Pathview: an R/Bioconductor package for pathway-based data integration and visualization (original) (raw)
Abstract
Summary: Pathview is a novel tool set for pathway-based data integration and visualization. It maps and renders user data on relevant pathway graphs. Users only need to supply their data and specify the target pathway. Pathview automatically downloads the pathway graph data, parses the data file, maps and integrates user data onto the pathway and renders pathway graphs with the mapped data. Although built as a stand-alone program, Pathview may seamlessly integrate with pathway and functional analysis tools for large-scale and fully automated analysis pipelines.
Availability: The package is freely available under the GPLv3 license through Bioconductor and R-Forge. It is available at http://bioconductor.org/packages/release/bioc/html/pathview.html and at http://Pathview.r-forge.r-project.org/.
Contact: luo_weijun@yahoo.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The pathway-based approach has been widely used in high-throughput data analysis (Emmert-Streib and Glazko, 2011; Kelder et al., 2010; Khatri et al., 2012). It has been successfully and routinely applied to gene expression (both microarray and RNA-Seq) (Luo et al., 2009), genetic and GWAS (Wang et al., 2010), proteomic and metabolomics data (Perroud et al., 2006; Xia and Wishart, 2010). Compared with the individual gene/molecule-based approach, pathway analysis is more sensitive, consistent and informative (Luo et al., 2009).
R/Bioconductor has become a primary software environment for high-throughput data analysis and visualization (Gentleman et al., 2004). Numerous pathway analysis methods and data types are implemented in R/Bioconductor, yet there has not been a dedicated and established tool for pathway-based data integration and visualization.
In this article, we introduce a novel package called Pathview. We did a detailed comparison between Pathview and existing pathway tools in R/Bioconductor and other languages in Supplementary Table S2. Pathview provides three features that are rarely implemented well in other tools: (i) fully accessible and functional pathway visualization. It adheres to human readable pathway definitions and layouts like KEGG (Ogata et al., 1999). No previous KEGG base tools provide full graphics, including node/edge attribute modifications, node/edge labels, legends and color keys. (ii) Strong data integration capacity. It integrates and works with data of different types (different omic levels, literature and so forth), IDs, formats, attributes, species and so forth. As far as we know, no other tool provides such extensive data mapping and integration support. (iii) Easy to automate and integrate with pathway analysis tools. Only a few tools can be directly automated and fully integrated into pathway analysis pipelines (Supplementary Table S2: automated analysis column). None of these features are brand new, but surprisingly, few of the existing tools provide satisfactory functionality in these aspects.
2 MAIN FEATURES
2.1 Overall design
The Pathview package can be divided into four functional modules: Downloader, Parser, Mapper and Viewer, as depicted in Supplementary Figure S1. Most importantly, Pathview maps and renders user data on relevant pathway graphs.
2.2 Data visualization
Pathview generates both native KEGG view (Fig. 1b) and Graphviz (Ellson et al., 2002) view (Fig. 1a) for pathways. Both graph styles adhere to human readable pathway definition and layout, yet still allow proper modification and customization of node and edge attributes. KEGG view retains all pathway meta-data, i.e. spatial and temporal information, tissue/cell types, inputs, outputs and connections. This is important for readability and interpretation of pathway biology. Graphviz view provides better control of node and edge attributes, better view of pathway topology and better understanding of the pathway analysis statistics. The different workflows for these two types of view are merged in Pathview (Supplementary Fig. S1). This keeps the user interface simple and consistent.
Fig. 1.
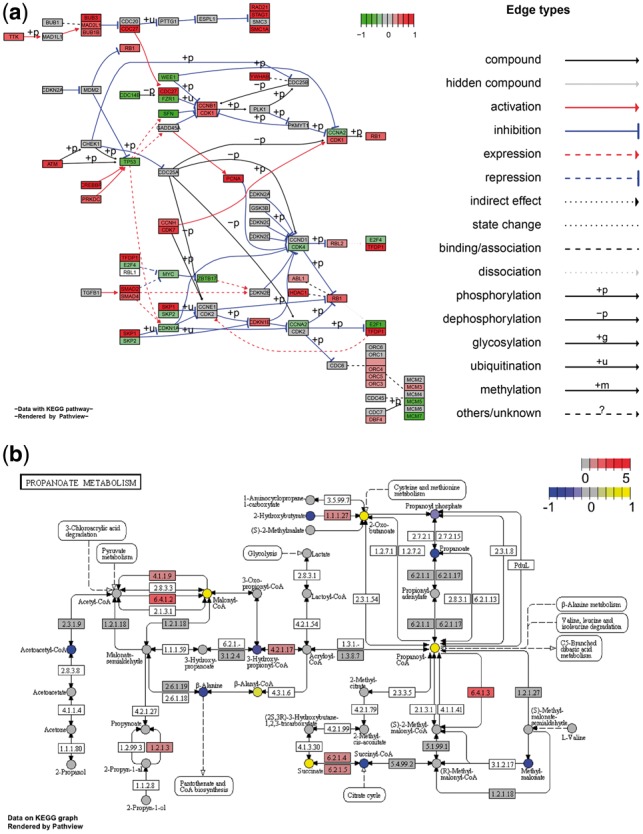
Example Pathview graphs: (a) Graphviz view on a canonical signaling pathway (hsa04110 Cell cycle) with gene data only, (b) KEGG view on a metabolic pathway (hsa00640 Propanoate metabolism) with both discrete gene data and continuous metabolite data integrated. The same graphs at a higher resolution or with a different color scheme are shown in Supplementary Figures S3 and S4
2.3 Data integration
Pathview provides strong support for data integration (Supplementary Table S1). It can be used to integrate, analyze and visualize a wide variety of biological data: gene expression, protein expression, metabolite level, genetic association, genomic variation, literature record and other data types mappable to pathways. Notably, it can be directly used for metagenomic data when the data are mapped to KEGG ortholog pathways. The integrated Mapper module maps 12 types of gene or protein IDs, and 21 types of compound or metabolite related IDs to standard KEGG gene or compound IDs, and also maps between these external IDs. For other types of IDs (for instance, Affymetrix microarray probe set IDs) not included in the common ID lists, Pathview’s auxiliary functions will map user data to pathways when users provide the ID mapping data manually. Pathview applies to pathways for over 2000 species, and species can be specified in multiple formats: KEGG code, scientific name or common name. In addition, Pathview works with different data attributes and formats, both continuous and discrete data (Fig. 1b and Supplementary Table S1), either in matrix or vector format, with single or multiple samples/experiments and so forth.
2.4 Automated and integrated analysis
Pathview is open source, fully automated and error-resistant. Therefore, it seamlessly fits in integrated pathway or gene set analysis workflows.
Pathview can be easily integrated with a wide variety of existing tools in or communicating to R/Bioconductor for high-throughput data analysis and pathway analysis. In the package vignette, we show an integrated analysis using Pathview with another Bioconductor package gage (Luo et al., 2009).
In automated pathway analysis, we frequently use heatmap, scatter plots or stacked line plots to view the perturbation patterns. These plots are simple and can be generated quickly in batches. However, they contain little information beyond the numeric changes. With Pathview, we can view molecular perturbations in intuitive and informative pathway contexts. Importantly, such graphs can be generated equally efficient as the classical scatter or line plots. This will greatly improve the analysis and interpretation of high-throughput molecular data.
KEGG XML data files frequently contain minor deficiencies, inconsistent, incomplete or even error records because of manual curation. These deficiencies adversely affect the parsing, mapping and rendering processes and automation. Pathview accommodates these deficiencies, corrects them or skips the problematic pathway with warning. For example, Pathview Parser corrects for the improper KEGG definition of enzyme-compound interactions by merging and resolving the conflicting ECrel record and associated reactions records (Supplementary Fig. S2). In normal cases, Pathview uses KEGGgraph package (Zhang and Wiemann, 2009) to parse KEGG XML data files.
3 DISCUSSION AND CONCLUSION
Pathview maps and renders user data onto pathway graphs, which are intuitive, informative and well annotated. It integrates and works with a large variety of data types, IDs, formats and attributes. Pathview can be easily combined with other tools for automated and efficient pathway analysis pipelines. Currently, Pathview works with all types and species of KEGG pathways. We plan to support pathways from Reactome (Croft et al., 2011), NCI Pathway Interaction and other databases based on needs in the future.
Funding: UNC general administration (to W.L. and C.B.).
Conflict of Interest: none declared.
Supplementary Material
Supplementary Data
REFERENCES
- Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson J, et al. Graphviz—open source graph drawing tools. In: Mutzel P, Jünger M, Leipert S, editors. Graph Drawing. Berlin Heidelberg: Springer; 2002. pp. 483–484. [Google Scholar]
- Emmert-Streib F, Glazko GV. Pathway analysis of expression data: deciphering functional building blocks of complex diseases. PLoS Comput. Biol. 2011;7:e1002053. doi: 10.1371/journal.pcbi.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder T, et al. Finding the right questions: exploratory pathway analysis to enhance biological discovery in large datasets. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, et al. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, et al. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud B, et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol. Cancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- Zhang JD, Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25:1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data