The Herpes Simplex Virus ICP0 RING Finger Domain Inhibits IRF3- and IRF7-Mediated Activation of Interferon-Stimulated Genes (original) (raw)
Abstract
Virus infection induces a rapid cellular response in cells characterized by the induction of interferon. While interferon itself does not induce an antiviral response, it activates a number of interferon-stimulated genes that collectively function to inhibit virus replication and spread. Previously, we and others reported that herpes simplex virus type 1 (HSV-1) induces an interferon -independent antiviral response in the absence of virus replication. Here, we report that the HSV-1 proteins ICP0 and vhs function in concert to disable the host antiviral response. In particular, we show that ICP0 blocks interferon regulatory factor IRF3- and IRF7-mediated activation of interferon-stimulated genes and that the RING finger domain of ICP0 is essential for this activity. Furthermore, we demonstrate that HSV-1 modifies the IRF3 pathway in a manner different from that of the small RNA viruses most commonly studied.
Infection of cells by viruses elicits a potent and immediate cellular response aimed at limiting virus replication and spread. The hallmark of this response is the production and secretion of interferons (IFNs), a family of pleiotropic cytokines with antiviral, antiproliferative, and immune modulatory functions (37, 45, 69, 79, 80, 86, 92). Once produced, IFNs α and β are secreted from virally infected cells, bind to their cognate receptors on surrounding cells, and initiate a cascade of phosphorylation events of specific Jak/Stat molecules, culminating in translocation of a phosphorylated Stat-containing complex into the nucleus. This complex binds the IFN-stimulated response element (ISRE) within the promoter region of interferon-stimulated genes (ISGs) and initiates gene transcription (44, 46, 98).
While IFNs α and β possess multiple activities, they were initially discovered and are best known for their antiviral properties. The classical IFN-induced antiviral pathways include the double-stranded RNA-dependent protein kinase R, 2′,5′-oligoadenylate synthetase, and Mx pathways, which lead to protein synthesis inhibition, RNA degradation, and inhibition of viral replication, respectively (31, 47, 77, 79). The interferon response can also induce apoptosis (see reference 31 for a review).
It has become clear over the last several years that ISGs can also be activated by viral infection or double-stranded RNA, a by-product of viral infection, in the absence of IFN production (2, 90, 94-96, 100). For example ISG56, an IFN-stimulated protein involved in translational regulation via binding to eukaryotic initiation factor 3 (33), has been shown to be upregulated by IFN, double-stranded RNA, or virus through independent pathways (34). Recent work has shown that IRF3, a member of the interferon regulatory factor (IRF) family of transcription factors (36, 53, 78, 88), can bind directly to ISRE elements and induce ISGs in the absence of IFN production (49, 96, 97, 99).
The current model of virus-induced IRF3 activation consists of five sequential steps: (i) phosphorylation by a cellular “virus-activated” kinase(s), (ii) a conformational change resulting in protein dimerization, (iii) translocation to the nucleus, (iv) association with CBP and/or p300 coactivators, and (v) ISRE binding (41, 49, 82). Virus infection activates the cellular DNA-dependent protein kinase DNA-PK, leading to the phosphorylation of IRF3 on Thr135 followed by IRF3 nuclear localization (38). However, the role of N-terminal IRF3 phosphorylation is unclear, as other studies have found that virus-mediated IRF3 dimerization, nuclear translocation, and cofactor binding require C-terminal phosphorylation (49, 51, 99). The cellular kinases TBK-1 and IKKɛ were found to phosphorylate IRF3 and IRF7 following infection with Sendai virus (29, 83). While multiple residues within the C-terminal region of IRF3 are phosphorylated (49, 99), to date only Ser396 has been found to be required for virus-mediated IRF3 activation in vivo (81).
Previously, we and others reported that herpes simplex virus type 1 (HSV-1), a large enveloped DNA virus, triggers a host cellular response characterized by the induction of a specific set of genes, primarily ISGs, resulting in the activation of an antiviral state in an IFN-independent fashion (60, 67). This cellular response is mediated by HSV-1 virion particles and is inhibited by virus replication, suggesting that a newly synthesized viral protein(s) inhibits the response. The related herpesvirus human cytomegalovirus (HCMV) induces expression of ISGs in both the presence and absence of viral gene expression, although significantly more ISGs are induced when viral gene expression is inhibited, again suggesting that a newly made virus product suppresses the host response (7, 100, 101).
Recently, the HCMV virion protein pp65 was shown to inhibit antiviral gene expression by antagonizing NF-κB and IRF1 pathways (6). With HSV-1, virus binding and penetration are essential for antiviral state induction (60, 71), whereas with HCMV, soluble glycoprotein B appears sufficient to trigger the response (5, 84). The IFN-independent induction of ISGs by HSV-1 and HCMV does not require the Jak-Stat IFN pathway and appears to involve IRF3 (5, 60, 66, 67, 71).
Recently, the HSV-1 immediate-early protein ICP0 was shown to block ISG induction in a proteasome-dependent fashion (14). The mechanism by which ICP0 blocks ISG induction remains to be elucidated and is the focus of this study. ICP0 is expressed very early in infection and performs a variety of functions (21). ICP0 is a promiscuous activator of both viral and cellular promoters and appears to function synergistically with another immediate-early protein, ICP4 (28, 74, 93). During viral infection, ICP0 has been found to associate with a number of cellular proteins, including elongation factor 1δ (39), cyclin D3 (40), kinetochore protein CENP-C (23), ubiquitin-specific protease 7 (USP7, also known as HAUSP) (26, 59), and PML, the prototypic member of nuclear domains known as ND10, PML bodies, or PODs (25, 55, 56).
Considering that many ND10 constituents, including PML, are upregulated by IFN and that a number of viruses, including HSV-1, localize to and subsequently disrupt ND10, one hypothesis has been that ND10 functions as “nuclear defense” sites (54, 57). Of particular interest is that ICP0 affects the SUMO-1 modification of ND10 components and their subsequent proteasome-mediated degradation (10, 24, 25, 55, 65). While a number of functional regions of ICP0 have been identified, an N-terminal cysteine/histidine motif termed the RING finger appears to be important for the biological activities of ICP0 (21). Although the RING finger domain resembles a zinc finger DNA binding domain, it does not appear to bind DNA (22, 27) and instead acts as a ubiquitin E3 ligase that is required for the degradation of several cellular proteins, including PML (4).
While ICP0 may function to counteract host defense mechanisms through disruption of ND10, we and others have shown that ICP0 renders HSV-1 relatively resistant to the effects of IFN, in part by counteracting an IFN-induced block to virus transcription (35, 61, 63). In the current study, we set out to address the mechanism by which HSV-1 blocks the IFN-independent antiviral response initiated following entry of virus particles. We have found that ICP0 and vhs, the virion host shut-off protein, counteract the cellular IFN-independent antiviral response at early and late times postinfection, respectively. ICP0 specifically blocks IRF3- and IRF7-mediated cellular responses and requires the RING finger for this activity. Since HSV-1 does not appear to activate the IRF3 pathway in the same fashion as the small RNA viruses most commonly studied, the precise mechanism of ICP0 inhibition of IRF3 activity remains to be determined. These studies, however, expand our knowledge on both cellular antiviral responses to virus infection and the countermeasures adopted by viruses to ensure their successful replication and spread.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic lung (HEL) fibroblasts and U2OS osteosarcoma cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, while Vero, E5 (13), V27 (73), and A549 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum. 293 HEK cells were maintained in α minimal essential medium supplemented with 10% fetal bovine serum. HSV-1 strains KOS (wild type), d22lacZ (ICP22−) (52), N38 (ICP47−) (91), ΔICP6 (ICP6−) (30), and Δsma (vhs−) (72) were grown in Vero cells, n212 and 7134 (ICP0−) (8), KM110 (ICP0−VP16−) (64), and FXE (ICP0 RING finger mutant) (18) were grown in U2OS cells, d120 (ICP4−) was grown on E5 cells (13), and 5dl1.2 (ICP27−) was grown on V27 cells (73). Newcastle disease virus (NDV, strain La Sota) and Sendai virus (strain Cantell) were kindly provided by E. Nagy and J. Hiscott, respectively. All HSV-1 infections utilized a multiplicity of infection of 10 PFU per cell (as determined in the cell line used for propagation) unless otherwise specified. NDV and Sendai virus were used at 40 hemagglutination activity units per 106 cells. UV inactivation was performed with a UV Stratalinker 2400 (Stratagene) for the length of time required to reduce viral titers by a factor of 105 (data not shown).
Creation and characterization of 7134Δsma (ICP0− vhs−) double mutant virus.
U2OS cells were coinfected with 5 PFU per cell of both 7134 and Δsma viruses. The ICP0 coding region in 7134 is replaced with the lacZ gene, allowing color selection with the addition of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) (100 μg per ml) in the overlay (1% agarose in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum). Individual blue plaques were isolated, and following amplification DNA was isolated from virally infected cells as previously described (64) for use in PCR and Southern blot analyses. PCR was used to detect an internal _Sma_I-_Sma_I deletion within the Δsma vhs gene as previously described (62).
Southern blot analysis to ensure that both copies of ICP0 retained the 7134 mutation was performed as previously described (8, 64). Briefly, an ICP0 fragment spanning the entire open reading frame was hybridized to DNA digested with _Pst_I and _Sst_I. ICP0 resides in a genomic _Pst_I fragment and is not cleaved by _Sst_I. lacZ, however, contains an internal _Sst_I site, and thus recombinant 7134 ICP0 is cleaved into two fragments, whereas wild-type ICP0 is not. Positive clones were plaque purified three times on U2OS cells, followed by a final round of PCR and Southern blot analysis.
RT-PCR and Western blot analysis.
Total cellular RNA was harvested with Trizol (Gibco-BRL) according to the manufacturer's instructions. For reverse transcription (RT)-PCR, aliquots (2 μg) were reverse transcribed with 200 ng of random hexamer primer and Superscript II reverse transcriptase (Invitrogen). PCR was subsequently performed as per the manufacturer's specifications with the following primers: human ISG56 forward and reverse (5′-ACGGCTGCCTAATTTACAGC-3′ and 5′-AGTGGCTGATATCTGGGTGC-3′) and human glyceraldehyde-3-phosphate dehydrogenase forward and reverse (5′-CGGAGTCAACGGATTTGGTCGTA-3′ and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′).
Preparation of whole-cell extracts and immunoblot analyses.
For preparation of whole-cell extracts, cells were washed twice and then harvested in cold phosphate-buffered saline, followed by centrifugation at 200 × g for 3 min at 4°C. Cell pellets were resuspended in whole-cell extract buffer [20 mM HEPES, pH 7.4, 100 mM NaCl, 10 mM β-glycerophosphate, 0.2% Triton X-100, 50 mM NaF, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, and 1X protease inhibitor cocktail (Sigma)], lysed on ice for 15 min, and then centrifuged for 10 min at 13,000 × g at 4°C. Extract concentrations were determined with a Bradford assay kit (Bio-Rad), and the indicated amounts of extracts were run on 9% polyacrylamide gels, transferred to nitrocellulose, and probed with antibodies specific to IRF3 (Santa Cruz SC-9082), IRF7 (Santa Cruz SC-9083), β-actin (Santa Cruz SC-1616), CBP (Santa Cruz SC-369), TBK1 (Santa Cruz M-375), ICP0 (Goodwin Institute), or ISG56 (provided by G. Sen) at a dilution of 1:1,000. To visualize DNA-PK, nuclear extracts were prepared as previously described (1), and 25 μg of extracts was run on gels containing 8% acrylamide and 0.07% bisacrylamide. Western blot analysis was performed with a 1:2,000 dilution of an anti-DNA-PK monoclonal antibody (provided by S. Lees-Miller).
Luciferase assays.
HEK 293 cells were transfected with pRLTK, RANTES-pGL3, or IFNB-pGL3 reporter constructs along with expression constructs encoding constitutively activated IRF3 (IRF3-5D) or IRF7 (IRF7-Δ247-467) (48-50). At 8 h posttransfection, cells infected with 40 hemagglutination activity units of Sendai virus per 106 cells or left uninfected, as indicated. Luciferase activity was analyzed 24 h posttransfection by the dual-luciferase reporter assay (Promega) as per the manufacturer's instructions. Relative luciferase activity was measured as the activation relative to the basal level of reporter gene in the presence of pFLAG-CMV2 vector after normalization with cotransfected activity. Values represent the mean ± standard deviation for three experiments.
RESULTS
ISG induction in response to virus particles is abolished by ICP0 and vhs.
Previously, we used microarray analysis to show that HSV-1 particles induce ISGs in an IFN-independent fashion (60). In these studies we found that ISG induction was masked following virus replication, yet we failed to identify a potential viral inhibitor using HSV-1 mutants individually deleted for an immediate-early gene product. Subsequently, DeLuca and colleagues determined that ICP0 was capable of inhibiting ISG induction through the use of HSV-1 mutants deleted for several immediate-early gene products along with an adenovirus expressing ICP0 (14). In our original study, we assayed mRNA accumulation by Northern blot analysis 18 to 24 h postinfection, because ISG mRNA was readily detectable at this time with either genetically inactivated virus (KM110) or UV-inactivated wild-type HSV-1.
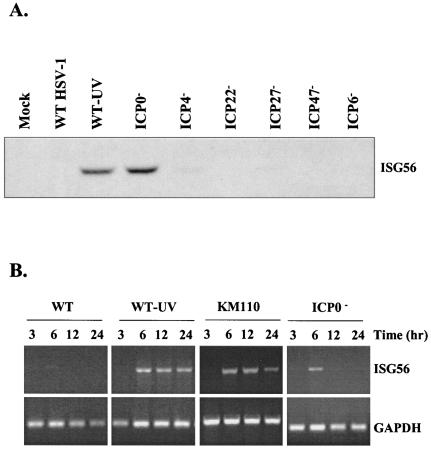
To reconcile our results with those of DeLuca and colleagues, we assayed ISG56 mRNA and protein levels at various times postinfection with a panel of single immediate-early mutant viruses in human embryonic lung (HEL) fibroblasts. As shown in Fig. 1A, aside from UV-inactivated wild-type virus, only the virus deleted for ICP0 demonstrated ISG56 protein accumulation at 12 h postinfection, in accordance with the work of DeLuca and colleagues. Interestingly, RT-PCR analysis with the ICP0 mutant n212 consistently yielded ISG56 message at early but not late times postinfection (Fig. 1B), in accordance with our previous results. Similar results were obtained with two additional ICP0 mutants, 7134 (refer to Fig. 2) and dlX3.1 (75) (data not shown), where ISG56 mRNA accumulation peaked at 6 h postinfection and then declined rapidly. In the absence of virus gene expression, using either UV-inactivated HSV-1 or genetically inactivated KM110, we observed sustained ISG56 mRNA levels at late times postinfection. These data suggest that in the absence of ICP0, ISG56 mRNA accumulates at early times postinfection but subsequently disappears due to the presence of an additional virus-induced activity.
FIG. 1.
Expression of ISG56 is only observed in the absence of ICP0 expression. (A) Western blot analysis of whole-cell extracts (40 μg) of HEL fibroblasts harvested 12 h postinfection with the indicated viruses. The ISG56 protein is only visible following infection with UV-inactivated wild-type virus (strain KOS, WT-UV) or an ICP0 null mutant (n212). (B) RT-PCR analysis of total RNA harvested from HEL fibroblasts at various times postinfection with the indicated viruses. In the absence of ICP0, transient ISG56 mRNA accumulation is seen, whereas in the absence of HSV-1 replication (UV-inactivated KOS or genetically inactivated KM110), sustained ISG56 mRNA accumulation is observed. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
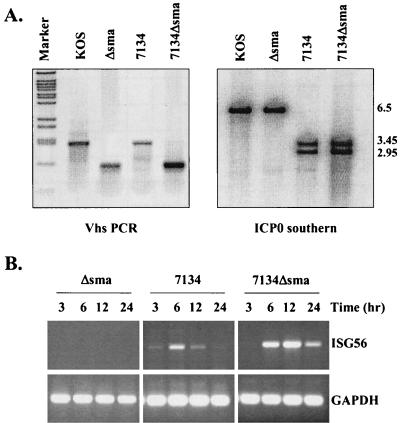
FIG. 2.
Deletion of vhs and ICP0 results in sustained ISG56 mRNA accumulation. (A) Construction of a double mutant virus deleted for vhs and ICP0. U2OS cells were coinfected with Δsma and 7134 to create a double mutant virus as outlined in Materials and Methods. (Left panel) PCR analysis was used to confirm the internal _Sma_I-_Sma_I fragment deletion that defines the vhs Δsma mutation. _Bst_EII-cut lambda DNA was used as a marker. (Right panel) Southern blot analysis with a radiolabeled ICP0 probe was used to distinguish wild-type ICP0 and 7134 ICP0 sequences based on the presence of a unique _Sst_I site within the lacZ gene that replaces the ICP0 sequence in the 7134 mutation. (B) RT-PCR analysis of total RNA harvested from HEL fibroblasts at various times postinfection with the indicated viruses. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
HSV-1 particles contain within their tegument the virion host shut-off protein (vhs) that either is, or is part of, an RNase that causes the degradation of both viral and cellular mRNAs (15, 72, 87). Thus, we assayed whether this protein was responsible for the decline in ISG56 mRNA levels at later times postinfection. For this experiment, we created a double mutant virus by recombining the 7134 ICP0 null virus (8) with the Δsma vhs-deleted virus (72) (Fig. 2). Due to an internal deletion of a _Sma_I-_Sma_I fragment, the Δsma and wild-type vhs alleles are easily distinguishable by PCR analysis, while Southern blot analysis distinguishes wild-type and 7134 ICP0 alleles based on the introduction of an _Sst_I restriction site within the lacZ gene that replaces ICP0 coding sequences in the 7134 mutant (8) (Fig. 2A).
An RT-PCR time course analysis of HEL fibroblasts infected with Δsma, 7134, or 7134Δsma was performed in order to determine the contributions of ICP0 and vhs activities in the accumulation of ISG message. As demonstrated above with n212 (Fig. 1B), ISG56 mRNA accumulation following infection with 7134 was transient, peaking at approximately 6 h postinfection (Fig. 2B). In contrast, sustained ISG56 mRNA was observed upon deletion of both ICP0 and vhs, indicating that vhs activity contributes to the loss of ISG56 mRNA at late times postinfection. This vhs-induced activity was only observed in the absence of ICP0, consistent with the observation that ICP0 prevents ISG induction following virus entry (14). Accordingly, we failed to detect ISG56 mRNA accumulation in fibroblasts infected with Δsma, as this viral mutant contains wild-type ICP0. Similar results were found by RT-PCR with primers specific for other ISGs originally identified by microarray analysis (data not shown).
ICP0 RING finger inhibits IRF3- and IRF7-mediated ISG induction.
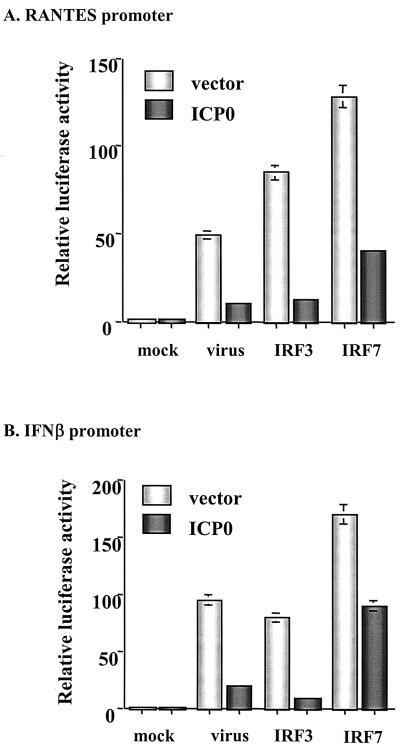
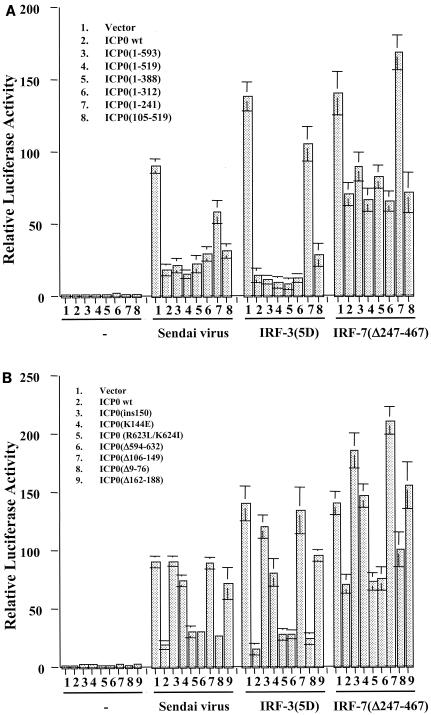
IFN-independent ISG induction following virus infection may occur through the activation of the constitutively expressed transcription factor IRF3 due to its ability to bind to the ISRE element within ISG promoters (32, 36). In addition, Preston and colleagues showed that in the absence of virus gene expression, HSV-1 induces the DNA binding activity of IRF3 (71). In a series of transient-transfection assays with a luciferase reporter under the control of two different ISRE-containing ISG promoters, RANTES and IFN-β, we found that expression of ICP0 significantly dampened ISG promoter activation in response to Sendai virus infection and expression of constitutively activated forms of both IRF3 [IRF-3(5D)] and IRF7 [IRF-7(Δ247-467)] (48-50) (Fig. 3). In all transient transfection assays, a dose-response curve was performed with the ICP0 plasmid (data not shown).
FIG. 3.
ICP0 blocks virus-, IRF3-, and IRF7-mediated activation of ISG-specific promoters. Luciferase assays in 293 cells were used to measure activation of the RANTES (A) or IFN-β (B) promoters in response to infection with the paramyxovirus Sendai virus or expression of constitutively activated forms of IRF3 [IRF-3(5D)] and IRF7 [IRF-7(Δ247-467)] in the presence or absence of full-length ICP0. Results represent the mean ± standard deviation of three independent experiments.
To determine which region of ICP0 was responsible for this inhibition, constructs containing ICP0 deletion, truncation, and point mutations were utilized (Fig. 4). We first assayed a series of ICP0 truncation mutants for their ability to inhibit the activation of the IFN-β promoter by Sendai virus or constitutively activated forms of IRF3 and IRF7. As shown in Fig. 5A, successive truncation of the C terminus to residue 312 had no effect on the inhibitory activity of ICP0. However, the truncation mutant 1-241 failed to inhibit activation of the IFN-β promoter by virus, IRF3, or IRF7. Along with the observation that mutant 105-519 effectively repressed IFN-β promoter activation, these data suggest that the region between residues 105 and 312 is important for ICP0-mediated inhibition.
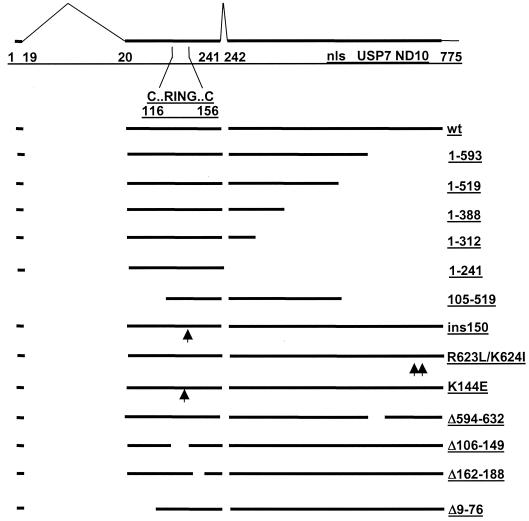
FIG. 4.
Schematic representation of ICP0 functional domains and mutations used in this study. The uppermost line depicts the 775-amino-acid ICP0 open reading frame (thick line) with the positions of the introns (thin angled lines) and the amino acid coordinates at the intron-exon junctions marked. These sequences are contained within the ICP0 expression plasmid p111 (19). Also marked are the positions of the first and last cysteine residues of the RING finger, the nuclear localization signal (nls) (residues 501 to 510) (17), the USP7 interaction domain (residues 594 to 633) (58), and the sequences required for self-multimerization and efficient localization at ND10 (residues 633 to 720) (11, 55, 58). Deletion mutants 1-519, 1-388, and 1-312 express C-terminally truncated proteins and were constructed by utilizing the _Mlu_I, _Xma_I, and _Nru_I restriction sites at the positions of these 3′ truncations. Mutant 105-519 expresses the coding region between the _Xho_I and _Mlu_I sites. Mutant _ins_150 contains a 4-amino-acid insertion in codon 150 (p110F1) (19), mutant K144E has a lysine to glutamic acid substitution in the helix region of the RING finger (16), and deletion mutants Δ594-632, Δ106-149, Δ162-188, and Δ9-76 have the stated deletions (p110D12, p110FXE, p110D22, and p110D1, respectively) (17). Truncation mutant 1-593 has stop codons inserted into codon 594 (p110E52X) (58). Finally, mutant 1-241 contains only the sequences encoded in exons 1 and 2 (p110262) (25). Mutants Δ106-149, Δ162-188, and _ins_150 have lost ubiquitin E3 ligase activity both in vitro and in vivo, while mutant K144E retains in vitro activity but appears to have reduced activity in vivo (4, 20).
FIG. 5.
ICP0 RING finger is both necessary and sufficient to block virus-, IRF7-, and IRF3-mediated ISG promoter activation. Luciferase assays in 293 cells were used to measure the activation of the IFN-β promoter following Sendai virus infection or expression of constitutively activated IRF3 [IRF-3(5D)] and IRF7 [IRF-7(Δ247-467)] in the presence or absence of full-length and mutant forms of ICP0 (refer to Fig. 4). Results represent the mean ± standard deviation of three independent experiments.
Within this region of ICP0 is the RING finger motif, which has been shown to be responsible for many of the biological activities of ICP0. The RING finger is a zinc-stabilized fold that requires a histidine and several cysteine residues (of which the first and last are at residues 116 and 156) to form a structure that has catalytic E3 ligase activity (3, 4, 20). To assess the role of the RING finger motif in inhibiting IRF3- and IRF7-mediated IFN-β promoter activation, we next tested a series of ICP0 mutants harboring deletions, insertions, and point mutations both within and exterior to the RING finger domain (Fig. 5B).
Deletion mutant Δ106-149 (the lesion in virus FXE) removes several of the essential metal-coordinating residues and thus abolishes the RING finger activity, while the mutation in mutant ins150 affects a critical alpha helix within the RING fold with similar functional consequences (4, 20). Mutation K144E lies within the same helix and causes reduced catalytic activity in transfected and infected cells (20). Deletion Δ162-188 removes sequences that are part of the RING domain fold (3) and produces a phenotype that is indistinguishable in all available assays from that of a core RING deletion (20). These four ICP0 mutants failed to inhibit virus-, IRF3-, and IRF7-mediated activation of the IFN-β promoter. Consistent with the data in Fig. 5A, deletions (Δ9-76 and Δ594-632) and point mutations (R623L/K624I) outside of the RING finger region had no adverse affect on the inhibitory activity of ICP0. Similar results were obtained with the RANTES promoter construct (data not shown).
ICP0 does not induce the degradation of known IRF3 pathway components.
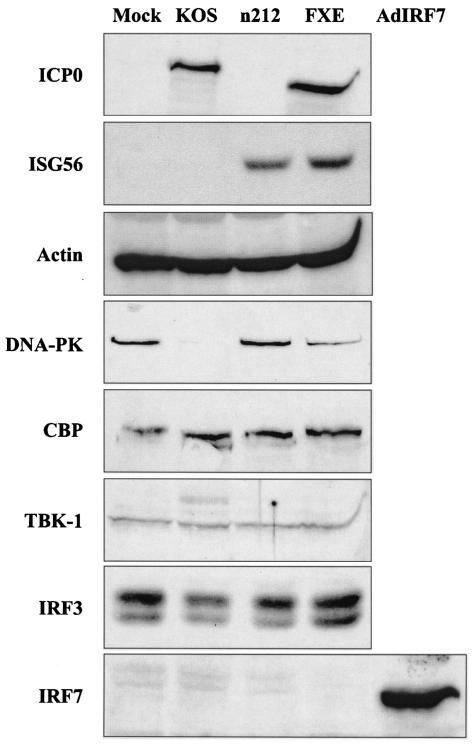
DeLuca and colleagues demonstrated that the proteasome inhibitor MG132 prevented ICP0 from inhibiting ISG induction (14), suggesting that the function of ICP0 is to induce the proteasomal degradation of a member(s) of the ISG induction pathway. To determine whether ICP0 blocks ISG induction by inducing the degradation of known IRF3 pathway components, we determined the levels of endogenous TBK-1, IRF3, IRF7, and CBP protein following infection of HEL fibroblasts for 10 h with wild-type HSV-1 (strain KOS), n212, and the RING finger mutant FXE (Fig. 6). Due to the Δ106-149 deletion of ICP0 within the FXE virus, the ICP0 protein migrates slightly faster than its wild-type counterpart (top panel). ISG56 protein was readily observed following infection with n212 and FXE, consistent with previous results (Fig. 1A and 5B, respectively). Actin served as a control for protein loading and nonspecific protein degradation, while DNA-PK served as a positive control for ICP0-mediated degradation, as this nuclear protein was shown to be specifically degraded by wild-type ICP0 (43).
FIG. 6.
ICP0 fails to degrade known components of the IRF3 pathway. Western blot analysis of whole-cell extracts (40 μg) or nuclear extracts (25 μg, for DNA-PK) of HEL fibroblasts harvested 10 h postinfection with the indicated viruses. Wild-type ICP0 fails to induce the degradation of IRF3, CBP, or TBK-1, components of the cellular antiviral pathway, whereas DNA-PK is specifically degraded by wild-type but not mutant ICP0. In the absence of wild-type ICP0, ISG56 protein is observed, whereas expression of IRF7 is only detected with a recombinant adenovirus expressing human IRF7 (AdIRF7). Actin was used as an internal loading control.
We failed to detect a significant decrease in the level of endogenous TBK-1, one of two kinases shown to phosphorylate IRF3 following virus infection (29, 83). As expression of the second kinase, IKKɛ, is thought to be restricted to activated leukocytes (70), we did not test for endogenous levels of this kinase in HEL fibroblasts. However, we did observe that expression of wild-type ICP0 along with IKKɛ in a transient transfection assay failed to decrease levels of the IKKɛ protein (data not shown). We also failed to detect a decrease in the endogenous levels of IRF3 and CBP. IRF7 is not constitutively expressed in cells, and we failed to detect significant induction of this protein at the time of harvest (10 h postinfection). However, in subsequent time course analyses, we frequently detected low levels of IRF7 at late times (12 to 24 h) postinfection with wild-type HSV-1 (data not shown). Infection with an adenovirus overexpressing IRF7 was used as a positive control for IRF7 detection. Thus, ICP0 does not appear to block ISG induction by inducing the degradation of known IRF3 pathway constituents. Further work is required to identify the mechanism of ICP0 action with respect to the proteasome and ISG inhibition.
HSV-1 does not induce the hyperphosphorylation of IRF3.
In the current model of virus-mediated IRF3 activation, viral infection induces the phosphorylation on multiple C-terminal serine and threonine residues of IRF3, followed by nuclear translocation and proteasome-mediated degradation (49, 82, 99). These studies have primarily utilized small, related RNA viruses such as Sendai virus and Newcastle disease virus, members of the paramyxovirus family of negative, single-stranded RNA viruses, in transformed cell lines such as A549 lung epithelial cells. In the previous experiment, we failed to detect significant degradation of IRF3 following HSV-1 infection.
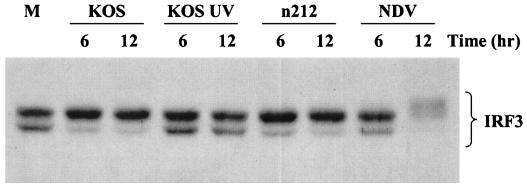
To determine if replicating HSV-1 induces the hyperphosphorylation of IRF3, we performed Western blot analysis on whole-cell extracts harvested from A549 cells and HEL fibroblasts infected with KOS, UV-inactivated KOS, n212, and NDV. Results from A549 cells are presented in Fig. 7. Newcastle disease virus induced the hyperphosphorylation of IRF3 between 6 and 12 h postinfection, illustrated by the appearance of slower-migrating species of IRF3. We also noted the accompanying degradation of IRF3 following hyperphosphorylation in response to Newcastle disease virus infection, as previously reported in the literature (49). Following HSV-1 infection, however, we failed to detect a similar level of IRF3 hyperphosphorylation. Instead, we routinely observed a shift from the lower to the upper form of IRF3. While this shift was routinely observed in both A549 cells and HEL fibroblasts, the shift was consistently easier to detect in A549 cells and was primarily observed at later times (18 to 24 h postinfection) in HEL fibroblasts (data not shown).
FIG. 7.
HSV-1 fails to hyperphosphorylate IRF3. Western blot analysis of whole-cell extracts (40 μg) of A549 cells harvested 6 and 12 h postinfection with the indicated viruses. Endogenous IRF3 is present as two species and becomes hyperphosphorylated and subsequently degraded following infection with the paramyxovirus Newcastle disease virus (NDV). A shift from the lower to the upper form of IRF3 is readily observed following infection with wild-type and ICP0 null viruses but not in the absence of virus gene expression.
The apparent lack of hyperphosphorylation characteristic of small RNA viruses was not due to the action of ICP0, since n212 failed to demonstrate a similar level of IRF3 phosphorylation. Of interest, although UV-inactivated KOS consistently failed to elicit a similar shift from the lower to the upper form of IRF3, n212 mediated this posttranslational modification with kinetics similar to the wild-type virus. A similar shift was observed with three additional ICP0 mutant viruses (data not shown). Thus, HSV-1 replication induces a cellular function that is distinct from that mediated by RNA viruses but does not rely on the activity of ICP0. Studies are currently under way to delineate the mechanism by which HSV-1 modifies IRF3 following productive infection.
DISCUSSION
Our current working hypothesis of HSV-1 pathogenesis is that incoming virus particles stimulate an antiviral response that induces the direct induction of ISGs through the activation of IRF3 (12). Following virus replication, HSV-1 abolishes this cellular response through the activities of ICP0 and vhs. At early times postinfection, ICP0 appears to specifically inhibit the IRF3 pathway via the RING finger domain, while vhs presumably functions in a nonspecific fashion later in infection. Since vhs mutants that produce ICP0 fail to accumulate detectable levels of ISG mRNA, while ICP0 mutants that produce vhs transiently accumulate ISG mRNA, it is likely that ICP0 represents the primary mechanism utilized by HSV-1 to counteract the cellular antiviral response to incoming virus particles. A related herpesvirus, HCMV, appears to function in a similar fashion. Using microarray analysis, Shenk and colleagues demonstrated that while both wild-type and UV-inactivated HCMV induce ISGs, significantly more ISGs were induced following infection with UV-inactivated virus, indicating that virus replication produces a protein that functions to suppress the host antiviral response (100, 101). Furthermore, both HSV-1 and HCMV particles were found to induce the DNA binding activity of IRF3 (66, 71).
ICP0 is a multifunctional protein to which diverse activities have been ascribed (21). With respect to virus pathogenesis and the antiviral response, ICP0 appears to play an important role in both IFN-dependent and -independent pathways. Classical antiviral pathways involve the production of IFNs, primarily IFN-α and IFN-β. Along with ICP34.5, ICP0 renders HSV-1 relatively resistant to the effects of these IFNs, in part by overcoming an IFN-induced block to virus transcription (35, 61, 63). The cellular antiviral response to incoming particles, however, was shown to be IFN independent (60, 67).
Recent studies with the IRF family of transcription factors have found that IRF3 and IRF7 are involved not only in virus-induced production of type I IFNs but also in the direct induction of ISGs through binding to ISRE elements in ISG promoters (36, 88). Given the observations that HSV-1 particles induce IRF3 DNA-binding activity and that ICP0 blocks ISG induction, it was not surprising that ICP0 was found to specifically block IRF3- and IRF7-mediated activation of two ISRE-containing ISG promoters. The ICP0 inhibitory effect on activated IRF7 was not as great as that observed following virus infection or with activated IRF3. Since IRF3 is constitutively expressed while IRF7 induction is dependent on IFN signaling (88), the preferential ability of ICP0 to inhibit IRF3 would be predicted given its apparent role in overcoming early antiviral responses, including those not mediated by IFN.
However, it remains to be determined exactly how ICP0 blocks IRF3-mediated antiviral responses. The initial work of DeLuca and colleagues found that ICP0 functions via a proteasome-dependent pathway (14). Known components of the virus-induced IRF3 pathway, including TBK-1, IKKɛ, IRF3, and CBP, were not degraded following the production of ICP0. During these studies, however, we discovered that HSV-1 does not appear to activate the IRF3 pathway in the same fashion as the RNA viruses most commonly studied. First, several groups studying paramyxovirus infection have reported that virus replication is required for IRF3 and/or IRF7 C-terminal hyperphosphorylation and subsequent activation (85, 89). Not only does HSV-1 replication mask ISG induction, we failed to observe IRF3 hyperphosphorylation following infection with either wild-type or ICP0 null viruses. In contrast, we routinely observed a shift from the lower band to the upper band of the endogenous IRF3 doublet, which has been observed previously in the context of N-terminal phosphorylation of IRF3.
Work from the laboratories of Hiscott and Howley demonstrated that N-terminal phosphorylation occurs in response to stress stimuli and involves DNA-PK (38, 82), a cellular kinase involved in DNA double-strand break repair and V(D)J recombination (42). However, the role of N-terminal IRF3 phosphorylation during virus infection remains to be determined, as the majority of published data suggest that C-terminal IRF3 phosphorylation is essential (49, 51, 81, 99).
Interestingly, ICP0, through its RING finger domain, has been found to induce the proteasome-dependent degradation of DNA-PK (43, 68). It is unclear at this time, however, how ICP0, DNA-PK, and IRF3 may be related, particularly with respect to the IFN-independent antiviral state. Also unclear are the differences in the activation of the IRF3 pathway between large DNA and small RNA viruses. Specifically, although we failed to detect IRF3 hyperphosphorylation characteristic of infection with paramyxoviruses, replicating HSV-1 appeared to posttranslationally modify IRF3 in some fashion in an ICP0-independent manner. Additionally, in the transient transfection assays, ICP0 was able to block activation of two ISRE-containing ISG promoters in response to paramyxovirus infection. Thus, although HSV-1 and paramyxovirus infection may differentially modify components of the IRF3 pathway, a common event in this pathway is inhibited by ICP0.
While the precise mechanism of blocking IRF3-mediated activation remains to be elucidated, it is clear that the RING finger motif of ICP0 is critical. While sequences in the C-terminal region of ICP0, including the nuclear localization signal, the USP7-binding domain and the ND10 localization region, are important for ICP0 function, the N-terminal RING finger domain appears to be important for most biological activities of ICP0 (21). In this study, we found that the region containing the RING finger domain appears to be both necessary and sufficient for the ICP0-mediated suppression of the IRF3 pathway, at least within our experimental design.
Although similar to zinc finger DNA binding motifs, the ICP0 RING finger fails to bind DNA in solution, either alone or in the context of full-length ICP0 (22, 27) and it is now clear that the RING finger of ICP0 constitutes a typical ubiquitin E3 ligase domain (4). The RING finger is essential, however, for localization to and the subsequent disruption of ND10 (25, 55). Since many ND10 constituents are upregulated by IFN, one hypothesis is that ND10 serve as an intracellular defense mechanism and that ICP0 functions to overcome this defense. Recent work with mouse fibroblasts that are deficient in PML has indicated that, in contrast to the PML-positive control cells, pretreatment of PML−/− cells with IFN had little effect on the replication of ICP0-negative HSV-1 (9). Of interest, CBP has been found to localize to ND10 via its interaction with PML (76).
Although ICP0 induces the proteasome-dependent degradation of both SUMO-1 modified and unmodified forms of PML, the levels of CBP appear to remain constant throughout infection. We have found, however, that following infection with wild-type virus or transfection with full-length ICP0, the punctate nuclear staining of CBP, characteristic of ND10 constituents, changes to a diffuse nuclear staining (K. Laryea and K. Mossman, unpublished data). This difference in nuclear localization is not observed, however, with ICP0 null or RING finger mutant virus infection. How these observations collectively fit in to the mechanism of ICP0 suppression of the cellular antiviral response is unclear at the moment, but warrants further investigation.
In conclusion, we have shown that HSV-1 responds to the IRF3-mediated cellular response to incoming virus particles by subverting the IRF3 pathway through an ICP0-specific mechanism. This mechanism involves the ICP0 RING finger and the proteasome but does not appear to induce the degradation of known signaling components. In addition, the RNase activity associated with vhs further ensures that ISG mRNA fails to accumulate, due to its ability to nondiscriminately degrade both viral and cellular mRNA. Thus, HSV-1 elicits at least two mechanisms to ensure successful virus replication in light of the rapid induction of an immune response, characterized by the direct upregulation of both intracellular and secreted immune modulatory functions. These studies serve to increase our knowledge of both the immune response to pathogen infection and the strategies used by viruses to overcome these barriers and will ultimately permit the design of effective therapeutics to suppress viral infection.
Acknowledgments
We thank E. Nagy, J. Hiscott, S. Lees-Miller, and G. Sen for reagents and D. Cummings and K. Laryea for technical assistance.
The Canadian Institutes of Health Research sponsored this work. R.L. was supported by a Fonds de la Recherche en sante du Quebec (FRSQ) Chercheur Boursier. The Medical Research Council supports the work in the laboratory of R.D.E.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19**:**2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, S. K., G. T. Leonaard, T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270**:**19624-19629. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, P. N., B. Luisi, A. Milner, M. Elliott, and R. Everett. 1994. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237**:**201-211. [DOI] [PubMed] [Google Scholar]
- 4.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76**:**841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19**:**3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100**:**11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75**:**12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63**:**4570-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77**:**7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18**:**935-941. [DOI] [PubMed] [Google Scholar]
- 11.Ciufo, D. M., M. A. Mullen, and G. S. Hayward. 1994. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J. Virol. 68**:**3267-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. 78**:**1685-1696. [DOI] [PMC free article] [PubMed]
- 13.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56**:**558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76**:**2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleoltyic cleavage of target RNAs in cell extracts. J. Virol. 73**:**7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R., P. O'Hare, D. O'Rourke, P. Barlow, and A. Orr. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol. 69**:**7339-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J. Mol. Biol. 202**:**87-96. [DOI] [PubMed] [Google Scholar]
- 18.Everett, R. D. 1989. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J. Gen. Virol. 70**:**1185-1202. [DOI] [PubMed] [Google Scholar]
- 19.Everett, R. D. 1987. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 6**:**2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74**:**9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22**:**761-770. [DOI] [PubMed] [Google Scholar]
- 22.Everett, R. D., P. Barlow, B. Luisi, G. Hope, A. Orr, A. Milner, and D. Lyon. 1993. A novel arrangement of zinc binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J. Mol. Biol. 234**:**1038-1047. [DOI] [PubMed] [Google Scholar]
- 23.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18**:**1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72**:**6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13**:**5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett, R. D., M. Meredith, and A. Orr. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73**:**417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate-early polypeptide Vmw110. Nucleic Acids Res. 19**:**6155-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett, R. D., C. M. Preston, and N. D. Stow. 1991. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication, p. 49-76. In E. K. Wagner (ed.), The control of herpes virus gene expression. CRC Press Inc., New York, N.Y.
- 29.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4**:**491-496. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein, D. J., and S. K. Weller. 1988. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology 166**:**41-51. [DOI] [PubMed] [Google Scholar]
- 31.Goodbourne, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81**:**2341-2364. [DOI] [PubMed] [Google Scholar]
- 32.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-Stimulated genes. J. Virol. 76**:**5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19**:**6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267**:**209-219. [DOI] [PubMed] [Google Scholar]
- 35.Harle, P., B. S. Jr., D. J. Carr, and W. P. Halford. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293**:**295-304. [DOI] [PubMed] [Google Scholar]
- 36.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19**:**1-13. [DOI] [PubMed] [Google Scholar]
- 37.Joklik, W. K. 1990. Interferons, p. 383-410. In B. N. Fields and D. M. Knipe (ed.), Virology, 2nd ed. Raven Press Ltd., New York, N.Y.
- 38.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 99**:**2818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71**:**1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawaguchi, Y., C. V. Sant, and B. Roizman. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71**:**7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20**:**4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, S. H., and C. H. Kim. 2002. DNA-dependent protein kinase complex: a multifunctional protein in DNA repair and damage checkpoint. Mol. Cell 13**:**159-166. [PubMed] [Google Scholar]
- 43.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type I transactivator ICP0. J. Virol. 70**:**7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leonard, W. J. 2001. Role of Jak kinases and STATs in cytokine signal transduction. Int. J. Hematol. 73**:**271-277. [DOI] [PubMed] [Google Scholar]
- 45.LePage, C., P. Genin, M. G. Baines, and J. Hiscott. 2000. Interferon activation and innate immunity. Rev. Immunogenet. 2**:**374-386. [PubMed] [Google Scholar]
- 46.Levy, D. E. 1995. Interferon induction of gene expression through the Jak-Stat pathway. Semin. Virol. 6**:**181-189. [Google Scholar]
- 47.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12**:**143-156. [DOI] [PubMed] [Google Scholar]
- 48.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19**:**959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18**:**2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275**:**34320-34327. [DOI] [PubMed] [Google Scholar]
- 51.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19**:**2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73**:**5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237**:**1-14. [DOI] [PubMed] [Google Scholar]
- 54.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20**:**660-667. [DOI] [PubMed] [Google Scholar]
- 55.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75**:**1223-1233. [DOI] [PubMed] [Google Scholar]
- 56.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate-early gene 1 product (ICP0). J. Gen. Virol. 74**:**2679-2690. [DOI] [PubMed] [Google Scholar]
- 57.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129**:**278-287. [DOI] [PubMed] [Google Scholar]
- 58.Meredith, M., A. Orr, M. Elliott, and R. Everett. 1995. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology 209**:**174-187. [DOI] [PubMed] [Google Scholar]
- 59.Meredith, M., A. Orr, and R. Everett. 1994. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology 200**:**457-469. [DOI] [PubMed] [Google Scholar]
- 60.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75**:**750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74**:**2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74**:**6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76**:**1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mossman, K. L., and J. R. Smiley. 1999. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J. Virol. 73**:**9726-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73**:**5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18**:**3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-response genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81**:**2215-2218. [DOI] [PubMed] [Google Scholar]
- 68.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73**:**650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pestka, S., J. A. Langer, K. C. Zoon, and C. E. Samuel. 1987. Interferons and their actions. Annu. Rev. Biochem. 56**:**727-777. [DOI] [PubMed] [Google Scholar]
- 70.Peters, R. T., and T. Maniatis. 2001. A new family of IKK-related kinases may function as I kappa B kinase kinases. Biochim. Biophys. Acta 1471**:**M57-62. [DOI] [PubMed] [Google Scholar]
- 71.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75**:**8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Read, G. S., B. M. Bradley, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptides. J. Virol. 67**:**7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J. Virol. 64**:**1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 1043-1107. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 75.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61**:**829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salomoni, P., and P. P. Pandolfi. 2002. The role of PML in tumor suppression. Cell 108**:**165-170. [DOI] [PubMed] [Google Scholar]
- 77.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14**:**778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato, M., T. Taniguchi, and N. Tanaka. 2001. The interferon system and interferon regulatory factor transcription factors-studies from gene knockout mice. Cytokine Growth Factor Rev. 12**:**133-142. [DOI] [PubMed] [Google Scholar]
- 79.Sen, G. C. 2001. Viruses and interferon. Annual Rev. Microbiol. 55**:**255-281. [DOI] [PubMed] [Google Scholar]
- 80.Sen, G. C., and R. M. Ransohoff. 1993. Interferon-induced antiviral actions and their regulation. Adv. Virus Res. 42**:**57-102. [DOI] [PubMed] [Google Scholar]
- 81.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for In vivo activation of interferon regulatory factor 3 In response to virus and double-stranded RNA. J. Biol. Chem. 278**:**9441-9447. [DOI] [PubMed] [Google Scholar]
- 82.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276**:**355-363. [DOI] [PubMed] [Google Scholar]
- 83.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300**:**1148-1151. [DOI] [PubMed] [Google Scholar]
- 84.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98**:**7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276**:**8951-8957. [DOI] [PubMed] [Google Scholar]
- 86.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67**:**227-264. [DOI] [PubMed] [Google Scholar]
- 87.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69**:**6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19**:**623-655. [DOI] [PubMed] [Google Scholar]
- 89.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76**:**3659-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tiwari, R. K., J. Kusari, and G. C. Sen. 1987. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 6**:**3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Umene, K. 1986. Conversion of a fraction of the unique sequences to part of the inverted repeats in the S component of the herpes simplex virus type 1 genome. J. Gen. Virol. 67**:**1035-1048. [DOI] [PubMed] [Google Scholar]
- 92.Uze, G., G. Lutfalla, and K. E. Mogensen. 1995. α and β interferons and their receptor and their friends and relations. J. Interferon Cytokine Res. 15**:**3-26. [DOI] [PubMed] [Google Scholar]
- 93.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acid Res. Mol. Biol. 51**:**123-165. [DOI] [PubMed] [Google Scholar]
- 94.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206**:**901-910. [DOI] [PubMed] [Google Scholar]
- 95.Wathelet, M. G., I. M. Clauss, J. Content, and G. A. Huez. 1988. Regulation of two interferon-inducible human genes by interferon, poly(rI). poly(rC) and viruses. Eur. J. Biochem. 174**:**323-329. [DOI] [PubMed] [Google Scholar]
- 96.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1**:**507-518. [DOI] [PubMed] [Google Scholar]
- 97.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18**:**1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Williams, B. R. G., and S. J. Haque. 1997. Interacting pathways of interferon signaling. Semin. Oncol. 24**:**S9-70-S9-77. [PubMed] [Google Scholar]
- 99.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17**:**1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu, H., J. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95**:**14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu, H., J. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94**:**13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]