) was generated by Imgenex Corporation (San Diego, CA). Adenovirus used to over express PLF was generated as described by (Litvin et al., 2006; Zhu et al., 2009). Adenovirus overexpressing β-galactosidase (AdLacZ) or GFP (AdGFP) was used as a control (Litvin et al., 2006; Zhu et al., 2009).
Primary osteoblast cultures
Parietal calvaria were isolated from 2 day old neonatal rats and the periosteum and cranial sutures were removed to reduce contamination with non-osteoblast cells (Litvin et al., 2006; Xu et al., 2000). Pieces of calvaria were then subjected to five sequential digestions with 2% collagenase-P/0.25% trypsin of 5, 15 (X2), and 25 (X2) min at 37°C in a shaking water-bath. Cells from the first two digestions were discarded to remove non-osteoblast cells. Osteoblast-enriched cell populations were obtained from the 3rd to 5th digestions. These cells were plated in 10 mm2 dishes at a concentration of 5 × 105 cells in Earle’s Minimal Essential Medium ((Sigma, St. Louis, MO) supplemented with 10% fetal calf serum (FCS) and penicillin and streptomycin. Cells were incubated at 37°C with 5% CO2, and the media was changed every 3 days until they were 80% confluent. Stocks were made and frozen in liquid nitrogen for future use.
Treatment of primary osteoblasts with recombinant TNFα, MIP-2 or MIP3a
Primary osteoblasts were plated at a density of 1 × 104 cells/well in EMEM media containing 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin in 12-well plates. Serum was withdrawn the next day and total protein was collected from cells after treatment with increasing concentrations of TNFα (R& D systems), MIP-2 or MIP3a for 48 hrs (Biosource, Camarillo CA, USA).
Osteoblast proliferation
The effect of PLF over expression and knock down mediated by adenovirus was examined using the CyQUANT cell proliferation assay, bromodeoxyuridine incorporation and RT-PCR of cell cycle genes.
CyQUANT Cell Proliferation Assay
Primary osteoblasts were plated in 24-well plates at a concentration of 1 × 104 cells/well in EMEM media containing 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Twenty-four hours later, cells were infected with AdPLF or AdLacZ or AdsiPLF (50 moi) and 72 h post-infection and plates were frozen at −80°C for lysis of cells. Proliferation was assessed using the CyQUANT Cell Proliferation Assay Kit (Invitrogen, Carlsbad, CA). The CyQuant GR dye shows strong fluorescence when bound to cellular nucleic acids. For the assay, 200 μl of the CyQuant GR solution was added to each well (for each treatment, n = 6) and incubated for 5 min at room temperature. Fluorescence was measured at an emission wavelength of 520 nm and excitation wavelength of 480 nm on the Wallac Plate Reader (Perkin-Elmer, Waltham, MA).
The effect of rPLF or recombinant Periostin protein (rPN; R&D systems) on proliferation was also assessed using the CyQUANT and 5-bromo-2-deoxyuridine (BrdU) incorporation assay. Primary osteoblasts were plated at a concentration of 1 × 104 cells/well as described above. Serum was withdrawn the next day, followed by rPLF treatment at a concentration of 0, 1, 10, 50 and 100ng/ml and rPN treatment at a concentration of 100ng/ml. 48 hours post-treatment cells were frozen at −80°C and proliferation was assessed using the CyQUANT Cell Proliferation Assay Kit as described above or the BrdU assay as below.
Bromodeoxyuridine (BrdU) incorporation assay
Primary osteoblasts were cultured in 96 well dishes at a concentration of 2,500 cells/well. Serum was withdrawn the next day, followed by rPLF or rPN treatment for 48 hrs at the concentrations described above. BrdU (Sigma, St. Louis, MO) was added at a concentration of 10mM to the cultures for next 18 hrs. Cultures were then fixed and treated with monoclonal anti-BrdU (Sigma, St. Louis, MO) followed by incubation with goat anti-mouse IgG horseradish peroxidase (HRP) conjugate (Promega, Madison, WI). Wells were washed with PBS and incubated with tetramethylbenzidine (TMB) substrate (KPL; Gaithersburg, MD) until a blue color developed. Wells were subsequently quenched with stop solution (KPL; Gaithersburg, MD) and absorbance was measured at 450 nm on an iMark Microplate Absorbance Reader (Bio-Rad, Hercules CA).
MC3T3-E1 Cells Adhesion assay
MC3T3-E1 mouse osteoblast cell line obtained from ATCC (Manasass, VA) was propagated in Alpha Minimum Essential Medium αMEM) with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained at 37°C. For adhesion assay 96 well plates were coated with poly-lysine (0.10mg/ml, Sigma, St. Louis, MO), fibronectin (5ug/ml, Sigma, St. Louis, MO)), and PLF conditioned media or GFP conditioned media. Conditioned media was obtained as follows: osteoblasts were infected with AdPLF (Zhu et al., 2009), cultured for 72 hours and serum free media collected. Media was tested to show elevated levels of PLF (Zhu et al., 2009). Plates were stored at 4°C until the next day. The solution was removed and wells were blocked with 1% BSA/PBS, for 1 hr at room temperature. 80% confluent plates of MC3T3 cells were trypsinized, and were either left untreated or incubated with anti-β1 integrin (10ug/ml, Santa Cruz Biotechnology, Santa Cruz, CA) at 37°C for 30 mins. Cells were then plated at a concentration of 2× 105 cells/well in the coated 96 well dish in αMEM with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin and incubated for 2 hrs at 37°C. Media was removed and dishes rinsed with PBS. Following fixation with 4% paraformaldehyde, cells were stained with 1% Methylene Blue (in 0.01M Borate Buffer pH= 8.5) for 30 min at RT, and eluted with a 1:1 Ethanol −0.1 M HCl solution for 20 mins. Absorbance was read at wavelength 620⌊ on an ELISA reader.
Osteoblast differentiation
Primary osteoblasts were plated in 12-well plates at a concentration of 1 × 104 cells/well in EMEM media with 10% FBS. Once the cells were 80% confluent, they were infected with AdPLF, AdLacZ or AdsiPLF (50 moi) or treated with rPLF and rPN (100ng/ml). Differentiation factors were added on day 5 (50 μg/ml ascorbic acid and 100 mM β-glycerolphosphate) and each time the media was changed. Markers of differentiation were assessed as described below. Total mRNA was collected for qRT-PCR. Alkaline phosphatase staining and activity was assessed on day 7, while calcium levels and mineral deposition (von Kossa staining) were assessed on day 21. All experiments were repeated in triplicate.
Alkaline phosphatase
Staining
Cells were washed with PBS and fixed with Citrate-Acetone-Formaldehyde fixative as per the manufacturers recommendation (Sigma-Aldrich Diagnostics, St. Louis, MO) for 30 sec. Cells were washed with double distilled water (ddH2O) for 45 sec, incubated in staining solution that included sodium nitrite, FRV-Alkaline solution, napthol AS-B alkaline solution (Sigma-Aldrich Diagnostics, St. Louis, MO) for 15 min at room temperature and protected from light. The cells were rinsed with ddH2O and allowed to air dry. A reddish-pink color indicated positively stained osteoblasts.
Activity
Cells were washed with PBS and lysed in 500 μl of 0.2% Triton X-100 in distilled water by shaking for 20 min at room temperature. A 50 μl aliquot of the cell lysate was mixed with 150 μl of working solution containing 200 μL Assay Buffer, 5 μL Mg Acetate (final concentration 5 mM) and 2 μL _p_-Nitrophenyl phosphate (pNPP) liquid substrate (final concentration 10 mM). Plates were read at 405λ at time zero and again at four minutes as per the manufacturer’s protocol (QuantiChromTM Alkaline Phosphatase Assay Kit, BioAssay Systems, Hayward, CA) Alkaline phosphatase activity (substrate release nmol/min) was normalized to total protein content. Total protein content was determined on identical duplicate cultures using BCA protein assay kit (Pierce).
Calcium deposition
Cells were rinsed with cold PBS and hydrolyzed in 400 μl 0.5 N HCl. Cell extracts were mixed with calcium binding reagent (o-Cresolphthalein Complexone and 8-Hydroxyquinoline, Sigma, St. Louis, MO) in 250 nM 2-Amino-2-Methyl-1, 3 Propanediol buffer and optical density measured at 575 nm using a iMark Microplate Absorbance Reader (Bio-Rad, Hercules CA). Concentration of calcium ions was calculated by comparison to a standard of known calcium density using the equation (A575 sample/A575 standard) × (concentration of standard) and data was expressed as mg calcium/well.
von Kossa staining and quantification of nodule area
Cells were washed with Hanks balanced salt solution and incubated in 3% silver nitrate solution under ultra-violet light for 45 min, washed in ddH2O, and incubated with 3% sodium thiosulfate for 5 min. Following this treatment cells were washed with ddH2O, counterstained with fast green for 5 min, washed again in ddH2O and air dried. For measurement of mineralized (von Kossa stained) nodule area, wells were quantified using a microscope (Nikon Corporation, Melville, NY) interfaced with an analog camera and a bioquantification software system (Bioquant Osteo II, Nashville, TN) using methods described in (Abdelmagid et al., 2007; Zhu et al., 2008). Briefly, nodules were encircled carefully with the Irregular Region of Interest Option (iROI) tool. Then, color thresholds were selected based on a pre-defined mean level of von Kossa staining using the thresholding tool. Next, a field count of the encircled nodule was generated using the Video Count Area array, which counted the number of pixels in the field that are at or above the defined threshold of von Kossa staining multiplied by the area of a pixel at the selected magnification. The Video Count Area array was also used to count all pixels within the encircled nodule. Finally, the mineralized nodule area was calculated by dividing the videocount of pixels that are at or above the pre-defined threshold of von Kossa staining by the total number of pixels in the chosen ROI. This determination was made at four different locations per well, and three wells per group, for each condition.
Reverse Transcriptase Quantitative Polymerase Chain Reaction (RT- qPCR)
Primary osteoblasts were cultured in 10 mm2 petri plates in EMEM with 10% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin at a concentration of 5×105 cells/well. Serum was withdrawn on day 1, 3, 5 and 11, rPLF (100ng/ml) was added on day 2, 4, 6 and 12 and total RNA was collected on day 4, 6, 8 and 14 for Cdk4, Runx2, ALP and osteocalcin (OC) quantification, respectively. One mg of total RNA was reverse transcribed into cDNA (SuperScript First strand, Invitrogen, Carlsbad, CA). Using the Taqman Gene Expression Assay system (Applied Biosystems, Foster City, CA), RT-qPCR was performed in triplicate on the ABI 7500 Real-Time PCR system (Applied Biosystems). All experiments were repeated three times, and the data was normalized to GAPDH, used as the house keeping gene.
Statistical Analyses
Univariate ANOVA was used in all cases to compare any significant changes between control and treatment group (Prism). All post hoc analyses were carried out using the Bonferroni test for multiple comparisons in which p values are adjusted to account for the number of comparisons performed in an ANOVA. An adjusted p value of < 0.05 was considered significant for all analyses.
RESULTS
Several cytokines and chemokines increase in serum and musculoskeletal tissues with high repetition high force task
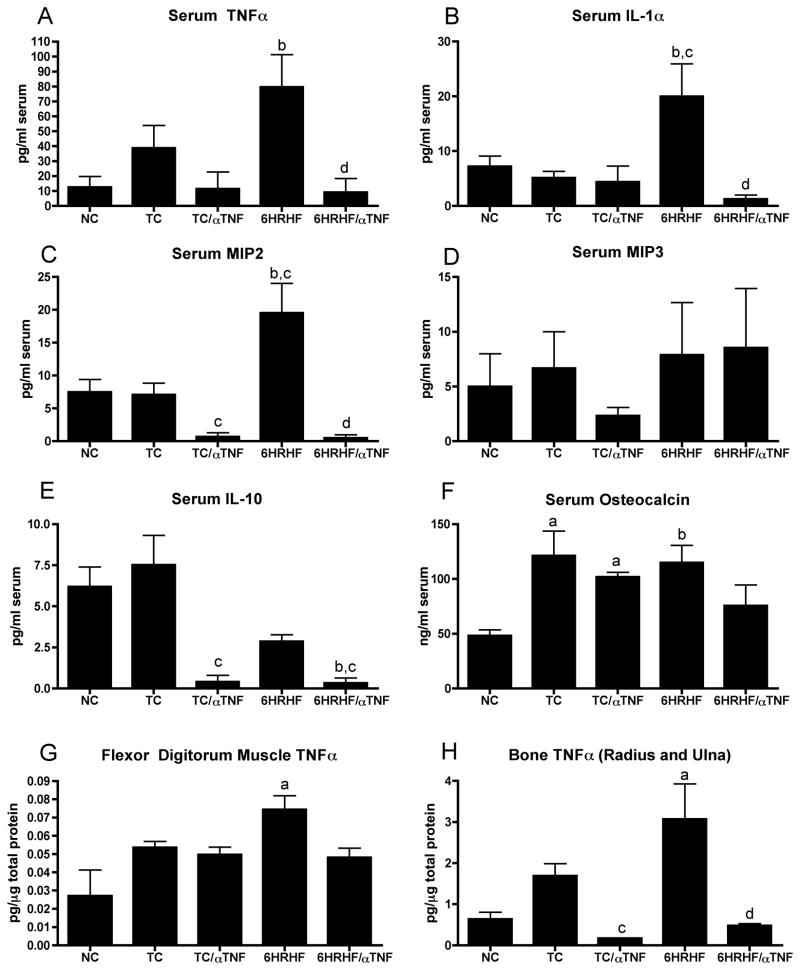
Previous studies using our repetitive reaching model revealed an increase in pro- inflammatory cytokines like tumor necrosis factor alpha (TNFα), interleukin 1alpha (IL-1α), and chemokines, like macrophage inflammatory proteins (MIP2 and MIP3a), in response to a high repetition negligible force task for 8 weeks (Barbe et al., 2008), and TNFα and MIP3, in response to a moderate repetition high force task for 6 weeks {Elliott, 2009 #10}. In this study, we observed that TNFα IL-1α and MIP2, were significantly elevated in the serum of animals that performed a high repetition high force task for 6 weeks (6HRHF; Fig 1A–C, p<0.05 for each), compared to NC. IL-1α and MIP2 were also significantly elevated in serum of 6 week HRHF rats compared to TC (Fig 1A–C, p<0.05 for each). However, MIP3a and IL-10 serum levels were not increased in 6HRHF animals compared to NC or TC (Fig 1D and E). Serum levels of TNFα, IL-1α, MIP2 and IL-10 were not significantly higher in TC rats compared to NC rats. There were also no detectable levels of IL-2, IL-4, IL-6 or GMCSF in serum of any group, and no significant serum increases above NC levels for IL-1β, IFNγ, CINC2a, MCP1 or RANTES (data not shown).
Figure 1. Anti-TNF intervention for 2 weeks attenuated increased cytokines/chemokines and osteocalcin levels in high repetition high force (HRHF) task animals.
Bar graphs showing A,B) tumor necrosis factor alpha (TNFα) and interleukin 1 alpha (IL-1α) levels in serum, C,D) chemokines macrophage inflammatory protein 2 and 3a (MIP2 and MIP3a) levels in serum, E) anti-inflammatory cytokine, IL-10, in serum, F) osteocalcin, a bone formation marker, levels in serum, F) TNFα levels in forelimb flexor digitorum muscles, and G) TNFα levels in forelimb radius and ulna bones,. Data shown for normal controls (NC), trained controls with and without anti-TNF treatment (TC/aTNF and TC, respectively), and 6 week HRHF animals with or without anti-TNF treatment (6HRHF/aTNF and 6HRHF, respectively), in the last 2 weeks before euthanasia. ap<0.01 compared to NC; bp<0.05 compared to NC; cp<0.05 compared to TC; dp<0.01 compared to 6HRHF.
Since serum TNFα underwent the highest fold change from control levels (6 fold change), we explored its levels in forelimb flexor muscles and bones, tissues previously shown to have elevated TNFα with repetitive task performance {Barbe, 2008 #15; Elliott, 2009 #10}. In this study, we observed that TNFα was significantly elevated in forelimb flexor digitorum muscle and bones (radius/ulna) of 6HRHF animals (Fig 1G, H, p<0.001 for each), compared to NC. As with serum, TNFα was not significantly increased in forelimb muscles and bones in TC rats compared to NC rats (Fig 1A–E).
Anti-TNF administration differentially affects expression of cytokines and chemokines in the WMSD model
Based on these results, and since TNFα is a key cytokine that regulates inflammation (Elliott et al., 2009b; Guo et al., 2008; Kaneki et al., 2006), we examined the effects of blocking TNFα in our model. For blocking, we used a rat specific antibody that binds both the soluble and membrane bound form of TNFα. The anti-TNF was administered to animals during the last 2 weeks of the 6 week task regimen, and was observed to effectively block task-induced increases of TNFα, IL-1α and MIP2 in the serum of treated 6HRHF versus untreated 6HRHF rats (Fig 1A–C, p<0.05 or p<0.01 for each). Also, treated 6HRHF rats had similar levels of TNFα, IL-1α and MIP2 as NC rats (Fig 1A–C). Serum MIP3a levels remained unaffected (Fig 1D). Interestingly, anti-TNF treatment also decreased serum IL-10 levels in TC/αTNF and 6HRHF/αTNF animals, compared to untreated TC animals (Fig 1E, p<0.05 for each), and in 6HRHF/αTNF compared to NC (p<0.05). TNFα levels in flexor digitorum muscles in these same animals were markedly but not significantly reduced in treated versus untreated 6-week HRHF rats (Fig 1G). In contrast, TNFα levels in radius/ulna in these same animals were significantly reduced in treated versus untreated 6HRHF rats (Fig 1H, p<0.001). Treated 6HRHF rats had similar levels of TNFα in radius/ulna as NC rats. Anti-TNF also reduced TNFα levels in radius/ulna of TC/αTNF rats compared to untreated TC (p<0.05).
Anti-TNF does not affect osteocalcin expression in WMSD model
We have previously shown that osteocalcin (OC), a serum biomarker of bone formation, is elevated in response to HRHF task performance and is highest at week 6 in HRHF rats (Rani et al., 2009b). TNFα negatively regulates bone formation by increasing bone resorption, decreasing osteoblast-mediated bone formation, and increasing apoptosis in osteoblasts (Horowitz et al., 2001). Therefore, we examined the levels of OC with and without anti-TNF administration. Although both training and HRHF task performance increased osteocalcin levels (Fig 1F, p<0.01 in TC, and p<0.05 in 6HRHF, compared to NC), anti-TNF attenuated this increase despite continued task performance in 6HRHF/αTNF rats (osteocalcin levels were not significantly higher in these rats compared to NC).
Anti-TNF rescued grip strength but not forepaw mechanical sensation with HRHF
Performance of repetitive tasks in this model leads to increased macrophages and pro-inflammatory cytokines (IL-1α, IL-1β, and TNF-α in nerves and musculoskeletal tissues (Al-Shatti et al., 2005; Barbe et al., 2003; Barbe et al., 2008; Clark et al., 2004; Clark et al., 2003) that are associated with reduced grip strength and increased paw withdrawal thresholds to von Frey hair testing, the latter indicative of inflammation-induced cutaneous hypersensitivity {Elliott, 2009 #10; Barbe, 1992 #50; Fedorczyk, 2009 #3} although with continued task performance, cutaneous hyposensitivity developed in the HRHF animals as median nerve function declined due to fibrotic nerve compression and axonal demyelination (Clark et al., 2004). Therefore, in this study we assessed paw withdrawal threshold response and grip strength in animals with or without anti-TNF administration. Treatment attenuated declines in grip strength in TC/αTNF animals (Fig 2A; TC/αTNF rats were not significantly different from NC and were p<0.05 compared to untreated TC, which were significantly decreased from NC rats at p<0.001). Anti-TNF treatment also improved grip strength in 6HRHF/αTNF animals compared to 6HRHF (Fig 2A, p<0.001), although they were still significantly less than NC (Fig 2A, p<0.001). Cutaneous hypersensitivity (decreased withdrawal thresholds responses) was already present after the 5-week training period in TC rats (Fig 2B, p<0.001 compared to NC), and remained so with performance of the HRHF task for 6 weeks. Anti-TNF treatment in the last two weeks before euthanasia did not return cutaneous sensitivity to normal control levels (Fig 2B; p<0.001 compared to NC).
Figure 2. Anti-TNF intervention for 2 weeks improved grip strength significantly but not ameliorate forepaw mechanical hypersensitivity in high repetition high force (HRHF) task animals.
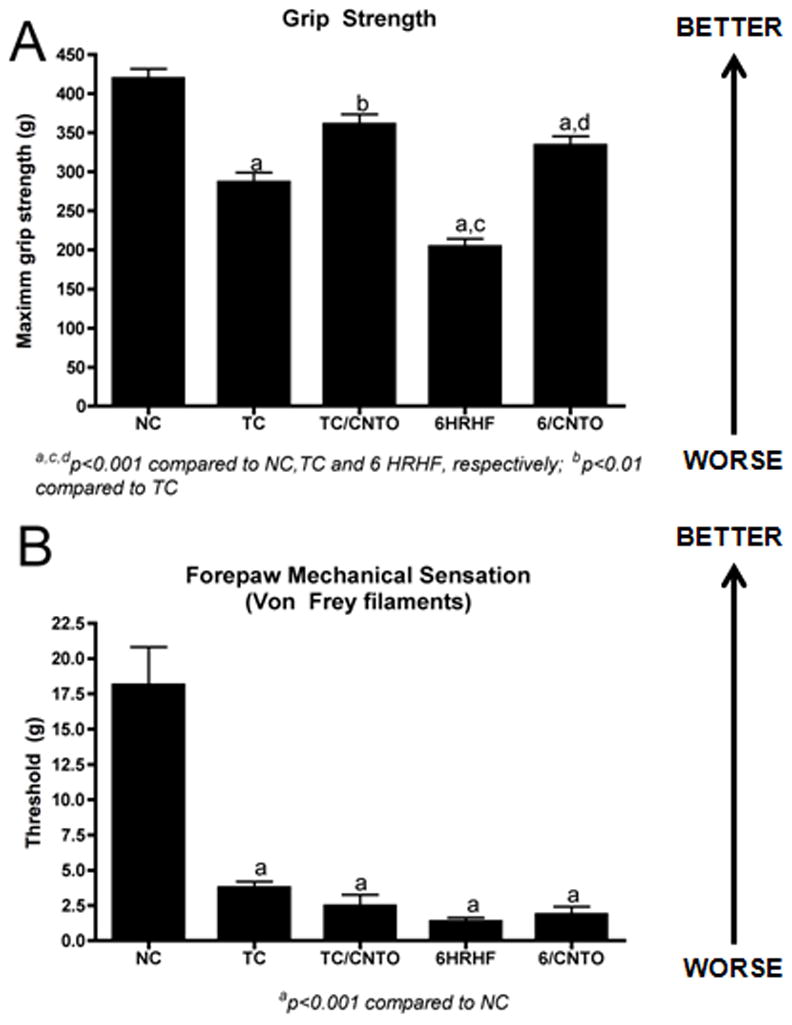
A) Forelimb grip strength and B) cutaneous mechanical sensitivity using Von Frey test (withdrawal threshold) is shown for normal controls (NC), trained controls (TC) with or without anti-TNF treatment (TC/aTNF and TC, respectively), and 6 week HRHF animals with or without anti-TNF treatment (6HRHF/aTNF and 6HRHF, respectively), in the last 2 weeks before euthanasia. ap<0.001 compared to NC; bp<0.05 compared to TC; cp<0.001 compared to TC; dp<0.001 compared to 6HRHF.
PLF is down regulated but CTGF levels remain unaffected by anti-TNF administration
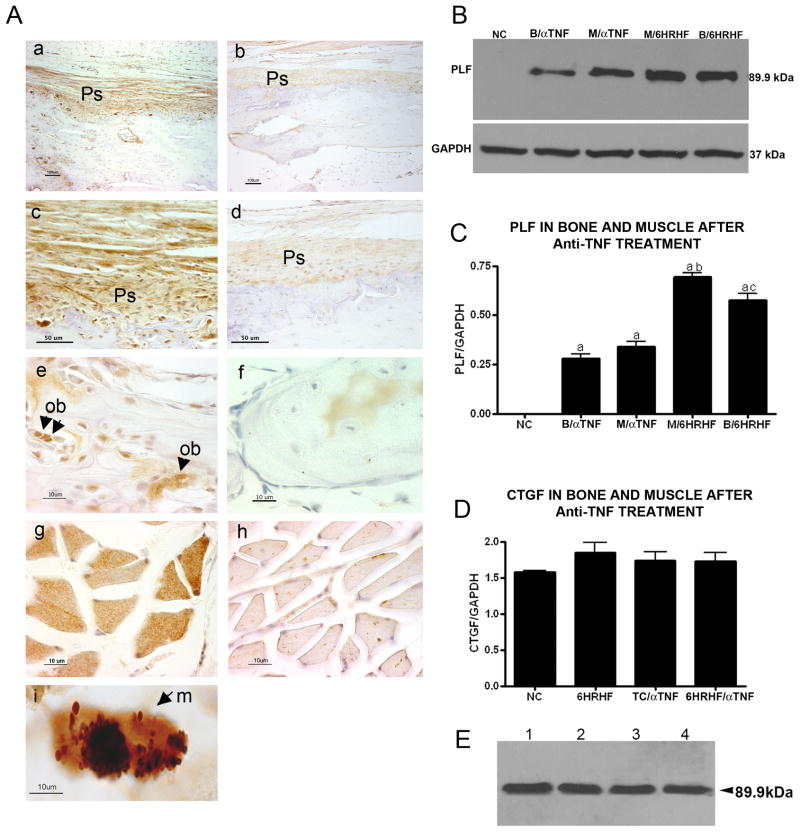
In our previous studies (Rani et al., 2009a; Rani et al., 2009b), we observed that PLF was up regulated in multiple tissues after performance of a HRHF task, with the highest levels in forelimb bones in 6-week HRHF rats. Since serum and tissues of 6-week HRHF rats showed significant increases in several pro-inflammatory cytokines and chemokines (Fig 1), and since anti-TNF treatment attenuated these increases, we investigated if anti-TNF treatment altered PLF levels in musculoskeletal tissues. After anti-TNF treatment, PLF expression decreased significantly in radius and ulna periosteum (Fig 3Aa–d shows radius, compare a to b, and c to d), in osteoblasts in these same bones (Fig 3Ae,f shows radius, compare e to f), and in flexor digitorum muscles (Fig 3A, compare g to h). Western blot analysis of proteins from forelimb bones (radius and ulna combined, Fig 3B) and flexor digitorum muscles (Fig 3C) of a second cohort of animals confirmed a decrease in PLF levels, (Fig 3B&C: p<0.001 compared to NC). In contrast, levels of another matricellular protein, connective tissue growth factor (CTGF), showed no changes after anti-TNF treatment (Fig 3D). In Figure 3E we show that recombinant PLF expressed in bacteria migrated at 90kD when analyzed by western blot and probed with anti-PLF specific antibody. The recombinant protein will be used in experiments described below.
Figure 3. Anti-TNF intervention lowered PLF levels in forelimb bones and flexor digitorum muscles of high repetition high force (HRHF) task animals.
A) Immunolocalization of PLF in forelimb bones (a–f) or flexor digitorum muscles (g–i) of 6 wk HRHF task animals with or without anti-TNF treatment. Sections were immunoreacted with PLF antibody; brown color indicates positive staining; hematoxylin (a nuclear stain) was used as a counterstain. PLF immunoreactivity was detected in radial periosteum (a–d) and osteoblasts (e–f, arrowheads), and flexor digitorum muscle fibers (g–h, arrowhead in g shows higher staining). PLF was also detected in macrophages (small arrow) in 6 wk HRHF task animals (i). (B,C) Total protein collected from radial/ulna bones (B) and flexor digitorum muscles (M) of normal controls (NC), 6 week HRHF animals with no anti-TNF treatment (B/HRHF and M/HRHF), and 6 week HRHF with anti-TNF treatment (B/ aTNF and M/ aTNF), and were resolved by 10% SDS PAGE and blots were probed with anti-PLF; GAPDH was used as a loading control. Experiments were repeated three times and bar graphs of the ratio of PLF/GAPDH are shown. a,b,cp<0.01 compared to NC, M/ aTNF and B/ aTNF, respectively. (D) Total protein collected from flexor digitorum muscles of normal control (NC), trained control with anti-TNF (TC/ aTNF), and 6 wk HRHF animals with or without anti-TNF (6HRHF/ aTNF and 6HRHF, respectively) was resolved by 10% SDS PAGE and probed with anti-CTGF antibody; GAPDH was used as a loading control. Experiments were repeated three times and bar graphs of the ratio of PLF/GAPDH are shown. (E) Western blot showing the single band for recombinant PLF made in E. coli, numbers 1–4 refer to the different clones examined for protein expression. Ps= Perisoteum; Ob= Osteoblast.
PLF expression is downstream of TNFα, MIP-2 and MIP 3a
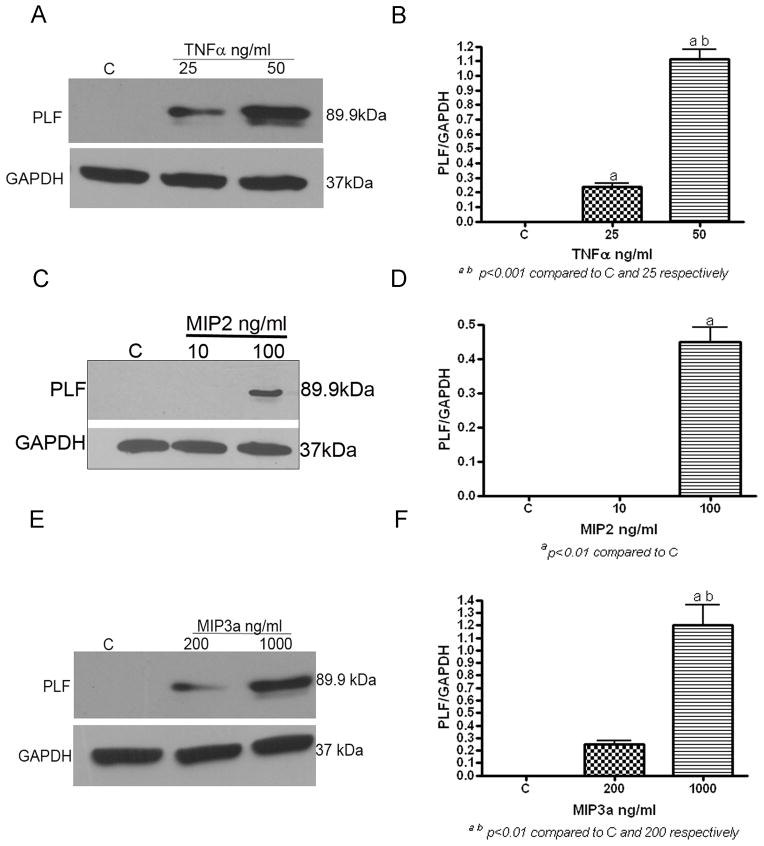
Since inflammatory cytokines and chemokines were increased by 6 weeks of HRHF task performance, as was PLF (Figs 1 & 3), and since treatment with anti-TNF reduced PLF levels (Fig 3), we examined whether PLF might be downstream of TNFα and MIP-2, and perhaps even MIP3a since there was a decrease (albeit insignificant) in MIP3a after anti-TNF treatment in TC rats. Therefore, we examined PLF expression by western blot analysis in vitro, in primary osteoblasts after treatment with TNFα, MIP-2 and MIP 3a. Treatment of primary osteoblasts with physiological doses of TNFα for 48 hours, resulted in dose dependent up regulation of PLF (Fig 4A & B, p<0.001). Similarly treatment with chemokines like MIP-2 and MIP3 also lead to a dose dependent up regulation of PLF in these cultures (Fig 4C & D, p<0.01; Fig 4E & F, p<0.001, respectively).
Figure 4. TNFα, MIP2 and MIP3a treatment increased PLF levels in primary osteoblasts culture in vitro.
A–F) Primary osteoblasts were serum starved prior to treatment with TNFα**,** MIP2 or MIP3a at varying doses as indicated, or were untreated (C), then total protein was collected for western blot analysis 48 hrs after treatment. Total protein was resolved on 10% SDS-PAGE and blots probed with anti-PLF; GAPDH was used a loading control (A, C & E). Experiments were repeated three times and bar graphs of the ratio of PLF/GAPDH are shown (B, D & F). ap<0.01 compared to C; bp<0.01 compared to lower dose cytokine treatment.
PLF promotes primary osteoblast proliferation
PLF increases in response to a HRHF task regimen, is localized in bone to osteoblasts and osteocytes, and has highest expression coincident with highest levels of a serum biomarker of bone formation (Fig 1 & 2, and (Rani et al., 2009b)). We have shown previously that adenoviral mediated over expression of PLF in vivo in rat femurs and in vitro in primary osteoblasts resulted in increased proliferation and differentiation of these cells (Zhu et al., 2009). These findings suggest that PLF acts as an anabolic factor in bone formation. Also, adenovirus mediated over expression of PLF results in elevated intracellular levels and secretion to the extracellular milieu (Litvin et al., 2006; Zhu et al., 2009), making it difficult to decipher whether PLF renders its effects as a cytoplasmic protein, as a secreted protein, or both. Thus, to dissect whether the effects of PLF on bone formation is a result of inside-out signaling as a cytoplasmic protein or a result of outside-in signaling as a secreted protein, we resorted to in vitro cell culture techniques. The role of PLF as a secreted protein was investigated using recombinant PLF, whereas its role as a cytoplasmic and secreted protein was examined by adenoviral over expression. Primary osteoblasts in culture were either treated with rPLF or with AdPLF, and various markers of proliferation and differentiation were assessed.
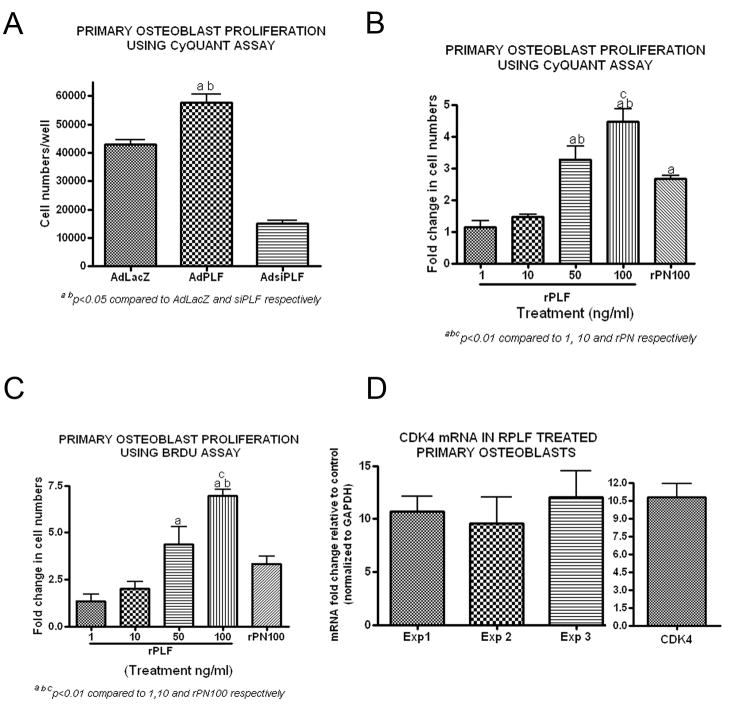
Using the CyQUANT method, primary osteoblasts infected with AdPLF showed increased proliferation compared to control cells infected with AdLacZ and decreased proliferation when PLF was knocked down using AdsiPLF (Fig 5A, p<0.05 and 0.001 respectively). Proliferation also increased significantly when primary osteoblasts were treated with rPLF (1–100ng/ml) (Fig 5B). This effect was dose dependent with increasing amounts of rPLF resulting in greater proliferation as evidenced by higher fold changes in cell numbers from untreated control levels. Recombinant periostin (PN; 100ng/ml) also enhanced proliferation, although at significantly lower levels than rPLF at the same dose (Fig. 5B, p<0.01). These findings were confirmed using a BrdU incorporation assay for assessment for proliferation (Fig. 5C, p<0.01). Effects of rPLF on proliferation were also assessed by examining mRNA levels of Cyclin Dependent Kinase 4 (CDK4), a master regulator of the cell cycle. Treatment of primary osteoblasts with rPLF (100ng/ml) resulted in a tenfold increase in CDK4 mRNA levels compared to controls (Fig 5D).
Figure 5. PLF promotes primary osteoblast proliferation and increases CDK4 mRNA levels in primary osteoblast in vitro.
A) Primary rat osteoblasts were infected with AdLacZ (expression of β galactosidase by adenovirus, control), AdPLF (over-expression of PLF by adenovirus), or AdsiPLF (adenoviral expression of small interfering RNA directed towards PLF, Knockdown) at a moi of 50. 72 h post-infection, proliferation was assessed by CyQUANT Cell Proliferation Assay Kit. Cell numbers were based on the fluorescence output (emission wavelength 520 nm). ap<0.05 compared to AdLacZ; bp<0.001 compared to AdsiPLF. B) Primary rat osteoblasts were treated with recombinant PLF (rPLF, 1–100ng/ml), recombinant Periostin (rPN, 100ng/ml), or were untreated. 48 h post-treatment, proliferation was assessed by CyQUANT Cell Proliferation Assay Kit. Cell numbers were based on the fluorescence output (emission wavelength 520 nm). Bar graphs represent fold change over untreated controls. a,b,cp<0.01 compared to 1 and 10 ng/ml rPLF, and rPN100, respectively. C) Primary rat osteoblasts were treated with recombinant PLF (rPLF, 1–100ng/ml), recombinant Periostin (rPN, 100ng/ml) or were untreated. 24 h post-treatment, BrdU was added to the cell cultures for additional 18 h and proliferation was assessed by ELISA performed on cell lysates and absorbance read at 495⌊. Bar graphs represent fold change over untreated control. a,b,cp<0.01 compared to 1 and 10 ng/ml rPLF, and rPN100, respectively. D) Primary rat osteoblasts were treated with recombinant PLF (100ng/ml) or were untreated. 24 h post-treatment, total RNA was collected and reverse transcribed to cDNA. Reverse Transcriptase quantitative Polymerase Chain Reaction (RT-qPCR) was used to assess the changes in CDK4 mRNA level between untreated controls and rPLF treated cultures. Experiments were repeated 3 times and values are expressed as fold change over control. Data was normalized using GAPDH mRNA levels. Right panel: Representative bar graph for pooled values from experiments 1–3.
PLF promotes adhesion
Adhesion is of fundamental importance to regulating the cell cycle, cell survival and cell differentiation. Periostin and βig-H3, members of the fasciclin family of proteins, mediate their effects on cell spreading, adhesion, migration and proliferation through interaction with integrins (Gillan et al., 2002; Horiuchi et al., 1999; Li et al., 2009; Thapa et al., 2005). We have also observed that PLF, a member of the same family of proteins, interacts with β1 integrin in cardiac cells (Litvin et al., 2005). To determine whether an interaction of PLF with β1 affects adhesion of osteoblasts, which in turn would regulate osteoblast proliferation and differentiation in vitro, we assessed adhesion of MC3T3 osteoblast-like cells on plates coated with poly-L-lysine (PL; used as a negative control), fibronectin (FN; used as a positive control), media conditioned with GFP (adenoviral control) or PLF. PLF promoted adhesion of MC3T3 cells but the process was significantly abrogated when cells were treated with anti-β1 integrin prior to plating (Fig. 6A, p<0.001).
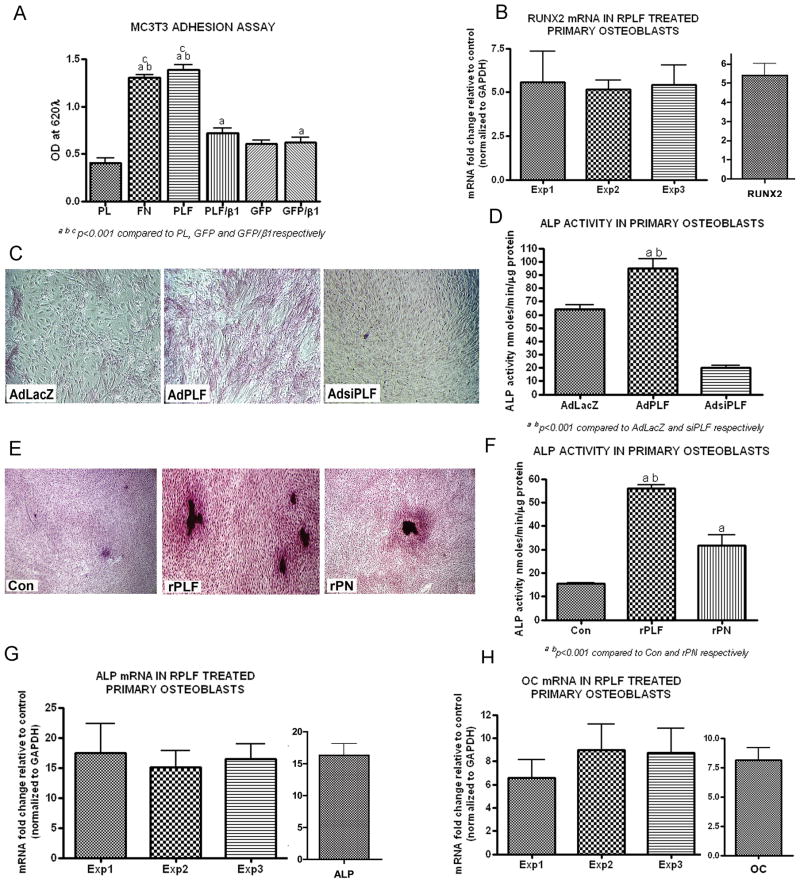
Figure 6. PLF promotes MC3T3-E1 osteoblast-like cell adhesion and differentiation of primary osteoblast cells in vitro.
A) MC3T3-E1 osteoblast-like cells were either left untreated or treated with anti-β1 integrin and plated in 96 well dishes coated with Poly-L-Lysine (PL; 0.10mg/ml), Fibronectin (FN; 5μg/ml), Periostin-like-factor conditioned media (PLF; 200μl/well) or green fluorescent protein conditioned media (GFP; 200μl/well) for 2 h at 37°C. Cells were fixed with 4% paraformaldehyde in PO4 buffer and stained with 1% methylene blue. Absorbance was read at 620⌊ on an ELISA plate reader. a,b,cp<0.01 compared to PL, GFP and GFP/β1, respectively. B) Primary rat osteoblasts were treated with recombinant PLF (100ng/ml), or were untreated. 48 h post-treatment, total RNA was collected and reverse transcribed to cDNA. Reverse Transcriptase quantitative Polymerase Chain Reaction (RT-qPCR) was used to assess the changes in RUNX2 mRNA levels between untreated controls and rPLF treated cultures. Experiments were repeated 3 times and values expressed as fold change over control data. Data was normalized using GAPDH mRNA levels. Right panel: Representative bar graph for pooled values from experiments 1–3. C) Primary rat osteoblasts were infected on day 2 of culture with AdLacZ (expression of β-galactosidase by adenovirus, control), AdPLF (over-expression of PLF by adenovirus), or AdsiPLF (adenoviral expression of siRNA directed towards PLF, knockdown) at a moi of 50. Cells were stained on day 7 for alkaline phosphatase (ALP); positive staining was red. D) ALP activity was assessed on parallel cultures, using p-nitrophenol as the substrate. a,bp<0.001 compared to AdLacZ and AdsiPLF, respectively. E) Primary rat osteoblasts were either untreated (Con) or treated with recombinant PLF (rPLF, 100ng/ml) or recombinant Periostin (rPN, 100ng/ml). Cells were stained on day 7 for ALP. F) ALP activity was assessed on parallel cultures. a,bp<0.001 and rPN, respectively. G) Rat primary osteoblasts were treated with recombinant PLF (100ng/ml), or were untreated. 48 h post-treatment, total RNA was collected and reverse transcribed to cDNA. Real Time Polymerase Chain Reaction (RT-PCR) was used to assess the changes in ALP mRNA levels between untreated controls and recombinant PLF treated cultures. Experiments were repeated 3 times and values expressed as fold change over controls. Data was normalized using GAPDH mRNA levels. Right panel: Representative bar graph for pooled values from experiments 1–3. H) Rat primary osteoblasts were treated with recombinant PLF (100ng/ml), or were untreated. 48 h post-treatment, total RNA was collected and reverse transcribed to cDNA. Real Time Polymerase Chain Reaction (RT-PCR) was used to assess the changes in osteocalcin (OC) mRNA level between untreated controls and recombinant PLF treated cultures. Experiments were repeated 3 times and values expressed as fold change over control. Data was normalized using GAPDH mRNA levels. Right panel: Representative bar graph for pooled values from experiments 1–3.
PLF promotes osteoblast differentiation
Osteoblast differentiation is a multi-step event, marked by the expression of different genes at different phases of the process. It is well established that osteoblast differentiation is divided into three stages and that these stages are a continuum that occur over a 21 to 28 day period in vitro. The stages are as follows: osteoblasts proliferate from the time of plating up to approximately day 7. Once they reach confluence they begin matrix production, which continues until approximately day 14 at which time the matrix becomes mineralized (days 14 and 28) (Owen et al., 1990). Several early, intermediate or late markers of differentiation can distinguish these stages. In this study, the effect of rPLF on Runt related transcription factor (Runx2/cbfa1; an early marker of osteoblast commitment and differentiation) was assessed on day 4; alkaline phosphatase, an intermediate marker, was assessed on day 7; while osteocalcin, von Kossa staining and calcium deposition, each late markers of differentiation, were evaluated on day 21.
We found that Runx2 mRNA levels, determined by quantitative RT-PCR, were 5 fold higher in osteoblasts cultured with rPLF than controls (Fig 6B). Adenoviral over expression of PLF promoted alkaline phosphatase staining and activity in primary osteoblasts compared to AdLacZ infected controls (Fig. 6C & D, p<0.001 respectively). However, adenoviral mediated silencing of PLF significantly reduced both alkaline phosphatase staining and activity (Fig 6C & D). In response to treatment with rPLF or rPN, alkaline phosphatase staining and activity increased significantly in primary osteoblasts compared to untreated controls (Fig 6E & F). However, the intensity of ALP staining and activity were considerably higher with rPLF than with rPN (Fig 6E & F, p<0.001). The results obtained by ALP staining and activity measurement were confirmed by RT- qPCR. Primary osteoblasts treated with rPLF showed a fifteen-fold increase in ALP mRNA levels from control levels (Fig 6G). Additionally, the late differentiation marker osteocalcin (OC) mRNA was assessed by RT- qPCR and demonstrated a sevenfold increase in rPLF treated cultures from control levels (Fig 6H).
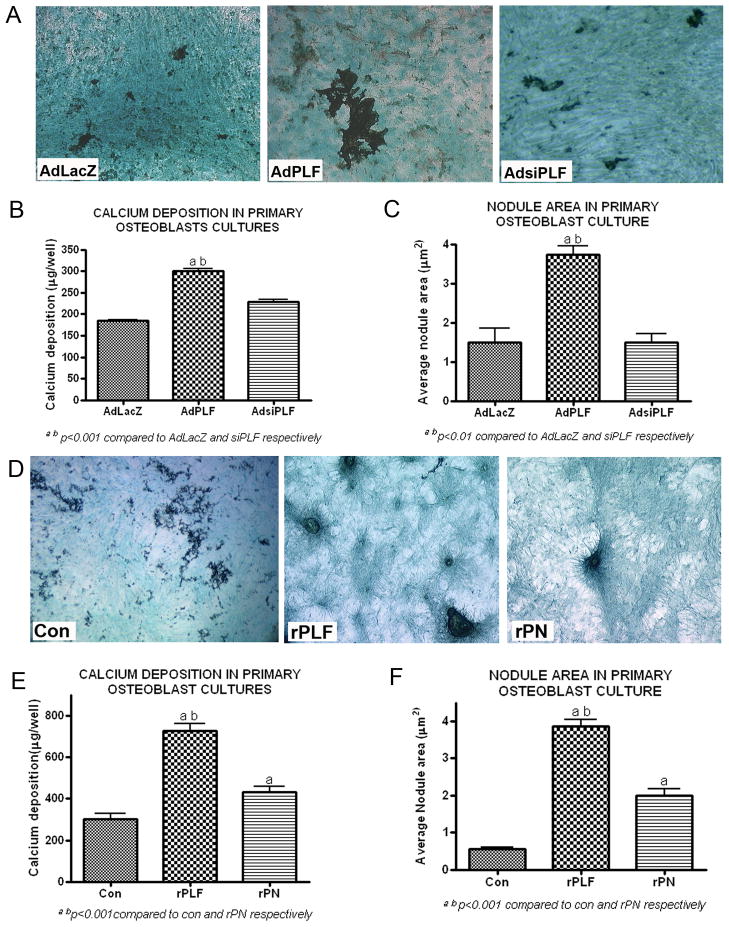
Primary osteoblast cultures infected with AdPLF showed higher amount of mineral deposition when stained with von Kossa compared to AdLacZ and AdsiPLF infected cells (Fig 7A). Calcium deposition and average nodule size were also markedly higher in AdPLF infected cultures compared to controls (Fig 7B & C, p<0.001 and 0.01, respectively). Similarly, primary osteoblasts treated with rPLF demonstrated higher amounts of mineral deposition than controls as assessed with von Kossa stain, higher calcium deposition, and increased nodule surface area (Fig 7D, E and F, respectively; p<0.001 each). In accordance with our previous data, treatment of osteoblasts with rPN resulted in an increase in von Kossa stain, calcium deposition and nodule surface area compared to controls, although not as high as with rPLF (Fig 7D, E and F, respectively; p<0.001 each).
Figure 7. PLF over-expression or recombinant PLF enhanced calcium deposition and mineralization in rat primary osteoblasts on day 21 of culture.
A–C) Primary rat osteoblasts were infected with AdLacZ (control), AdPLF (over-expression) or AdsiPLF (knockdown; siRNA against PLF) at a moi of 50. (A) Cells were stained with von Kossa for mineral deposition and counterstained with light green. (B) Bar graph of amount of calcium deposited per well. (C) Bar graph of average nodule area. a,bp<0.001 compared to AdLacZ and AdsiPLF, respectively. D–F) Rat primary osteoblasts were treated with recombinant PLF (100ng/ml), recombinant Perisotin (rPN-100ng/ml) or were untreated. (D) Cells were stained with von Kossa and light green. (E) Bar graph of amount of calcium deposited per well. (F) Bar graph of average nodule area. a,bp<0.001 and rPN, respectively.
DISCUSSION
We show here that performance of a HRHF task for 6 weeks results in moderate serum and musculoskeletal inflammatory responses that coincide with moderate increases of a bone formation marker, osteocalcin, and increased PLF, a matricellular protein that we have previously identified as a possible marker of adaptive bone remodeling. We also report here that use of a TNFα antibody in vivo attenuated not only the increased serum and musculoskeletal TNFα, but also serum IL-1α and MIP2a, as well as inflammation-induced nociceptor related behaviors in the 6 week HRHF animals.
Our observed increase of serum TNFα in 6-week HRHF animals was similar in level (Mean 79.53 ± SEM 21.72) as that seen in past studies from our laboratory examining the effects of performing lower demand tasks, e.g. as a high repetition low force task for 6 weeks (Mean 94.04 ± SEM 36.53) or a moderate repetition high force task for 6 weeks (Mean 75.36 ± SEM 12.55) (Barbe et al., 2008; Elliott et al., 2009b). However, it is important to note that levels of serum TNFa in 6-week HRHF rats in this study were 7 fold less than in 12-week HRHF rats (Mean 612 + SEM 225) that were not part of this study since the focus of this study was anabolic tissue changes rather than catabolic tissue changes known to be induced by high levels of TNFa. Our observed increase in IL-1α in HRHF rats was an expected result since IL-1α is known to be up regulated by TNFα in several cell types (Le et al., 1987). TNFα is also known to induce its effects on many downstream genes via the NF-κB pathway, which in turn leads to expression of multiple genes. NF-κB-regulated genes encode proteins that play important roles in inflammation and immune responses. These proteins include proinflammatory cytokines, such as IFN-β, IL-2, and IL-6 (Baldwin, 1996; Drouet et al., 1991; Zhang et al., 1995); and chemokines, such as monocyte chemoattractant protein-1, IL-8, RANTES, and macrophage-inflammatory protein (MIP) 3 and 1α- (Genin et al., 2000; Grove and Plumb, 1993; Stein and Baldwin, 1993; Sugita et al., 2002; Ueda et al., 1997). In this study, we did not observe any changes in serum MIP3a levels in response to anti-TNF treatment, although MIP3a is a downstream gene of TNFα, as shown in Hela cells (Sugita et al., 2002). We have previously observed increased MIP3a after performance of a lower demand high repetition negligible force task (Barbe et al., 2008) but not after performance of a moderate repetition high force task (Elliott et al., 2009b). The reason behind differences in MIP3a induction in our studies is unknown. Perhaps we missed the increase in MIP3a due to our sampling of serum only in week 6 in this study. In contrast, although MIP2 serum levels have not been reported to be affected by TNFα levels, MIP2 levels not only increased temporally with TNFα, but anti-TNF treatment down-regulated MIP2 levels in 6 week HRHF animals. Anti-TNF treatment also effectively blocked task-induced increases of TNFα levels in serum and bone. However, task-induced increases of TNFα in muscle, though reduced, were not down regulated significantly by anti-TNF treatment, perhaps due to the short administration of the anti-TNF drug.
It is well known that TNFα negatively regulates bone formation by increasing osteoclast formation, differentiation and activity and by decreasing osteoblast function (Kaneki et al., 2006; Zhang et al., 2001), therefore one would expect to see increased bone formation when TNF signaling is blocked. In our model we observed that serum osteocalcin, a biomarker of bone formation, was not significantly affected by anti-TNF treatment. Although, this may be due to short duration of anti-TNF administration, the significant increases in trained control animals are more indicative of a training induced increase in bone formation.
Grip strength and cutaneous mechanical sensation are behavioral indicators of underlying tissue inflammation and/or injury. Decreases in grip strength can be indicative of discomfort (and has been termed an indicator of movement-related hyperalgesia), nerve or muscle injury, or motor dysfunction resulting from performance of the repetitive reaching task. Studies by Kehl and colleagues (Kehl and Fairbanks, 2003; Kehl et al., 2003; Kehl et al., 2000) show that direct injection of inflammatory cytokines into muscles results in decreased grip strength. Our results support this finding in that we observed that grip strength was rescued by anti-TNF treatment. We also used graded Von Frey monofilaments to test paw withdrawal thresholds in response to palmar stimulation, a sensory test that can be used as an indicator of both underlying tissue inflammation and nerve injury (Clark et al, 2004; Elliott et al, 2009). In this report, forepaw mechanical hypersensitivity was evident in not only task rats but also trained control rats indicating that inflammatory responses had already been elicited in palmar tissues and/or in the median nerve during the initial 5-week training period of 10 min/day, 5 days/week. The anti-TNF treatment during the last 2 weeks of HRHF task performance did not attenuate this hypersensitivity in either the trained control or the HRHF groups. This was actually not unexpected since TNFα plays a role in the development of mechanical hypersensitivity and its exogenous application to nerves induces this pain behavior (Igarashi et al., 2000). Also, anti-TNFα treatment reduces mechanical hypersensitivity only if administered early before central nervous system sensitization but not after (Murata et al., 2005; Sasaki et al., 2007). Previous studies from our lab showed that central nervous system changes in the form of increased nociceptive neurotransmitters and receptors in the spinal cord are present by week 6 of performance of moderate demand tasks (Elliott et al., 2009a; Elliott et al., 2009b; Elliott et al., 2008) and may also have occurred before week 6 with performance of this higher demand task. Studies are underway in our lab to examine the onset of this central sensitization in rats performing the HRHF task, and the effectiveness of various interventions in preventing this change.
When the levels of PLF in bone and muscle of animals were examined after anti-TNF administration, a significant decrease was observed by both immuhohistochemistry and western blot analysis. This finding suggests that PLF may be downstream of TNFα. Interestingly, the levels of CTGF, another matricellular protein, did not change in response to after anti-TNF, indicating that abrogating TNFα signaling does not affect the expression of all matricellular proteins universally. Our in vitro results confirmed that treatment with specific inflammatory cytokines and chemokines induce PLF in a dose dependent manner. This again suggests that PLF may be downstream of the inflammatory signaling cascade.
We have previously shown in both in vitro and in vivo studies that PLF acts as an anabolic molecule in bone formation (Zhu et al., 2009). In addition, in our repetitive loading model, PLF was associated temporally with adaptive bone remodeling in this and in a previous study (Rani et al., 2009b). Together, these findings indicate that PLF plays an important role in bone formation and remodeling in vivo. We also investigated the role of PLF in primary osteoblasts in vitro using adenoviral and recombinant PLF. Specifically, we investigated the role of PLF in osteoblast proliferation, adhesion and differentiation, since in vivo bone formation is a result of each of these processes. PLF over expression mediated by adenovirus, which leads to the production of PLF intracellularly before its secretion into the media, resulted in increased proliferation of primary osteoblasts, while adenoviral knockdown of PLF abrogated the proliferation process. Similar results were observed when cells were treated with recombinant PLF. These findings suggest that PLF mediated proliferation of cells is accomplished both as an intracellular molecule which signals inside-out of the cell, as well as a matricellular protein that signals outside-in.
Adhesion is another event that is required for differentiation of cells. PLF also lead to increased adhesion of MC3T3-E1 osteoblast like cells, via the β1 integrin receptor. Typically integrin ligand interactions occur via RGD domain. However, in the case of the fasciclin family of proteins, this interaction is mediated via the YH motif (a non RGD integrin binding domain). Since no RGD sequences are detected on PLF by sequence analysis, the PLF-integrin interaction also takes place most likely via the YH motif. PLF is known to interact with β1 integrin in cardiac cells and in primary osteoblasts (Litvin et al., 2006). Additionally, other members of this family including periostin and βIG- H3 also interact with different integrins to mediate adhesion and differentiation (Gillan et al., 2002; Horiuchi et al., 1999; Li et al., 2009; Thapa et al., 2005). In accordance with all these studies, we confirmed that adhesion of MC3T3 E1 osteoblast like cells is also mediated by β1 integrin since treating the cells with a specific antibody to β1 integrin abrogated the process of adhesion.
The differentiation of osteoblasts is a multistep process with involvement of numerous genes, some of which like Runx2, alkaline phosphatase and osteocalcin are so stage specific that they have been used as markers for a particular stage of differentiation. In this study we examined these three differentiation markers, both after adenoviral over expression of PLF and after treatment of cells with recombinant PLF. Similar to our observation with proliferation, PLF seemed to mediate the differentiation process both as an intracellular protein as well as an extracellular protein.
In conclusion, these findings suggest that an adaptive remodeling response occurs early with repetitive bone loading and that TNFα may be one of the inducing proteins. It is important to note though that TNFα is a potent inducer of bone resorption, can be cytotoxic at high levels, and, as shown in this study, plays a role in the development of one or more nociceptor-related behaviors which can be difficult to attenuate. Therefore, careful control of TNFα levels is probably needed for an appropriate balance of anabolic and catabolic tissue metabolism. Our current findings also indicate that PLF is downstream of the inflammation process and acts as an anabolic protein in bone formation processes present at 6 weeks of task performance. However, we have also shown that PLF levels decrease in bones with continued task performance (Rani et al, 2009a) to 12 weeks, the same time frame as a 7 fold increase in serum TNFalpha, Although we have more to investigate concerning the catabolic effects of chronic inflammation and performance of high demand repetitive tasks on bone, our current results suggest that moderate levels of inflammatory cytokines might be beneficial to the anabolic stage of bone loading. These results also suggest that PLF administration or up-regulation might be a useful means of achieving bone formation and/or adaptive bone remodeling.
References
- Abdelmagid SM, Barbe MF, Arango-Hisijara I, Owen TA, Popoff SN, Safadi FF. Osteoactivin acts as downstream mediator of BMP-2 effects on osteoblast function. J Cell Physiol. 2007;210:26–37. doi: 10.1002/jcp.20841. [DOI] [PubMed] [Google Scholar]
- Al-Shatti T, Barr AE, Safadi FF, Amin M, Barbe MF. Increase in inflammatory cytokines in median nerves in a rat model of repetitive motion injury. J Neuroimmunol. 2005;167:13–22. doi: 10.1016/j.jneuroim.2005.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott JA, Nuglozeh E, Rico MC, Arango-Hisijara I, Odgren PR, Safadi FF, Popoff SN. Connective tissue growth factor (CTGF/CCN2) is a downstream mediator for TGF-beta1-induced extracellular matrix production in osteoblasts. J Cell Physiol. 2007;210:843–52. doi: 10.1002/jcp.20917. [DOI] [PubMed] [Google Scholar]
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–83. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5:329–39. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- Barbe MF, Barr AE, Gorzelany I, Amin M, Gaughan JP, Safadi FF. Chronic repetitive reaching and graspingresults in decreased motor performance and widespread tissue responses in a rat model of MSD. J Orthop Res. 2003;21:167–76. doi: 10.1016/S0736-0266(02)00086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe MF, Elliott MB, Abdelmagid SM, Amin M, Popoff SN, Safadi FF, Barr AE. Serum and tissue cytokines and chemokines increase with repetitive upper extremity tasks. J Orthop Res. 2008;26:1320–6. doi: 10.1002/jor.20674. [DOI] [PubMed] [Google Scholar]
- Barr AE, Safadi FF, Gorzelany I, Amin M, Popoff SN, Barbe MF. Repetitive, negligible force reaching in rats induces pathological overloading of upper extremity bones. J Bone Miner Res. 2003;18:2023–32. doi: 10.1359/jbmr.2003.18.11.2023. [DOI] [PubMed] [Google Scholar]
- Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- Bornstein P. Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J Cell Biol. 1995;130:503–6. doi: 10.1083/jcb.130.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P. Matricellular proteins: an overview. J Cell Commun Signal. 2009 doi: 10.1007/s12079-009-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–16. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- Brigstock DR. Connective tissue growth factor (CCN2, CTGF) and organ fibrosis: lessons from transgenic animals. J Cell Commun Signal. 2009 doi: 10.1007/s12079-009-0071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–83. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BD, Al-Shatti TA, Barr AE, Amin M, Barbe MF. Performance of a high-repetition, high-force task induces carpal tunnel syndrome in rats. J Orthop Sports Phys Ther. 2004;34:244–53. doi: 10.2519/jospt.2004.34.5.244. [DOI] [PubMed] [Google Scholar]
- Clark BD, Barr AE, Safadi FF, Beitman L, Al-Shatti T, Amin M, Gaughan JP, Barbe MF. Median nerve trauma in a rat model of work-related musculoskeletal disorder. J Neurotrauma. 2003;20:681–95. doi: 10.1089/089771503322144590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter P, Leoni P, Abraham D. Connective tissue growth factor: structure-function relationships of a mosaic, multifunctional protein. Growth Factors. 2008;26:80–91. doi: 10.1080/08977190802025602. [DOI] [PubMed] [Google Scholar]
- Drouet C, Shakhov AN, Jongeneel CV. Enhancers and transcription factors controlling the inducibility of the tumor necrosis factor-alpha promoter in primary macrophages. J Immunol. 1991;147:1694–700. [PubMed] [Google Scholar]
- Elliott MB, Barr AE, Barbe MF. Spinal substance P and neurokinin-1 increase with high repetition reaching. Neurosci Lett. 2009a;454:33–7. doi: 10.1016/j.neulet.2009.01.037. [DOI] [PubMed] [Google Scholar]
- Elliott MB, Barr AE, Clark BD, Amin M, Amin S, Barbe MF. High force reaching task induces widespread inflammation, increased spinal cord neurochemicals and neuropathic pain. Neuroscience. 2009b;158:922–31. doi: 10.1016/j.neuroscience.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MB, Barr AE, Kietrys DM, Al-Shatti T, Amin M, Barbe MF. Peripheral neuritis and increased spinal cord neurochemicals are induced in a model of repetitive motion injury with low force and repetition exposure. Brain Res. 2008;1218:103–13. doi: 10.1016/j.brainres.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorczyk JM, Barr AE, Rani S, Gao HG, Amin M, Amin S, Litvin J, Barbe MF. Exposure-dependent increases in IL-1beta, substance P, CTGF, and tendinosis in flexor digitorum tendons with upper extremity repetitive strain injury. J Orthop Res. 2009 doi: 10.1002/jor.20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-kappa B and IFN-regulatory factor transcription factors. J Immunol. 2000;164:5352–61. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–64. [PubMed] [Google Scholar]
- Goetsch SC, Hawke TJ, Gallardo TD, Richardson JA, Garry DJ. Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol Genomics. 2003;14:261–71. doi: 10.1152/physiolgenomics.00056.2003. [DOI] [PubMed] [Google Scholar]
- Grove M, Plumb M. C/EBP, NF-kappa B, and c-Ets family members and transcriptional regulation of the cell-specific and inducible macrophage inflammatory protein 1 alpha immediate-early gene. Mol Cell Biol. 1993;13:5276–89. doi: 10.1128/mcb.13.9.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Yamashita M, Zhang Q, Zhou Q, Chen D, Reynolds DG, Awad HA, Yanoso L, Zhao L, Schwarz EM, Zhang YE, Boyce BF, Xing L. Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins. J Biol Chem. 2008;283:23084–92. doi: 10.1074/jbc.M709848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF, Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–49. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- Horowitz MC, Xi Y, Wilson K, Kacena MA. Control of osteoclastogenesis and bone resorption by members of the TNF family of receptors and ligands. Cytokine Growth Factor Rev. 2001;12:9–18. doi: 10.1016/s1359-6101(00)00030-7. [DOI] [PubMed] [Google Scholar]
- Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96:3540–5. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Drissi MH, O’Keefe RJ, Schwarz EM. A rapid multiparameter approach to study factors that regulate osteoclastogenesis: demonstration of the combinatorial dominant effects of TNF-alpha and TGF-ss in RANKL-mediated osteoclastogenesis. Calcif Tissue Int. 2003;73:584–93. doi: 10.1007/s00223-003-0059-8. [DOI] [PubMed] [Google Scholar]
- Igarashi A, Nashiro K, Kikuchi K, Sato S, Ihn H, Fujimoto M, Grotendorst GR, Takehara K. Connective tissue growth factor gene expression in tissue sections from localized scleroderma, keloid, and other fibrotic skin disorders. J Invest Dermatol. 1996;106:729–33. doi: 10.1111/1523-1747.ep12345771. [DOI] [PubMed] [Google Scholar]
- Igarashi A, Okochi H, Bradham DM, Grotendorst GR. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell. 1993;4:637–45. doi: 10.1091/mbc.4.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Kikuchi S, Shubayev V, Myers RR. 2000 Volvo Award winner in basic science studies: Exogenous tumor necrosis factor-alpha mimics nucleus pulposus-induced neuropathology. Molecular, histologic, and behavioral comparisons in rats. Spine (Phila Pa 1976) 2000;25:2975–80. doi: 10.1097/00007632-200012010-00003. [DOI] [PubMed] [Google Scholar]
- Jugdutt BI, Palaniyappan A, Uwiera RR, Idikio H. Role of healing-specific-matricellular proteins and matrix metalloproteinases in age-related enhanced early remodeling after reperfused STEMI in dogs. Mol Cell Biochem. 2009;322:25–36. doi: 10.1007/s11010-008-9936-9. [DOI] [PubMed] [Google Scholar]
- Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, Boyce BF, Xing L. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281:4326–33. doi: 10.1074/jbc.M509430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A, Satoh K, Masamune A, Hirota M, Kimura K, Umino J, Hamada S, Satoh A, Egawa S, Motoi F, Unno M, Shimosegawa T. Periostin, secreted from stromal cells, has biphasic effect on cell migration and correlates with the epithelial to mesenchymal transition of human pancreatic cancer cells. Int J Cancer. 2008;122:2707–18. doi: 10.1002/ijc.23332. [DOI] [PubMed] [Google Scholar]
- Kehl LJ, Fairbanks CA. Experimental animal models of muscle pain and analgesia. Exerc Sport Sci Rev. 2003;31:188–94. doi: 10.1097/00003677-200310000-00006. [DOI] [PubMed] [Google Scholar]
- Kehl LJ, Hamamoto DT, Wacnik PW, Croft DL, Norsted BD, Wilcox GL, Simone DA. A cannabinoid agonist differentially attenuates deep tissue hyperalgesia in animal models of cancer and inflammatory muscle pain. Pain. 2003;103:175–86. doi: 10.1016/s0304-3959(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Kehl LJ, Trempe TM, Hargreaves KM. A new animal model for assessing mechanisms and management of muscle hyperalgesia. Pain. 2000;85:333–43. doi: 10.1016/S0304-3959(99)00282-1. [DOI] [PubMed] [Google Scholar]
- Kudo Y, Ogawa I, Kitajima S, Kitagawa M, Kawai H, Gaffney PM, Miyauchi M, Takata T. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res. 2006;66:6928–35. doi: 10.1158/0008-5472.CAN-05-4540. [DOI] [PubMed] [Google Scholar]
- Kudo Y, Siriwardena BS, Hatano H, Ogawa I, Takata T. Periostin: novel diagnostic and therapeutic target for cancer. Histol Histopathol. 2007;22:1167–74. doi: 10.14670/HH-22.1167. [DOI] [PubMed] [Google Scholar]
- Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–9. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- Le JM, Weinstein D, Gubler U, Vilcek J. Induction of membrane-associated interleukin 1 by tumor necrosis factor in human fibroblasts. J Immunol. 1987;138:2137–42. [PubMed] [Google Scholar]
- Li G, Jin R, Norris RA, Zhang L, Yu S, Wu F, Markwald RR, Nanda A, Conway SJ, Smyth SS, Granger DN. Periostin mediates vascular smooth muscle cell migration through the integrins alphanubeta3 and alphanubeta5 and focal adhesion kinase (FAK) pathway. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie-Venema H, Eralp I, Markwald RR, van den Akker NM, Wijffels MC, Kolditz DP, van der Laarse A, Schalij MJ, Poelmann RE, Bogers AJ, Gittenberger-de Groot AC. Periostin expression by epicardium-derived cells is involved in the development of the atrioventricular valves and fibrous heart skeleton. Differentiation. 2008;76:809–19. doi: 10.1111/j.1432-0436.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- Lindner V, Wang Q, Conley BA, Friesel RE, Vary CP. Vascular injury induces expression of periostin: implications for vascular cell differentiation and migration. Arterioscler Thromb Vasc Biol. 2005;25:77–83. doi: 10.1161/01.ATV.0000149141.81230.c6. [DOI] [PubMed] [Google Scholar]
- Litvin J, Blagg A, Mu A, Matiwala S, Montgomery M, Berretta R, Houser S, Margulies K. Periostin and periostin-like factor in the human heart: possible therapeutic targets. Cardiovasc Pathol. 2006;15:24–32. doi: 10.1016/j.carpath.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Litvin J, Selim AH, Montgomery MO, Lehmann K, Rico MC, Devlin H, Bednarik DP, Safadi FF. Expression and function of periostin-isoforms in bone. J Cell Biochem. 2004;92:1044–61. doi: 10.1002/jcb.20115. [DOI] [PubMed] [Google Scholar]
- Litvin J, Zhu S, Norris R, Markwald R. Periostin family of proteins: therapeutic targets for heart disease. Anat Rec A Discov Mol Cell Evol Biol. 2005;287:1205–12. doi: 10.1002/ar.a.20237. [DOI] [PubMed] [Google Scholar]
- Ma T, Miyanishi K, Suen A, Epstein NJ, Tomita T, Smith RL, Goodman SB. Human interleukin-1-induced murine osteoclastogenesis is dependent on RANKL, but independent of TNF-alpha. Cytokine. 2004;26:138–44. doi: 10.1016/j.cyto.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Minamitani C, Tokuda H, Adachi S, Matsushima-Nishiwaki R, Yamauchi J, Kato K, Natsume H, Mizutani J, Kozawa O, Otsuka T. p70 S6 kinase limits tumor necrosis factor-alpha-induced interleukin-6 synthesis in osteoblast-like cells. Mol Cell Endocrinol. 2009 doi: 10.1016/j.mce.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Murata Y, Olmarker K, Takahashi I, Takahashi K, Rydevik B. Effects of selective tumor necrosis factor-alpha inhibition to pain-behavioral changes caused by nucleus pulposus-induced damage to the spinal nerve in rats. Neurosci Lett. 2005;382:148–52. doi: 10.1016/j.neulet.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Nakajima A, Seki N, Okawa A, Kato M, Moriya H, Amizuka N, Einhorn TA, Yamazaki M. Gene expression of periostin in the early stage of fracture healing detected by cDNA microarray analysis. J Orthop Res. 2004;22:520–5. doi: 10.1016/j.orthres.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Norris RA, Borg TK, Butcher JT, Baudino TA, Banerjee I, Markwald RR. Neonataland adult cardiovascular pathophysiological remodeling and repair: developmental role of periostin. Ann N Y Acad Sci. 2008;1123:30–40. doi: 10.1196/annals.1420.005. [DOI] [PubMed] [Google Scholar]
- Ochi H, Hara Y, Tagawa M, Shinomiya K, Asou Y. The roles of TNFR1 in lipopolysaccharide-induced bone loss: Dual effects of TNFR1 on bone metabolism via osteoclastogenesis and osteoblast survival. J Orthop Res. 2009 doi: 10.1002/jor.21028. [DOI] [PubMed] [Google Scholar]
- Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–30. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- Puglisi F, Puppin C, Pegolo E, Andreetta C, Pascoletti G, D’Aurizio F, Pandolfi M, Fasola G, Piga A, Damante G, Di Loreto C. Expression of periostin in human breast cancer. J Clin Pathol. 2008;61:494–8. doi: 10.1136/jcp.2007.052506. [DOI] [PubMed] [Google Scholar]
- Rani S, Barbe MF, Barr AE, Litvin J. Induction of periostin-like factor and periostin in forearm muscle, tendon, and nerve in an animal model of work-related musculoskeletal disorder. J Histochem Cytochem. 2009a;57:1061–73. doi: 10.1369/jhc.2009.954081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S, Barbe MF, Barr AE, Litvin J. Periostin-like-factor and Periostin in an animal model of work-related musculoskeletal disorder. Bone. 2009b;44:502–12. doi: 10.1016/j.bone.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DJ, Haddadin RI, Kang MH, Oh DJ. Matricellular proteins in the trabecular meshwork. Exp Eye Res. 2009;88:694–703. doi: 10.1016/j.exer.2008.11.032. [DOI] [PubMed] [Google Scholar]
- Ritchlin CT, Haas-Smith SA, Li P, Hicks DG, Schwarz EM. Mechanisms of TNF-alpha-and RANKL-mediated osteoclastogenesis and bone resorption in psoriatic arthritis. J Clin Invest. 2003;111:821–31. doi: 10.1172/JCI16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Yu CY, Dai M, Tam C, Loda M, Auclair D, Chen LB, Elias A. Elevated serum periostin levels in patients with bone metastases from breast but not lung cancer. Breast Cancer Res Treat. 2003;77:245–52. doi: 10.1023/a:1021899904332. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Kikuchi S, Konno S, Sekiguchi M, Watanabe K. Anti-TNF-alpha antibody reduces pain-behavioral changes induced by epidural application of nucleus pulposus in a rat model depending on the timing of administration. Spine (Phila Pa 1976) 2007;32:413–6. doi: 10.1097/01.brs.0000255097.18246.bc. [DOI] [PubMed] [Google Scholar]
- Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev. 2008;19:133–44. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Stanton LW, Garrard LJ, Damm D, Garrick BL, Lam A, Kapoun AM, Zheng Q, Protter AA, Schreiner GF, White RT. Altered patterns of gene expression in response to myocardial infarction. Circ Res. 2000;86:939–45. doi: 10.1161/01.res.86.9.939. [DOI] [PubMed] [Google Scholar]
- Stein B, Baldwin AS., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol Cell Biol. 1993;13:7191–8. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Kohno T, Yamamoto K, Imaizumi Y, Nakajima H, Ishimaru T, Matsuyama T. Induction of macrophage-inflammatory protein-3alpha gene expression by TNF-dependent NF-kappaB activation. J Immunol. 2002;168:5621–8. doi: 10.4049/jimmunol.168.11.5621. [DOI] [PubMed] [Google Scholar]
- Takanami I, Abiko T, Koizumi S. Expression of periostin in patients with non-small cell lung cancer: correlation with angiogenesis and lymphangiogenesis. Int J Biol Markers. 2008;23:182–6. doi: 10.1177/172460080802300308. [DOI] [PubMed] [Google Scholar]
- Thapa N, Kang KB, Kim IS. Beta ig-h3 mediates osteoblast adhesionand inhibits differentiation. Bone. 2005;36:232–42. doi: 10.1016/j.bone.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Furusato B, Takashima Y, Ravulapalli S, Dobi A, Srivastava S, McLeod DG, Sesterhenn IA, Ornstein DK, Shirasawa S. The increased expression of periostin during early stages of prostate cancer and advanced stages of cancer stroma. Prostate. 2009;69:1398–403. doi: 10.1002/pros.20988. [DOI] [PubMed] [Google Scholar]
- Ueda A, Ishigatsubo Y, Okubo T, Yoshimura T. Transcriptional regulation of the human monocyte chemoattractant protein-1 gene. Cooperation of two NF-kappaB sites and NF-kappaB/Rel subunit specificity. J Biol Chem. 1997;272:31092–9. doi: 10.1074/jbc.272.49.31092. [DOI] [PubMed] [Google Scholar]
- Xu J, Smock SL, Safadi FF, Rosenzweig AB, Odgren PR, Marks SC, Jr, Owen TA, Popoff SN. Cloning the full-length cDNA for rat connective tissue growth factor: implications for skeletal development. J Cell Biochem. 2000;77:103–15. doi: 10.1002/(sici)1097-4644(20000401)77:1<103::aid-jcb11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Fukushima H, Shin M, Katagiri T, Doi T, Takahashi T, Jimi E. TNF{alpha} represses BMP signaling by interfering with the DNA binding of smads through the activation of NF-{kappa}B. J Biol Chem. 2009 doi: 10.1074/jbc.M109.070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu M, Kitaura H, Fujimura Y, Eguchi T, Kohara H, Morita Y, Yoshida N. IL-12 inhibits TNF-alpha induced osteoclastogenesis via a T cell-independent mechanism in vivo. Bone. 2009;45:1010–6. doi: 10.1016/j.bone.2009.07.079. [DOI] [PubMed] [Google Scholar]
- Yoshioka N, Fuji S, Shimakage M, Kodama K, Hakura A, Yutsudo M, Inoue H, Nojima H. Suppression of anchorage-independent growth of human cancer cell lines by the TRIF52/periostin/OSF-2 gene. Exp Cell Res. 2002;279:91–9. doi: 10.1006/excr.2002.5590. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Broser M, Rom W. Activation of the interleukin 6 gene by Mycobacterium tuberculosis or lipopolysaccharide is mediated by nuclear factors NF IL 6 and NF-kappa B. Proc Natl Acad Sci U S A. 1995;92:3632. doi: 10.1073/pnas.92.8.3632d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Heulsmann A, Tondravi MM, Mukherjee A, Abu-Amer Y. Tumor necrosis factor-alpha (TNF) stimulates RANKL-induced osteoclastogenesis via coupling of TNF type 1 receptor and RANK signaling pathways. J Biol Chem. 2001;276:563–8. doi: 10.1074/jbc.M008198200. [DOI] [PubMed] [Google Scholar]
- Zhu S, Barbe MF, Amin N, Rani S, Popoff SN, Safadi FF, Litvin J. Immunolocalization of Periostin-like factor and Periostin during embryogenesis. J Histochem Cytochem. 2008;56:329–45. doi: 10.1369/jhc.7A7321.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Barbe MF, Liu C, Hadjiargyrou M, Popoff SN, Rani S, Safadi FF, Litvin J. Periostin-like-factor in osteogenesis. J Cell Physiol. 2009;218:584–92. doi: 10.1002/jcp.21633. [DOI] [PubMed] [Google Scholar]