Exosomal Signaling during Hypoxia Mediates Microvascular Endothelial Cell Migration and Vasculogenesis (original) (raw)
Abstract
Vasculogenesis and angiogenesis are critical processes in fetal circulation and placental vasculature development. Placental mesenchymal stem cells (pMSC) are known to release paracrine factors (some of which are contained within exosomes) that promote angiogenesis and cell migration. The aims of this study were: to determine the effects of oxygen tension on the release of exosomes from pMSC; and to establish the effects of pMSC-derived exosomes on the migration and angiogenic tube formation of placental microvascular endothelial cells (hPMEC). pMSC were isolated from placental villi (8–12 weeks of gestation, n = 6) and cultured under an atmosphere of 1%, 3% or 8% O2. Cell-conditioned media were collected and exosomes (exo-pMSC) isolated by differential and buoyant density centrifugation. The dose effect (5–20 µg exosomal protein/ml) of pMSC-derived exosomes on hPMEC migration and tube formation were established using a real-time, live-cell imaging system (Incucyte™). The exosome pellet was resuspended in PBS and protein content was established by mass spectrometry (MS). Protein function and canonical pathways were identified using the PANTHER program and Ingenuity Pathway Analysis, respectively. Exo-pMSC were identified, by electron microscopy, as spherical vesicles, with a typical cup-shape and diameters around of 100 nm and positive for exosome markers: CD63, CD9 and CD81. Under hypoxic conditions (1% and 3% O2) exo-pMSC released increased by 3.3 and 6.7 folds, respectively, when compared to the controls (8% O2; p<0.01). Exo-pMSC increased hPMEC migration by 1.6 fold compared to the control (p<0.05) and increased hPMEC tube formation by 7.2 fold (p<0.05). MS analysis identified 390 different proteins involved in cytoskeleton organization, development, immunomodulatory, and cell-to-cell communication. The data obtained support the hypothesis that pMSC-derived exosomes may contribute to placental vascular adaptation to low oxygen tension under both physiological and pathological conditions.
Introduction
Exosomes are secreted nanovesicles (30–100 nm diameter) formed through the inward budding of multivesicular bodies (MVBs) that traffic and transfect proteins, mRNAs and miRNAs into target cells [1]. The significance of exosomal signaling in diverse aspects of physiology and pathophysiology has only recently been recognized [2]. Exosomes have now been reported to display immunomodulatory activity [3], [4] containing molecules such as HLA-G5, B7–H1, B7–H3 [5] and syncytian-1 [6] from trophoblast cells, suppression and activation of natural killer cells and macrophages [7], [8]; promote cell migration and metastasis [9], [10], traffic hydrophobic mediators of cell differentiation [11] and viral proteins [12]; and promote allograft survival and induce donor specific allograft tolerance [13]. Of particular relevance to this study, exosomes released from progenitor cells stimulate: endothelial cell migration [14]; tissue vascularization and angiogenesis [15], [16]; induce cell proliferation [17]; and are cardioprotective of ischemia/reperfusion injury [18].
Mesenchymal stem cells are archetypal multipotent progenitor cells that display fibroblastic morphology and plasticity to differentiate into diverse cell types including: osteocytes, adipocytes and endothelial cells. MSCs are isolated from various sources including bone marrow (principal source), adipose tissue and placenta. Within the human placenta, MSC have been isolated from umbilical cord blood and chorionic villi [19], [20] displaying phenotypes comparable to those isolated from bone marrow, including surface antigen expression (CD45−, CD14−, CD19−, CD80+, CD86+, CD40+ and B7H2+) and the capacity to differentiate into multiple linages in vitro. MSC have been implicated in wound healing and display the ability to migrate to sites of injury and engage in tissue repair and regeneration of bone, cartilage, liver tissue or myocardial cells [21], [22]. MSC modulate immune responses in collagen disease, multiple sclerosis and transplants bone marrow and contribute to vasculogenesis, angiogenesis and endothelial repair [23], processes that are fundamental for tissue repair. MSC affect tissue repair through the release of paracrine mediators [24]–[27] including exosomes [28].
MSCs are present in the first trimester human placenta, however, their role in placental vascular development remains to be established. During early pregnancy, the placental vasculature develops under hypoxic conditions. During the first trimester, placental PO2 is ∼ 2.6% before placento-maternal perfusion is established. At around 12 weeks of pregnancy, the placenta is perfused with maternal blood and PO2 increases to ∼ 8% [29], [30]. There is a paucity of information about the role of MSC and, in particular, the release of exosomes from MSC during this critical period of vascular development. Of note, however, Hofmann et al., [31], recently proposed that exosomes may function as part of an oxygen sensing mechanism that promotes vasculogenesis and angiogenesis. We hypothesize that: (i) exosomes released by pMSC act paracellularly to promote cell migration and angiogenesis within the placental villus tree; and (ii) that the release of exosomes from pMSC is responsive to changes in oxygen tension.
The aim of this study, therefore, was to establish the effect of oxygen tension on the release of exosomes from pMSC; and the effects of pMSC-derived exosomes on the migration and angiogenic tube formation of human placental microvascular endothelial cells (hPMEC). pMSC-derived exosomes promote hPMEC cell migration and tube formation in vitro. The release and bioactivity of pMSC-derived exosomes is oxygen tension dependent. The data obtained are consistent with the hypothesis that pMSC-derived exosomes are released under hypoxic conditions and promote angiogenesis within the developing placenta.
Materials and Methods
First Trimester and Term Placental Collection
Tissue collection was approved by the Human Research Ethics Committees of the Royal Brisbane and Women’s Hospital, and the University of Queensland (HREC/09/QRBW/14). All experiments and data collection and analyses were conducted with an ISO 17025 and 21 CFR part 11 conforming laboratory environment. Written informed consent was obtained from women for the use of placental tissue for research purposes after clinically indicated termination of pregnancy in compliance with national research guidelines.
Isolation of Placental Mesenchymal Stem Cells
pMSC were isolated, from placental villi by enzymatic digestion using protocols adapted from Steigman & Fauza [32]. Briefly, placental chorionic villi (n = 6; 8–12 weeks gestational age) were separated from the remainder of the placenta unit and were washed in PBS. The villi were minced into small pieces and were transferred in to 50 ml tubes. The tissues were enzymatic digestion with dispase (2.4 U/ml) and collagenase (240 U/ml) made in PBS. The tissues were digested for 1 hr at 37°C on a rocker. The single cell suspension was then filtered through a 100 µm mesh into a new tube. The cells were centrifuged for 15 mins at 500×g at RT and the pellet was resuspended in 10 ml cDMEM. pMSC were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies™, Carlsbad, CA), supplemented with 10% fetal bovine serum, 100 IU/mL penicillin, and 100 µg/mL streptomycin (Life Technologies™), at 37°C with 5% CO2. pMSC were characterised by well-established cell surface markers. All cells used in this study were passaged less than 6.
MSC Differentiation Assays
The differentiation potential of placental villi MSC was established according to previously published methods [33]. For adipogenesis, 2×105 pMSC were seeded in 6 well plates until confluent and differentiation was induced by indomethacin (60 µM) dexamethasone (1 µm), insulin (5 µg/ml) and isobutylmethylxanthine (IBMX) (0.5 mM). After 21 days, cells were fixed with 10% formalin and stained with Oil Red O. Adipogenic differentiation was determined by the appearance of Oil Red O. Osteogenic differentiation was induced by culturing 3×105 cells in 6 well plates in the presence of osteogenic induction media containing dexamethasone (0.1 µM), β-glycerol phosphate (10 mM) and L-ascorbate-2-phosphate (0.2 mM) for 21 days. Cells were fixed in 10% formalin and stained with Alizarin red. Differentiation was determined by the appearance of red deposits, representing areas of mineralized calcium. All reagents were from Sigma-Aldrich. Staining for both adipogenic and osteogenic assays was visualized using bright field phase contrast microscopy.
Isolation of hPMEC
The effects of exosomes on endothelial cell migration and angiogenesis were assessed using human placenta microvascular endothelial cells (hPMEC). hPMEC were isolated as previously described [34]. In brief, chorionic villi obtained from placental tissue samples (∼4 cm3 of the chorionic villous) were digested with trypsin/EDTA (0.25/0.2%, 20 min, 37°C) followed by 0.1 mg/ml collagenase (2 h, 37°C, Type II Clostridium histolyticum; Boehringer, Mannheim, Germany) in medium 199 (M199, Gibco Life Technologies, Carlsbad, CA, USA) Digested tissue was resuspended in M199 containing 5 mM D-glucose, 20% newborn calf serum (NBCS), 20% fetal calf serum (FCS), 3.2 mM L-glutamine and 100 U/ml penicillin streptomycin (primary culture medium, PCM), and filtered through a 55 µm pore size Nylon mesh. Filtered cell suspension was transferred into a 1% gelatin-coated T25 culture flask for culture (37°C, 5% O2, 5% CO2) in PCM. After 5 days, confluent cells were trypsinized (trypsin/EDTA 0.25/0.2%, 3 min, 37°C) and subjected to CD31 (against platelet endothelial cell adhesion molecule 1, PECAM-1)-positive immunoselection using Dynabeads CD31 microbeads from MACS® (Miltenyi Biotech, Bergisch-Gladbach, Germany). Endothelial cells immunoselection was performed mixing anti-CD31 antibody-magnetic coated microbeads with the cell suspension (48×103 beads/ml cell suspension, 20 min, 4°C). Suspension medium was discarded and cells attached to the magnetic microbeads were collected and washed (3 times) in HBSS (37°C). CD31-coated microbead-attached cells were resuspended in PCM containing 10% NBCS and 10% FCS, and cultured under standard conditions (37°C, 5% CO2) until confluence in passage 3. Immunocytochemistry analysis established that more than 96% of cells in the endothelial preparations used in the present study, were positive for von Willebland Factor (vWF) and CD31 (data not shown).
Flow Cytometry
The expression of cell surface and intracellular antigens was assessed by flow cytometry (FACScalibur™, Becton Dickinson, San Jose, CA, USA). To identify intra-cellular antigens, cells were detached, blocked with 1% bovine serum albumin (BSA; Sigma, St. Louis, MO) in phosphate buffered saline (PBS, Life Technologies™) then fixed with 0.01% paraformaldehyde (PFA) (Sigma) and permeabilized with 0.5% Triton X-100. To characterize the expression of cell surface and intracellular antigens, cells were detached and blocked with 1% BSA and incubated with specific anti-human primary antibodies, either conjugated with PE, FITC or PE-Cy5 or unconjugated. For unconjugated antibodies, cells were subsequently washed with 1% BSA and incubated with secondary goat anti-murine IgM PE (Santa Cruz Biotechnology®, Santa Cruz, CA, USA). All samples were analyzed in triplicate by FACScalibur™ flow cytometry (Becton Dickinson). Positive controls were hESC and negative controls were IgG or IgM primary antibody-specific isotype controls.
Hypoxia
The effects of oxygen tension on the release of exosomes from pMSC were assessed by incubating cells for 48 h (in exosome-free culture medium) under an atmosphere of 5% CO2-balanced N2 to obtain 1%, 3% or 8% O2 (pO2 ∼6.75, ∼20.25 or ∼54 mmHg, respectively) in an automated PROOX 110-scaled hypoxia chamber (BioSpherics™, Lacona, NY, USA). Cell number and viability was determined after each experimental treatment by Trypan Blue exclusion and Countess® Automated cell counter (Life Technologies™). Proliferation data was collected for all the experimental conditions and in particular to assess the effects of proliferation hypoxic conditions using a real-time cell imaging system (IncuCyte™ live-cell ESSEN BioScience Inc, Ann Arbor, Michigan, USA). In all experiments, viability remained at >95%. Incubation media pO2 and pH were independently confirmed using a blood gas analyzer (Radiometer®, Brønshøj, Denmark) and NeoFox oxygen probe (Ocean Optics ™, Dunedin, FL, USA). HIF expression was used in Western blot analysis as a positive control for hypoxia in MSC (data not show).
Isolation and Purification of pMSC Exosomes
Exosomes were isolated from cell-free pMSC as previously described [35]. In brief, pMSC-conditioned media was centrifuged at 300×g for 15 min, 2000×g for 30 min, and 12000×g for 45 min to remove whole cells and debris. The resultant supernatant were passed through a 0.22 µm filter sterilize Steritop™ (Millipore, Billerica, MA, USA) and then centrifuged at 100,000×g for 75 min (Thermo Fisher Scientific Ins., Asheville, NC, USA, Sorvall, SureSpin™ 630/36, fixed angle rotor). The pellet was resuspended in PBS, washed and re-centrifuged (100,000×g, 75 min). The pellet was resuspended in PBS, layered on a cushion of 30% (w/v) sucrose and centrifuged at 110,000 g for 75 min. The fraction containing pMSC exosomes (∼3.5 ml, 1.1270 density using OPTi digital refractometer (Bellingham+Stanley Inc., Lawrenceville, GA, USA) was recovered with an 18-G needle and diluted in PBS, and then ultracentrifuged at 110 000×g of 70 min. Recovered exosomes were resuspended in 50 µl PBS and their protein contents were determined using the Bradford assay (Bio-Rad DC) [35]. Exosome samples (5 µl) were prepared by adding RIPA buffer (50 mM Tris, 1% Triton×100, 0.1% SDS, 0.5% DOC, 1 mM EDTA, 150 mM NaCl, protease inhibitor) directly to exosomes suspended in PBS and sonicated at 37°C for 15 s three times to lyse exosome membrane and solubilise the proteins. Bovine serum albumin (BSA) diluted in RIPA buffer and PBS mixture (1∶1) were prepared as protein standards (0, 200, 400, 600, 800, 1000, 1500 µg/mL). Standards and samples (exosomes) were transferred to 96-well plates and procedures outlined by the manufacture were followed. In brief, alkaline copper tartrate solution (BIO-RAD, USA) and dilute Folin Reagent (BIO-RAD, USA) were added to the samples and incubated for 15 min. The absorbance was read at 750 nm with Paradigm Detection Platform (Beckman Coulter, USA).
Transmission Electron Microscopy
The exosome fraction isolated by differential and buoyant density gradient centrifugation was assessed by transmission electron microscopy. Exosome pellets (as described above) were fixed in 3% (w/v) glutaraldehyde and 2% paraformaldehyde in cacodylate buffer, pH 7.3. Five microlitres of sample was then applied to a continuous carbon grid and negatively stained with 2% uranyl acetate. The samples were examined in an FEI Tecnai 12 transmission electron microscope (FEI™, Hillsboro, Oregon, USA).
Western Blot
Exosome proteins separated by polyacrylamide gel electrophoresis were transferred to Immobilon-®FL polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA) and probed with primary mouse monoclonal anti-CD63 (1∶2000), anti-CD81 (1∶1500) or anti-CD9 (1∶1500) as previously described [35] for specific exosome markers. Membranes were washed in Tris buffer saline Tween, and incubated (1 h) in TBST/0.2% BSA containing horseradish peroxidase-conjugated goat anti-mouse antibody. Proteins were detected by enhanced chemiluminescence with the SRX-101A Tabletop Processor (Konica Minolta, Ramsey, NJ, USA). The relative intensity of the bands was determined by densitometry using the GS-800 Calibrated Densitometer (Bio-Rad Laboratories, Hercules, CA, USA).
Migration and Tube Formation Assay
To assess the effect of exosomes on endothelial cell tube formation, hPMEC were cultured in 96 or 48-well culture plates (Corning Life Science, Tewksbury, MA, USA) according to the manufacturer’s instructions and visualized using a real-time cell imaging system (IncuCyte™ live-cell ESSEN BioScience Inc, Ann Arbor, Michigan, USA). Cells were imaged every hour to monitor treatment-induced cell migration, tube formation, confluence and morphologic changes. Cell migration was assessed by scratch assays, in which, hPMEC were grown to confluence and then a scratch was made using a 96-pin WoundMaker™. The wells were washed with PBS to remove any debris and incubated in the presence of 0 (control) 5, 10 or 20 µg protein/ml of pMSC-derived exosome isolated from cells cultured under 1%, 3% or 8% O2. Wound images were automatically acquired and registered by the IncuCyte™ software system. Typical kinetic updates were recorded at 2 h intervals for the duration of the experiment (48 h). The data were then analysed using an integrated metric: Relative Wound density. For the tube formation assay, 48-well culture plates on ice were incubated with 144 µl of chilled BD Matrigel matrix (10 mg/ml) per well at 37°C for 60 min. hPMEC (6×104) were resuspended in culture medium with the indicated concentration of pMSC-derived exosomes (5, 10 or 20 µg/ml) and incubated for up to 24 h at 37°C. The number of networks formed was determined using the IncuCyte™ system.
Proliferation Assay
A real-time imaging system (IncuCyteTM) was used to measure cell proliferation using non-label cell monolayer confluence approach. pMSC confluence was measure before and after the treatment (1%, 3% and 8% O2, 48 h). IncuCyteTM provide the capability to acquire high quality, phase-contrast images and an integrated confluence metric as a surrogate for cell number [36]. We used similar approach for to determine the effect of pMSC-derived exosomes on hPMEC proliferation during the migration assay.
Proteomic Analysis of Exosomes by Mass Spectrometry (MS)
Isolated exosomes were solubilised in 8 M urea in 50 mM ammonium bicarbonate, pH 8.5, and reduced with DTT for 1 h. Proteins were then alkylated in 10 mM iodoacetic acid (IAA) for 1 h in the dark. The sample was diluted to 1∶10 with 50 mM ammonium bicarbonate and digested with trypsin (20 µg) at 37°C for 18 h. The samples were desalted by solid phase extraction using a STAGE tip protocol (Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics). The eluted peptides were dried by centrifugal evaporation to remove acetonitrile and redissolved in Solvent A. The resulting peptide mixture was analysed by Liquid Chromatography (LC)/Mass Spectrometry (MS) LC-MS/MS on a 5600 Triple TOF mass spectrometer (AB Sciex, Framingham, U.S.A.) equipped with an Eksigent Nanoflow binary gradient HPLC system and a nanospray III ion source. Solvent A was 0.1% formic acid in water and solvent B was 0.1% fomic acid in acetonitrile. MS/MS spectra were collected using Information Dependent Acquisition (IDA) using a survey scan (m/z 350–1500) followed by 25 data-dependent product ion scans of the 25 most intense precursor ions. The data were searched using MASCOT and Protein Pilot search engines.
Functional Analysis of Exosome Proteome
Proteins identified by MS/MS were analyzed by PANTHER (Protein Analysis THrough Evolutionary Relationships; http://www.pantherdb.org). This software allows the prediction of classify proteins (and their genes) in order to facilitate high-throughput analysis. The classified proteins were classified according to their biological process and molecular function. Differentially expressed proteins were analyzed further by bioinformatic pathway analysis (Ingenuity Pathway Analysis [IPA]; Ingenuity Systems, Mountain View, CA; www.ingenuity.com).
Statistical Analysis
Data are represented as mean ± SEM, with n = 6 different cells culture (i.e. biological replicates) of pMSC isolated from first trimester pregnancies and n = 4 different cell cultures (i.e biological replicates) of hPMEC isolated from term placenta. Comparisons between two and more groups were performed by means of unpaired Student’s _t_-test and analysis of variance (ANOVA), respectively. If the ANOVA demonstrated a significant interaction between variables, post hoc analyses were performed by the multiple-comparison Bonferroni correction test. Statistical significance was defined at least p<0.05.
Results
Characterization of Exosome from Placental Mesenchymal Stem Cells
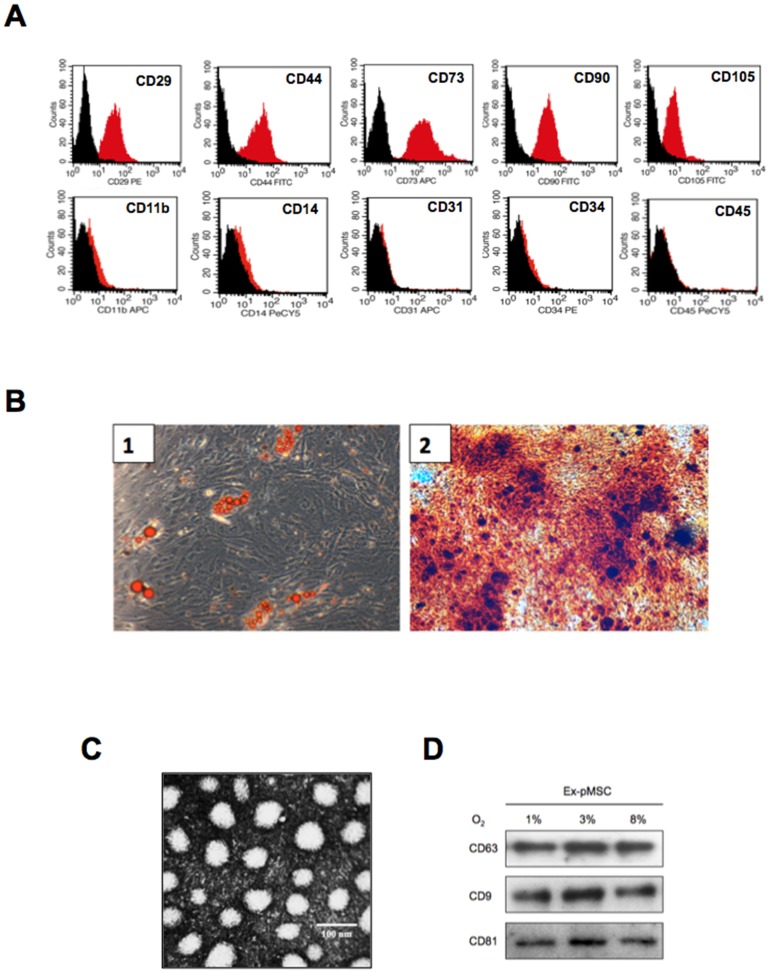
Cell surface protein expression by pMSC was characterized using flow cytometric analysis. pMSC were labelled with monoclonal antibodies specific for markers indicated in each histogram (Figure 1A). pMSC isolated from first trimester placental villi were positive for CD29+, CD44+, CD73+, CD90+, CD105+ (top panel) and negative for hematopoietic and endothelial markers: CD11b-, CD14−, CD31−, CD34−, CD45− (lower panel). When pMSC were stimulated under adipogenic and osteogenic conditions, they showed characteristic of adipocytes (Figure 1B1; formation of lipid vacuoles) and osteoblast cells (Figure 1B2; red deposits, representing areas of mineralized calcium), respectively. The exosomal particulate fraction isolated from pMSC was examined under transmission electron microscopy. Exosomes were identified as small vesicles between 40–100 nm in a cup-shaped form (Figure 1C). The particulate fraction was further characterized by the expression of specific exosome markers: CD63; CD9; and CD81 by Western blot analysis (Figure 1D).
Figure 1. Characterisation of exosomes from placental mesenchymal stem cell (pMSC).
Cells were isolated from chorionic villi obtained from first trimester pregnancy and cultured under standard conditions. Exosomes were isolated from pMSC supernatant as was indicated in Methods. (A) Representative flow cytometry histogram of pMSC labeled with positive markers such as CD29, CD44, CD73, CD90 and CD105 (top panel) or negative markers such as CD11b, CD14, CD31, CD34 and CD45 (bottom panel). Black solid peaks represent the isotype controls and the red solid peak represents the marker indicated. (B) Mulit differntiation potential of first trimester placental chorionic villi. 1, Adipogenesis was determined using oil red O staining of lipid droplets after 21 days in adipogenic media. 2, Osteogenesis was determined using alizarin red staining for the mineral matrix deposition after 21 days in osteogenic media. (C) Electron micrograph of exosomes isolated by ultracentrifuge from pMSC. (D) pMSC were exposed to 1%, 3% or 8% O2 during 48 hours and then exosomes proteins were isolated. Samples in each condition were analyzed by western blot after the separation of 20 ug of exosomes protein (same amount of exosome protein lead) for the presence of CD63, CD9 and CD81. In B, Scale bar 100 nm.
Effect of Oxygen Tension on Exosome Release
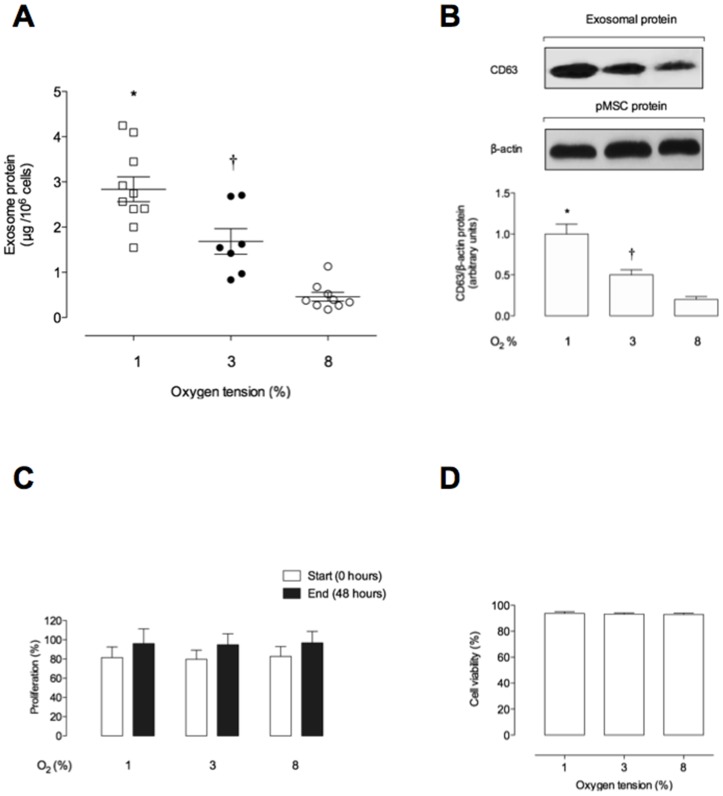
To determine the effects of oxygen tension on the release of exosomes from pMSC, cells were incubated under atmospheres of 1%, 3% or 8% O2 and the exosomes released were quantified (as total exosomal protein µg/106 pMSC). Under these conditions, exosomal protein release averaged 2.8±0.27, 1.6±0.28 and 0.46±0.01 µg protein/106 pMSC, respectively. Exosome release from pMSC was significantly inversely correlated to oxygen tension (ANOVA, p<0.001, n = 5; Figure 2A). Furthermore, the relative abundance of the specific exosome marker CD63 in this particulate fraction displayed a similar inverse correlation to oxygen tension, as assessed by Western blot (Figure 2B). During the time course of these experiments, cell proliferation was not significantly affected by oxygen tension (i.e. 1%, 3% or 8% O2) (Figure 2C). The effect of oxygen tension on exosome release was not associated with a decrease in cell viability (Figure 2D).
Figure 2. The level of pMSC-derived exosomes compared to low oxygen tension.
Exosomes were isolated from pMSC supernatant exposed to 1%, 3% or 8% oxygen per 48 h. (A) Levels of exosomes are presented as protein concentration from 1×106 pMSC cell. (B) Same volume of exosome pellet loaded and analyzed by western blot for CD63 and β-actin in exosome from pMSC and cells, respectively. Lower panel: CD63/β-actin ratio densitometries from data in top panel normalized to 1 in 1% O2. (C) Effect of low oxygen tension on pMSC proliferation. (D) Trypan blue dye exclusion test to show residual pMSC cell viability exposed to 1%, 3% or 8% O2. Values are mean ± SEM. In A and B, *P<0.001 versus all condition; † p<0.001 versus 8% O2.
Effect of pMSC-derived Exosomes on Cell Migration
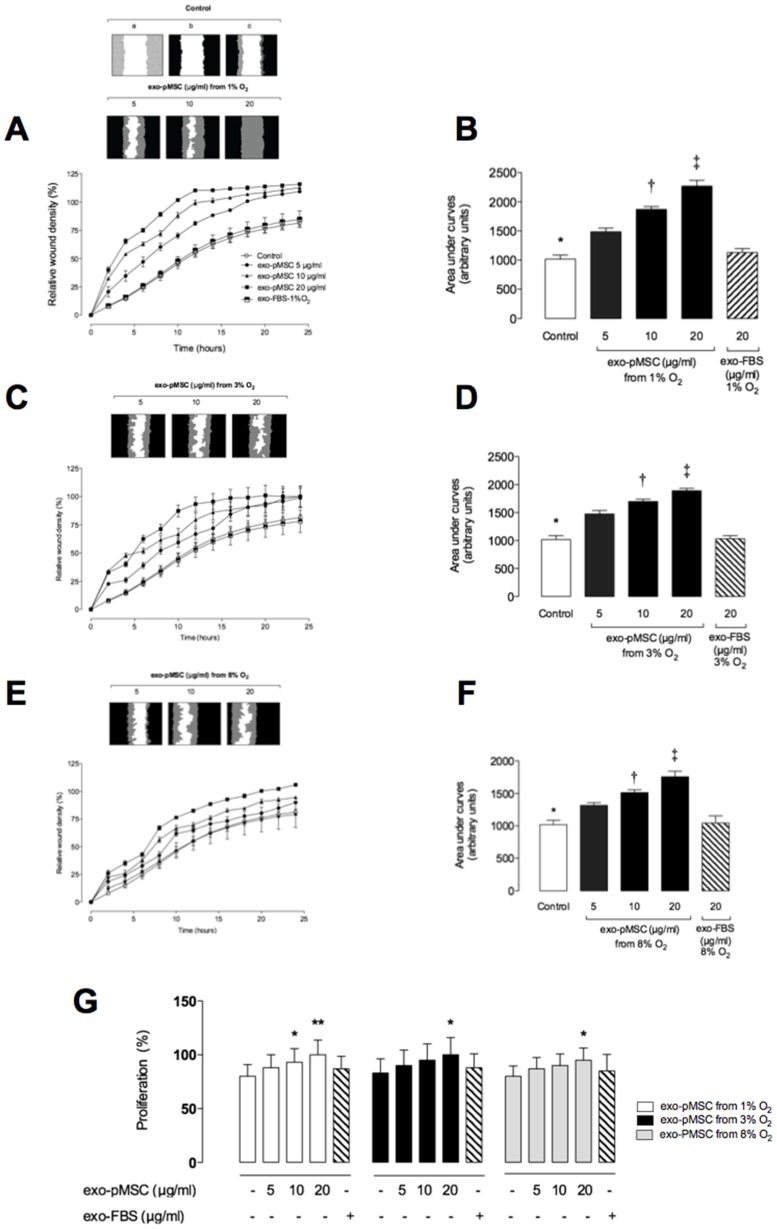
The effects of exosomes (5, 10 or 20 µg protein/ml) isolated from pMSC cultured under 1%, 3% or 8% O2 on hPMEC migration are presented in Figure 3 A, C and E. pMSC exosomes significantly increased hPMEC migration in a time- and dose-dependent manner (p<0.005, n = 6). In addition, the effect on hPMEC migration was greater when exosomes were prepared from cells cultured under low oxygen tensions. Using the IncuCyte live cell imaging enabled non-invasive system, the cell proliferation based on area metric (confluence) was measurement. Exosomes isolated from pMSC cultures under 1% O2 increased significantly the hPMEC proliferation in ∼1.18-fold and ∼1,25-fold with 10 µg/ml and 20 µg/ml, respectively (Figure 4B). Furthermore, exosomes isolated from pMSC cultured under 3% and 8% O2 increased hPMEC proliferation in ∼1.21-fold and ∼1,18-fold using 20 µg/ml, respectively. We did not find significant effect of FBS-derived exosome on hPMEC migration and proliferation. Half-maximal stimulatory time (ST50) and half-maximal stimulatory concentration (SC50) values are presented in Table 1.
Figure 3. Exosomes increases cell migration in hPMEC.
hPMEC were grown to confluence in complete media, wound were made using 96 well WoundMaker and culture in absence (○) or presence (• 5, ▴ 10 or ▪ 20 µg/ml) of exosomal protein obtained from pMSC exposed to different oxygen tension. (A) Top: a, hPMEC image immediately after wounding; b, Graphical representation showing the calculation of initial wound width; c, Graphical representation of cell migration at the midpoint of the experiment. Bottom: The time course of the concentration-dependent effect of exosomal protein from 1% O2 on hPMEC, (C) 3% O2 or (E) 8% O2. (B) Area under curves from data in A, (D) from data in C, (F) from data in E. (G) Effect of pMEC-derived exosomes on hPMEC proliferation. Data represent an n = 6 well each point. Values are mean ± SEM. In B, D and F: *p<0.005 versus all condition; † P<0.005 versus 5 µg/ml; ‡ p<0.005 versus 10 µg/ml. In G, *p<0.005 versus control (−) with exo-pMSC from 1%, 3% or 8% O2; **p<0.001 versus control (−) with exo-pMSC from 1% O2.
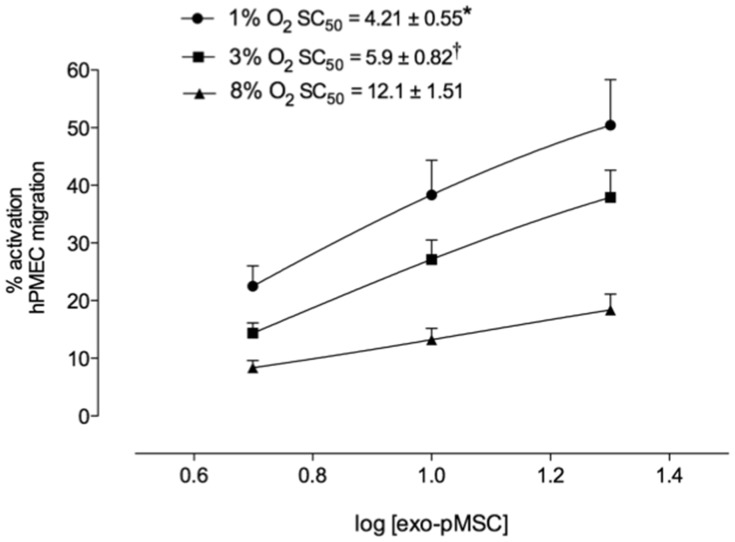
Figure 4. Concentration response of exosomes on hPMEC migration.
Activation analysis of exosomes effect on hPMEC migration. Concentration response of exosomal protein from pMSC exposed to 1% (•), 3% (▪) or 8% (▴) O2 on hPMEC migration. Insert: half-maximal stimulatory concentration (SC50) at 6 h. Data represent an n = 6 well each point. Values are mean ± SEM. Insert: *p<0.001 versus all condition; † p<0.005 versus 8% O2.
Table 1. Kinetic characteristic of exosome effects on hPMEC migration.
| Condition | Parameter | |
|---|---|---|
| Exosome [µg/ml) | ST 50 | |
| Control | – | 9.9±0.19 |
| 1% O2 | 5 | 7.9±0.19* |
| 10 | 4.0±0.18* | |
| 20 | 3.9±0.15*† | |
| 3% O2 | 5 | 8.0±0.25* |
| 10 | 6.2±0.30* | |
| 20 | 5.9±0.21* | |
| 8% O2 | 5 | 8.2±0.18* |
| 10 | 7.8±0.17* | |
| 20 | 6.4±0.21* |
Exosome activation was concentration dependent for each condition (half-maximal stimulatory concentration (SC 50) = 4.2±0,5 from 1% O2: versus 5.9±0.6 and 12±1.2 µg/ml from 3% and 8% O2, respectively) (Figure 4).
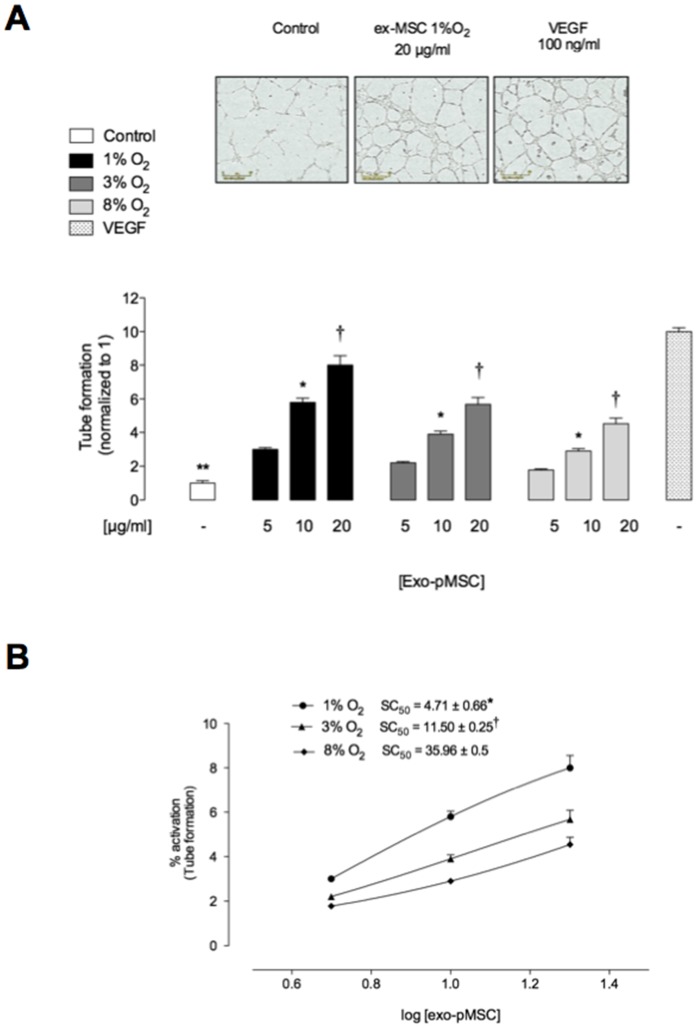
Effect of pMSC-derived Exosomes on in vitro Tube Formation
In vitro angiogenic tube formation assays were used as a surrogate endpoint to assess the angiogenic effects of pMSC-derived exosomes. pMSC-derived exosomes significantly increased tube formation by hPMEC in a dose- and time-dependent manner when compared to vehicle-treated cells (p<0.005, Figure 5A) and inversely correlated to oxygen tension. In addition, exosome-induced tube formation was significantly greater when exosomes were prepared from cells grown under low oxygen tensions (Figure 5A). Half-maximal stimulatory time was 4.71±0.66, 11.50±0.25 and 35.96±0.5 µg/ml for exosomes treatment from pMSC exposed to 1%, 3% and 8% oxygen, respectively.
Figure 5. Exosomes from hypoxia increases microvascular tube formation in a dose-dependent manner.
hPMEC were incubated in Matrigel in absence or presence of different exosomal protein concentration from pMSC exposed to 1%, 3% or 8% O2. (A) Quantitative analysis of the total tube formation. (B) Concentration response from data in A. insert: half-maximal stimulatory concentration (SC50) at 16 h. Values are mean ± SEM. In A, **p<0.001 versus all condition; *p<0.005 versus corresponding values in 5 µg/ml, † p<0.005 versus corresponding values in 10 or 5 µg/ml. In B, *P<0.005 versus all values; † p<0.005 versus values in 8% O2.
Proteomic Analysis of pMSC-derived Exosome
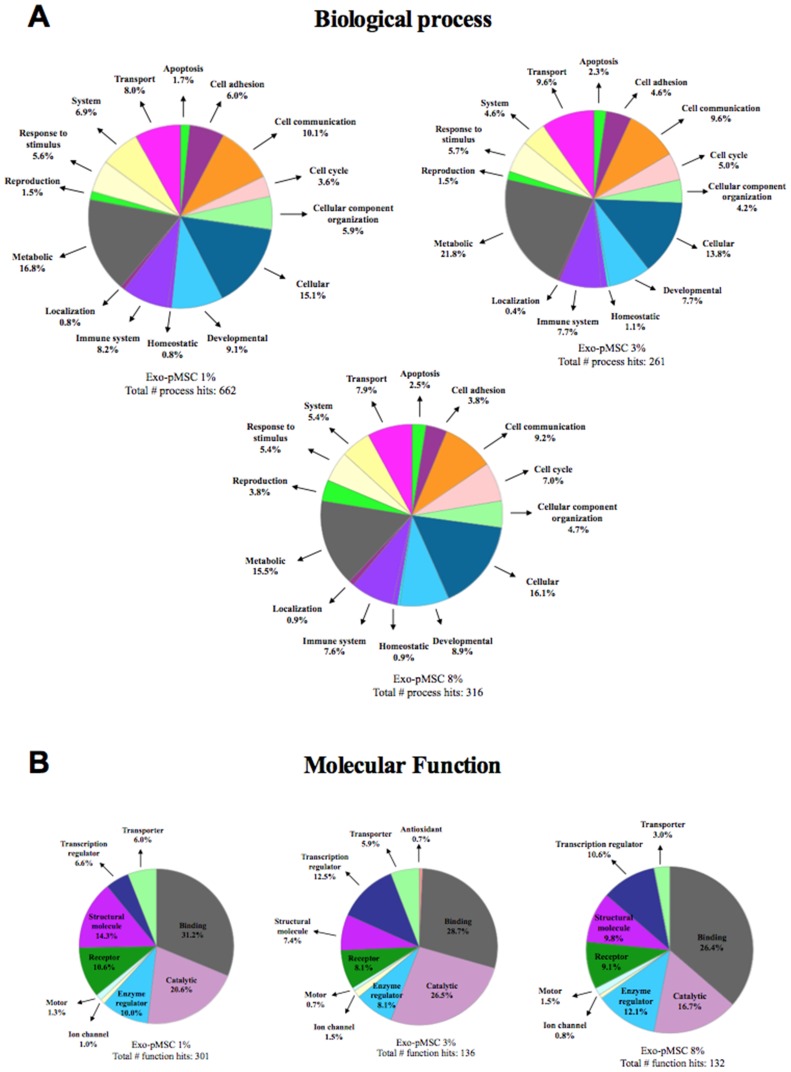
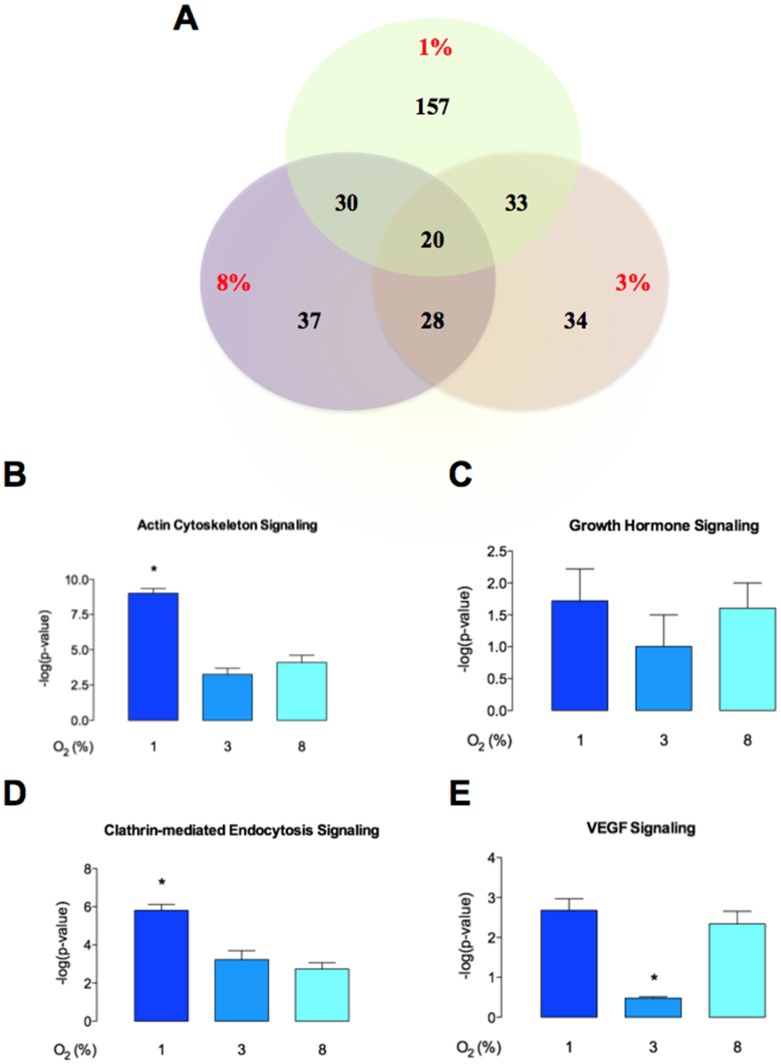
Mass spectrometry analysis identified over 200 exosomal proteins (Table 2). Data were subjected to ontology and pathway analysis using Panther and Gene Ontology algorithms and classified based on biological process and molecular function (Figure 6). In biological process, the most clusters identified were: cellular processes, cell communication, developmental and transport (Figure 6A). In molecular functions, the proteins related to binding and catalytic activity were the greatest recognized (Figure 6B). IPA analysis identified 157 proteins only present in exo-pMSC-1%O2 versus 34 and 37 individual proteins present in exo-pMSC-3%O2 and exo-pMSC-8%O2, respectively.(Figure 7A). Finally, the canonical pathways associated with our proteins defined by IPA Core analysis and related with cell migration were: actin cytoskeleton signaling, growth hormone signaling, clathrin-mediated endocytosis signaling, and VEGF signaling (Figure 7B-E). Furthermore, canonical pathways were associated with highest protein number in exosomes isolated from pMSC exposed to 1% O2 versus 3% and 8% O2.
Table 2. List of proteins identified in exosomes from pMSC exposed to different oxygen level.
| Exo-pMSC-1%O2 | ||||
|---|---|---|---|---|
| ID | Symbol | Entrez Gene Name | Location | Type(s) |
| A2MG_HUMAN | A2M | alpha-2-macroglobulin | Extracellular Space | transporter |
| ACTS_HUMAN | ACTA1 | actin, alpha 1, skeletal muscle | Cytoplasm | other |
| ACTB_HUMAN | ACTB | actin, beta | Cytoplasm | other |
| ACTN1_HUMAN | ACTN1 | actinin, alpha 1 | Cytoplasm | other |
| SAHH_HUMAN | AHCY | adenosylhomocysteinase | Cytoplasm | enzyme |
| FETUA_HUMAN | AHSG | alpha-2-HS-glycoprotein | Extracellular Space | other |
| AIM1_HUMAN | AIM1 | absent in melanoma 1 | Extracellular Space | other |
| ALBU_HUMAN | ALB | albumin | Extracellular Space | transporter |
| ALDOA_HUMAN | ALDOA | aldolase A, fructose-bisphosphate | Cytoplasm | enzyme |
| AMOL2_HUMAN | AMOTL2 | angiomotin like 2 | Plasma Membrane | other |
| ANXA1_HUMAN | ANXA1 | annexin A1 | Plasma Membrane | other |
| ANXA2_HUMAN | ANXA2 | annexin A2 | Plasma Membrane | other |
| ANXA5_HUMAN | ANXA5 | annexin A5 | Plasma Membrane | other |
| APOA1_HUMAN | APOA1 | apolipoprotein A-I | Extracellular Space | transporter |
| APOB_HUMAN | APOB | apolipoprotein B (including Ag(x) antigen) | Extracellular Space | transporter |
| APOC3_HUMAN | APOC3 | apolipoprotein C-III | Extracellular Space | transporter |
| APOE_HUMAN | APOE | apolipoprotein E | Extracellular Space | transporter |
| ARF5_HUMAN | ARF5 | ADP-ribosylation factor 5 | Cytoplasm | enzyme |
| ARHG2_HUMAN | ARHGEF2 | Rho/Rac guanine nucleotide exchange factor (GEF) 2 | Cytoplasm | other |
| ARPC3_HUMAN | ARPC3 | actin related protein 2/3 complex, subunit 3, 21 kDa | Cytoplasm | other |
| ASB18_HUMAN | ASB18 | ankyrin repeat and SOCS box containing 18 | unknown | other |
| ASH1L_HUMAN | ASH1L | ash1 (absent, small, or homeotic)-like (Drosophila) | Nucleus | transcription regulator |
| A16L1_HUMAN | ATG16L1 | autophagy related 16-like 1 (S. cerevisiae) | Cytoplasm | other |
| AT8B1_HUMAN | ATP8B1 | ATPase, aminophospholipid transporter, class I, type 8B, member 1 | Plasma Membrane | transporter |
| ATRN_HUMAN | ATRN | attractin | Extracellular Space | other |
| B3GN1_HUMAN | B3GNT1 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 | Cytoplasm | enzyme |
| BCL3_HUMAN | BCL3 | B-cell CLL/lymphoma 3 | Nucleus | transcription regulator |
| BCDO1_HUMAN | BCMO1 | beta-carotene 15,15′-monooxygenase 1 | Cytoplasm | enzyme |
| BEND5_HUMAN | BEND5 | BEN domain containing 5 | Cytoplasm | other |
| BMP1_HUMAN | BMP1 | bone morphogenetic protein 1 | Extracellular Space | peptidase |
| CJ118_HUMAN | C10orf118 | chromosome 10 open reading frame 118 | unknown | other |
| C1QT3_HUMAN | C1QTNF3 | C1q and tumor necrosis factor related protein 3 | Extracellular Space | other |
| CO3_HUMAN | C3 | complement component 3 | Extracellular Space | peptidase |
| CO5_HUMAN | C5 | complement component 5 | Extracellular Space | cytokine |
| CI114_HUMAN | C9orf114 | chromosome 9 open reading frame 114 | Nucleus | other |
| CALRL_HUMAN | CALCRL | calcitonin receptor-like | Plasma Membrane | G-protein coupled receptor |
| CAMP2_HUMAN | CAMSAP2 | calmodulin regulated spectrin-associated protein family, member 2 | unknown | other |
| CAND1_HUMAN | CAND1 | cullin-associated and neddylation-dissociated 1 | Cytoplasm | transcription regulator |
| CC147_HUMAN | CCDC147 | coiled-coil domain containing 147 | Extracellular Space | other |
| CB077_HUMAN | CCDC173 | coiled-coil domain containing 173 | unknown | other |
| CCD60_HUMAN | CCDC60 | coiled-coil domain containing 60 | unknown | other |
| CCD73_HUMAN | CCDC73 | coiled-coil domain containing 73 | unknown | other |
| CD44_HUMAN | CD44 | CD44 molecule (Indian blood group) | Plasma Membrane | enzyme |
| CD59_HUMAN | CD59 | CD59 molecule, complement regulatory protein | Plasma Membrane | other |
| CDCA2_HUMAN | CDCA2 | cell division cycle associated 2 | Nucleus | other |
| CFAH_HUMAN | CFH | complement factor H | Extracellular Space | other |
| CGRF1_HUMAN | CGRRF1 | cell growth regulator with ring finger domain 1 | unknown | other |
| CLCF1_HUMAN | CLCF1 | cardiotrophin-like cytokine factor 1 | Extracellular Space | cytokine |
| CNOT1_HUMAN | CNOT1 | CCR4-NOT transcription complex, subunit 1 | Cytoplasm | other |
| COG2_HUMAN | COG2 | component of oligomeric golgi complex 2 | Cytoplasm | transporter |
| COCA1_HUMAN | COL12A1 | collagen, type XII, alpha 1 | Extracellular Space | other |
| CO1A1_HUMAN | COL1A1 | collagen, type I, alpha 1 | Extracellular Space | other |
| CO1A2_HUMAN | COL1A2 | collagen, type I, alpha 2 | Extracellular Space | other |
| CO6A1_HUMAN | COL6A1 | collagen, type VI, alpha 1 | Extracellular Space | other |
| CO6A2_HUMAN | COL6A2 | collagen, type VI, alpha 2 | Extracellular Space | other |
| CO6A3_HUMAN | COL6A3 | collagen, type VI, alpha 3 | Extracellular Space | other |
| COMP_HUMAN | COMP | cartilage oligomeric matrix protein | Extracellular Space | other |
| CBPA1_HUMAN | CPA1 | carboxypeptidase A1 (pancreatic) | Extracellular Space | peptidase |
| SDF1_HUMAN | CXCL12 | chemokine (C-X-C motif) ligand 12 | Extracellular Space | cytokine |
| DEN1A_HUMAN | DENND1A | DENN/MADD domain containing 1A | Plasma Membrane | other |
| MYCPP_HUMAN | DENND4A | DENN/MADD domain containing 4A | Nucleus | other |
| DYH9_HUMAN | DNAH9 | dynein, axonemal, heavy chain 9 | Cytoplasm | other |
| DNJB4_HUMAN | DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | Nucleus | other |
| DSCAM_HUMAN | DSCAM | Down syndrome cell adhesion molecule | Plasma Membrane | other |
| EHD3_HUMAN | EHD3 | EH-domain containing 3 | Cytoplasm | other |
| EMAL6_HUMAN | EML6 | echinoderm microtubule associated protein like 6 | unknown | other |
| ENOA_HUMAN | ENO1 | enolase 1, (alpha) | Cytoplasm | transcription regulator |
| ENTP7_HUMAN | ENTPD7 | ectonucleoside triphosphate diphosphohydrolase 7 | Cytoplasm | enzyme |
| HYEP_HUMAN | EPHX1 | epoxide hydrolase 1, microsomal (xenobiotic) | Cytoplasm | peptidase |
| FA10_HUMAN | F10 | coagulation factor X | Extracellular Space | peptidase |
| F13A_HUMAN | F13A1 | coagulation factor XIII, A1 polypeptide | Extracellular Space | enzyme |
| THRB_HUMAN | F2 | coagulation factor II (thrombin) | Extracellular Space | peptidase |
| FA5_HUMAN | F5 | coagulation factor V (proaccelerin, labile factor) | Plasma Membrane | enzyme |
| F117A_HUMAN | FAM117A | family with sequence similarity 117, member A | unknown | transporter |
| F171B_HUMAN | FAM171B | family with sequence similarity 171, member B | unknown | other |
| F208B_HUMAN | FAM208B | family with sequence similarity 208, member B | unknown | other |
| FBLN1_HUMAN | FBLN1 | fibulin 1 | Extracellular Space | other |
| FBN1_HUMAN | FBN1 | fibrillin 1 | Extracellular Space | other |
| FIBA_HUMAN | FGA | fibrinogen alpha chain | Extracellular Space | other |
| FGF3_HUMAN | FGF3 | fibroblast growth factor 3 | Extracellular Space | growth factor |
| FLNA_HUMAN | FLNA | filamin A, alpha | Cytoplasm | other |
| FINC_HUMAN | FN1 | fibronectin 1 | Extracellular Space | enzyme |
| FRPD3_HUMAN | FRMPD3 | FERM and PDZ domain containing 3 | unknown | other |
| GALK1_HUMAN | GALK1 | galactokinase 1 | Cytoplasm | kinase |
| G3P_HUMAN | GAPDH | glyceraldehyde-3-phosphate dehydrogenase | Cytoplasm | enzyme |
| GSCR1_HUMAN | GLTSCR1 | glioma tumor suppressor candidate region gene 1 | Extracellular Space | other |
| GBG12_HUMAN | GNG12 | guanine nucleotide binding protein (G protein), gamma 12 | Plasma Membrane | enzyme |
| GRIN1_HUMAN | GPRIN1 | G protein regulated inducer of neurite outgrowth 1 | Plasma Membrane | other |
| GELS_HUMAN | GSN | gelsolin | Extracellular Space | other |
| TF2H1_HUMAN | GTF2H1 | general transcription factor IIH, polypeptide 1, 62 kDa | Nucleus | transcription regulator |
| HBB_HUMAN | HBB | hemoglobin, beta | Cytoplasm | transporter |
| HCN1_HUMAN | HCN1 | hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | Plasma Membrane | ion channel |
| HTR5A_HUMAN | HEATR5A | HEAT repeat containing 5A | unknown | other |
| H13_HUMAN | HIST1H1D | histone cluster 1, H1d | Nucleus | other |
| H2B1M_HUMAN | HIST1H2BM | histone cluster 1, H2bm | Nucleus | other |
| H31T_HUMAN | HIST3H3 | histone cluster 3, H3 | Nucleus | other |
| HMMR_HUMAN | HMMR | hyaluronan-mediated motility receptor (RHAMM) | Plasma Membrane | other |
| HS90A_HUMAN | HSP90AA1 | heat shock protein 90 kDa alpha (cytosolic), class A member 1 | Cytoplasm | enzyme |
| ENPL_HUMAN | HSP90B1 | heat shock protein 90 kDa beta (Grp94), member 1 | Cytoplasm | other |
| GRP78_HUMAN | HSPA5 | heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | Cytoplasm | enzyme |
| PGBM_HUMAN | HSPG2 | heparan sulfate proteoglycan 2 | Plasma Membrane | enzyme |
| HYDIN_HUMAN | HYDIN | HYDIN, axonemal central pair apparatus protein | unknown | other |
| ALS_HUMAN | IGFALS | insulin-like growth factor binding protein, acid labile subunit | Extracellular Space | other |
| IGHM_HUMAN | IGHM | immunoglobulin heavy constant mu | Plasma Membrane | transmembrane receptor |
| IGKC_HUMAN | IGKC | immunoglobulin kappa constant | Extracellular Space | other |
| IGSF8_HUMAN | IGSF8 | immunoglobulin superfamily, member 8 | Plasma Membrane | other |
| IP6K3_HUMAN | IP6K3 | inositol hexakisphosphate kinase 3 | Cytoplasm | kinase |
| ITB1_HUMAN | ITGB1 | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | Plasma Membrane | transmembrane receptor |
| ITIH2_HUMAN | ITIH2 | inter-alpha-trypsin inhibitor heavy chain 2 | Extracellular Space | other |
| ITIH3_HUMAN | ITIH3 | inter-alpha-trypsin inhibitor heavy chain 3 | Extracellular Space | other |
| IRK2_HUMAN | KCNJ2 | potassium inwardly-rectifying channel, subfamily J, member 2 | Plasma Membrane | ion channel |
| KI20B_HUMAN | KIF20B | kinesin family member 20B | Nucleus | enzyme |
| IMB1_HUMAN | KPNB1 | karyopherin (importin) beta 1 | Nucleus | transporter |
| K2C1_HUMAN | KRT1 | keratin 1 | Cytoplasm | other |
| K1C10_HUMAN | KRT10 | keratin 10 | Cytoplasm | other |
| K22E_HUMAN | KRT2 | keratin 2 | Cytoplasm | other |
| K1C39_HUMAN | KRT39 | keratin 39 | Cytoplasm | other |
| K1C9_HUMAN | KRT9 | keratin 9 | Cytoplasm | other |
| LAMB1_HUMAN | LAMB1 | laminin, beta 1 | Extracellular Space | other |
| LDB1_HUMAN | LDB1 | LIM domain binding 1 | Nucleus | transcription regulator |
| LG3BP_HUMAN | LGALS3BP | lectin, galactoside-binding, soluble, 3 binding protein | Plasma Membrane | transmembrane receptor |
| LHPL3_HUMAN | LHFPL3 | lipoma HMGIC fusion partner-like 3 | unknown | other |
| CQ054_HUMAN | LINC00469 | long intergenic non-protein coding RNA 469 | unknown | other |
| YA033_HUMAN | LOC339524 | uncharacterized LOC339524 | unknown | other |
| LONM_HUMAN | LONP1 | lon peptidase 1, mitochondrial | Cytoplasm | peptidase |
| LONF2_HUMAN | LONRF2 | LON peptidase N-terminal domain and ring finger 2 | unknown | other |
| LPAR6_HUMAN | LPAR6 | lysophosphatidic acid receptor 6 | Plasma Membrane | G-protein coupled receptor |
| LRP1_HUMAN | LRP1 | low density lipoprotein receptor-related protein 1 | Plasma Membrane | transmembrane receptor |
| TRFL_HUMAN | LTF | lactotransferrin | Extracellular Space | peptidase |
| LUM_HUMAN | LUM | lumican | Extracellular Space | other |
| LY75_HUMAN | LY75 | lymphocyte antigen 75 | Plasma Membrane | other |
| MACD1_HUMAN | MACROD1 | MACRO domain containing 1 | Cytoplasm | enzyme |
| MAP2_HUMAN | MAP2 | microtubule-associated protein 2 | Cytoplasm | other |
| MAST3_HUMAN | MAST3 | microtubule associated serine/threonine kinase 3 | unknown | kinase |
| MED16_HUMAN | MED16 | mediator complex subunit 16 | Nucleus | transcription regulator |
| MFGM_HUMAN | MFGE8 | milk fat globule-EGF factor 8 protein | Extracellular Space | other |
| MKLN1_HUMAN | MKLN1 | muskelin 1, intracellular mediator containing kelch motifs | Cytoplasm | other |
| MOES_HUMAN | MSN | moesin | Plasma Membrane | other |
| MTERF_HUMAN | MTERF | mitochondrial transcription termination factor | Cytoplasm | transcription regulator |
| MYH9_HUMAN | MYH9 | myosin, heavy chain 9, non-muscle | Cytoplasm | transporter |
| MYL6_HUMAN | MYL6 | myosin, light chain 6, alkali, smooth muscle and non-muscle | Cytoplasm | other |
| MYLK_HUMAN | MYLK | myosin light chain kinase | Cytoplasm | kinase |
| MY18B_HUMAN | MYO18B | myosin XVIIIB | Cytoplasm | other |
| NCTR1_HUMAN | NCR1 | natural cytotoxicity triggering receptor 1 | Plasma Membrane | transmembrane receptor |
| NID1_HUMAN | NID1 | nidogen 1 | Extracellular Space | other |
| NOL4_HUMAN | NOL4 | nucleolar protein 4 | Nucleus | other |
| NOTC3_HUMAN | NOTCH3 | notch 3 | Plasma Membrane | transcription regulator |
| NGBR_HUMAN | NUS1 | nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) | Cytoplasm | other |
| OGG1_HUMAN | OGG1 | 8-oxoguanine DNA glycosylase | Nucleus | enzyme |
| O51F1_HUMAN | OR51F1 | olfactory receptor, family 51, subfamily F, member 1 | Plasma Membrane | G-protein coupled receptor |
| ORC5_HUMAN | ORC5 | origin recognition complex, subunit 5 | Nucleus | other |
| PDIA1_HUMAN | P4HB | prolyl 4-hydroxylase, beta polypeptide | Cytoplasm | enzyme |
| PDIA3_HUMAN | PDIA3 | protein disulfide isomerase family A, member 3 | Cytoplasm | peptidase |
| F261_HUMAN | PFKFB1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | Cytoplasm | kinase |
| PGAM1_HUMAN | PGAM1 | phosphoglycerate mutase 1 (brain) | Cytoplasm | phosphatase |
| PI3R4_HUMAN | PIK3R4 | phosphoinositide-3-kinase, regulatory subunit 4 | Cytoplasm | kinase |
| KPYM_HUMAN | PKM | pyruvate kinase, muscle | unknown | kinase |
| PLCL1_HUMAN | PLCL1 | phospholipase C-like 1 | Cytoplasm | enzyme |
| PLMN_HUMAN | PLG | plasminogen | Extracellular Space | peptidase |
| PLPL8_HUMAN | PNPLA8 | patatin-like phospholipase domain containing 8 | Cytoplasm | enzyme |
| POSTN_HUMAN | POSTN | periostin, osteoblast specific factor | Extracellular Space | other |
| P2R3C_HUMAN | PPP2R3C | protein phosphatase 2, regulatory subunit B”, gamma | Cytoplasm | other |
| PREX1_HUMAN | PREX1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | Cytoplasm | other |
| PRP31_HUMAN | PRPF31 | PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae) | Nucleus | other |
| PSA3_HUMAN | PSMA3 | proteasome (prosome, macropain) subunit, alpha type, 3 | Cytoplasm | peptidase |
| PSA7L_HUMAN | PSMA8 | proteasome (prosome, macropain) subunit, alpha type, 8 | Cytoplasm | peptidase |
| PSB5_HUMAN | PSMB5 | proteasome (prosome, macropain) subunit, beta type, 5 | Cytoplasm | peptidase |
| PSB6_HUMAN | PSMB6 | proteasome (prosome, macropain) subunit, beta type, 6 | Cytoplasm | peptidase |
| PSB7_HUMAN | PSMB7 | proteasome (prosome, macropain) subunit, beta type, 7 | Cytoplasm | peptidase |
| PTX3_HUMAN | PTX3 | pentraxin 3, long | Extracellular Space | other |
| PUSL1_HUMAN | PUSL1 | pseudouridylate synthase-like 1 | unknown | enzyme |
| PZP_HUMAN | PZP | pregnancy-zone protein | Extracellular Space | other |
| ARIP4_HUMAN | RAD54L2 | RAD54-like 2 (S. cerevisiae) | Nucleus | transcription regulator |
| RFX8_HUMAN | RFX8 | regulatory factor X, 8 | unknown | other |
| RGPD3_HUMAN | RGPD5 (includes others) | RANBP2-like and GRIP domain containing 5 | Nucleus | other |
| RHPN2_HUMAN | RHPN2 | rhophilin, Rho GTPase binding protein 2 | Cytoplasm | other |
| RIMKA_HUMAN | RIMKLA | ribosomal modification protein rimK-like family member A | unknown | other |
| RN217_HUMAN | RNF217 | ring finger protein 217 | unknown | enzyme |
| RL35_HUMAN | RPL35 | ribosomal protein L35 | Cytoplasm | other |
| S10AB_HUMAN | S100A11 | S100 calcium binding protein A11 | Cytoplasm | other |
| SACS_HUMAN | SACS | spastic ataxia of Charlevoix-Saguenay (sacsin) | Nucleus | other |
| SALL4_HUMAN | SALL4 | sal-like 4 (Drosophila) | Nucleus | other |
| SDCB1_HUMAN | SDCBP | syndecan binding protein (syntenin) | Plasma Membrane | enzyme |
| SEM4G_HUMAN | SEMA4G | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G | Plasma Membrane | other |
| SEM6D_HUMAN | SEMA6D | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D | Plasma Membrane | other |
| SPB10_HUMAN | SERPINB10 | serpin peptidase inhibitor, clade B (ovalbumin), member 10 | Cytoplasm | other |
| ANT3_HUMAN | SERPINC1 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | Extracellular Space | other |
| PEDF_HUMAN | SERPINF1 | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 | Extracellular Space | other |
| A2AP_HUMAN | SERPINF2 | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 | Extracellular Space | other |
| SH3L3_HUMAN | SH3BGRL3 | SH3 domain binding glutamic acid-rich protein like 3 | Nucleus | other |
| S12A7_HUMAN | SLC12A7 | solute carrier family 12 (potassium/chloride transporters), member 7 | Plasma Membrane | transporter |
| S13A4_HUMAN | SLC13A4 | solute carrier family 13 (sodium/sulfate symporters), member 4 | Plasma Membrane | transporter |
| SL9C2_HUMAN | SLC9C2 | solute carrier family 9, member C2 (putative) | unknown | other |
| SNTB2_HUMAN | SNTB2 | syntrophin, beta 2 (dystrophin-associated protein A1, 59 kDa, basic component 2) | Plasma Membrane | other |
| SPAT7_HUMAN | SPATA7 | spermatogenesis associated 7 | unknown | other |
| STAB2_HUMAN | STAB2 | stabilin 2 | Plasma Membrane | transmembrane receptor |
| ST3L1_HUMAN | STAG3L1 | stromal antigen 3-like 1 | unknown | other |
| TBL2_HUMAN | TBL2 | transducin (beta)-like 2 | Plasma Membrane | other |
| TBPL1_HUMAN | TBPL1 | TBP-like 1 | Nucleus | transcription regulator |
| TBX20_HUMAN | TBX20 | T-box 20 | Nucleus | transcription regulator |
| TRFE_HUMAN | TF | transferrin | Extracellular Space | transporter |
| THA11_HUMAN | THAP11 | THAP domain containing 11 | Nucleus | other |
| TSP1_HUMAN | THBS1 | thrombospondin 1 | Extracellular Space | other |
| THY1_HUMAN | THY1 | Thy-1 cell surface antigen | Plasma Membrane | other |
| TIAR_HUMAN | TIAL1 | TIA1 cytotoxic granule-associated RNA binding protein-like 1 | Nucleus | transcription regulator |
| TIMP1_HUMAN | TIMP1 | TIMP metallopeptidase inhibitor 1 | Extracellular Space | other |
| TKT_HUMAN | TKT | transketolase | Cytoplasm | enzyme |
| TLN1_HUMAN | TLN1 | talin 1 | Plasma Membrane | other |
| TENA_HUMAN | TNC | tenascin C | Extracellular Space | other |
| TNAP3_HUMAN | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | Nucleus | enzyme |
| P53_HUMAN | TP53 | tumor protein p53 | Nucleus | transcription regulator |
| TPIS_HUMAN | TPI1 | triosephosphate isomerase 1 | Cytoplasm | enzyme |
| TPC12_HUMAN | TRAPPC12 | trafficking protein particle complex 12 | unknown | other |
| TITIN_HUMAN | TTN | titin | unknown | kinase |
| TTYH3_HUMAN | TTYH3 | tweety homolog 3 (Drosophila) | Plasma Membrane | ion channel |
| TBA1B_HUMAN | TUBA1B | tubulin, alpha 1b | Cytoplasm | other |
| TBB5_HUMAN | TUBB | tubulin, beta class I | Cytoplasm | other |
| TBB1_HUMAN | TUBB1 | tubulin, beta 1 class VI | Cytoplasm | other |
| TBB2A_HUMAN | TUBB2A | tubulin, beta 2A class IIa | Cytoplasm | other |
| TYK2_HUMAN | TYK2 | tyrosine kinase 2 | Plasma Membrane | kinase |
| UBQLN_HUMAN | UBQLNL | ubiquilin-like | unknown | other |
| UD2A3_HUMAN | UGT2A3 | UDP glucuronosyltransferase 2 family, polypeptide A3 | Plasma Membrane | enzyme |
| USMG5_HUMAN | USMG5 | up-regulated during skeletal muscle growth 5 homolog (mouse) | Cytoplasm | other |
| VAT1_HUMAN | VAT1 | vesicle amine transport protein 1 homolog (T. californica) | Plasma Membrane | transporter |
| CSPG2_HUMAN | VCAN | versican | Extracellular Space | other |
| VIME_HUMAN | VIM | vimentin | Cytoplasm | other |
| VTNC_HUMAN | VTN | vitronectin | Extracellular Space | other |
| WWC2_HUMAN | WWC2 | WW and C2 domain containing 2 | unknown | other |
| 1433E_HUMAN | YWHAE | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | Cytoplasm | other |
| ZN268_HUMAN | ZNF268 | zinc finger protein 268 | Nucleus | other |
| ZN510_HUMAN | ZNF510 | zinc finger protein 510 | Nucleus | other |
| ZN516_HUMAN | ZNF516 | zinc finger protein 516 | Nucleus | other |
| ZN599_HUMAN | ZNF599 | zinc finger protein 599 | unknown | other |
| ZN729_HUMAN | ZNF729 | zinc finger protein 729 | unknown | other |
| ZNF74_HUMAN | ZNF74 | zinc finger protein 74 | Nucleus | other |
| Exo-pMSC-3%O2 | ||||
| ID | Symbol | Entrez Gene Name | Location | Type(s) |
| ABCA1_HUMAN | ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | Plasma Membrane | transporter |
| MRP1_HUMAN | ABCC1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | Plasma Membrane | transporter |
| ACTB_HUMAN | ACTB | actin, beta | Cytoplasm | other |
| ADCK4_HUMAN | ADCK4 | aarF domain containing kinase 4 | Cytoplasm | kinase |
| FETA_HUMAN | AFP | alpha-fetoprotein | Extracellular Space | transporter |
| FETUA_HUMAN | AHSG | alpha-2-HS-glycoprotein | Extracellular Space | other |
| ALBU_HUMAN | ALB | albumin | Extracellular Space | transporter |
| ARHG2_HUMAN | ARHGEF2 | Rho/Rac guanine nucleotide exchange factor (GEF) 2 | Cytoplasm | other |
| BMR1B_HUMAN | BMPR1B | bone morphogenetic protein receptor, type IB | Plasma Membrane | kinase |
| BPTF_HUMAN | BPTF | bromodomain PHD finger transcription factor | Nucleus | transcription regulator |
| CJ118_HUMAN | C10orf118 | chromosome 10 open reading frame 118 | unknown | other |
| ERG28_HUMAN | C14orf1 | chromosome 14 open reading frame 1 | Cytoplasm | other |
| CH073_HUMAN | C8orf73 | chromosome 8 open reading frame 73 | unknown | other |
| CCD80_HUMAN | CCDC80 | coiled-coil domain containing 80 | Nucleus | other |
| MPIP1_HUMAN | CDC25A | cell division cycle 25 homolog A (S. pombe) | Nucleus | phosphatase |
| CDA7L_HUMAN | CDCA7L | cell division cycle associated 7-like | Nucleus | other |
| CDK13_HUMAN | CDK13 | cyclin-dependent kinase 13 | Nucleus | kinase |
| CNGA1_HUMAN | CNGA1 | cyclic nucleotide gated channel alpha 1 | Plasma Membrane | ion channel |
| COG2_HUMAN | COG2 | component of oligomeric golgi complex 2 | Cytoplasm | transporter |
| CODA1_HUMAN | COL13A1 | collagen, type XIII, alpha 1 | Plasma Membrane | other |
| CO1A1_HUMAN | COL1A1 | collagen, type I, alpha 1 | Extracellular Space | other |
| DIAC_HUMAN | CTBS | chitobiase, di-N-acetyl- | Cytoplasm | enzyme |
| DUPD1_HUMAN | DUPD1 | dual specificity phosphatase and pro isomerase domain containing 1 | unknown | enzyme |
| RNZ2_HUMAN | ELAC2 | elaC homolog 2 (E. coli) | Nucleus | enzyme |
| EPC2_HUMAN | EPC2 | enhancer of polycomb homolog 2 (Drosophila) | unknown | other |
| XPF_HUMAN | ERCC4 | excision repair cross-complementing rodent repair deficiency, complementation group 4 | Nucleus | enzyme |
| ERI2_HUMAN | ERI2 | ERI1 exoribonuclease family member 2 | unknown | other |
| ETS1_HUMAN | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | Nucleus | transcription regulator |
| EXTL2_HUMAN | EXTL2 | exostoses (multiple)-like 2 | Cytoplasm | enzyme |
| FA10_HUMAN | F10 | coagulation factor X | Extracellular Space | peptidase |
| THRB_HUMAN | F2 | coagulation factor II (thrombin) | Extracellular Space | peptidase |
| F117A_HUMAN | FAM117A | family with sequence similarity 117, member A | unknown | transporter |
| F168A_HUMAN | FAM168A | family with sequence similarity 168, member A | unknown | other |
| F208A_HUMAN | FAM208A | family with sequence similarity 208, member A | unknown | other |
| F210A_HUMAN | FAM210A | family with sequence similarity 210, member A | Cytoplasm | other |
| FBN1_HUMAN | FBN1 | fibrillin 1 | Extracellular Space | other |
| FGF18_HUMAN | FGF18 | fibroblast growth factor 18 | Extracellular Space | growth factor |
| FINC_HUMAN | FN1 | fibronectin 1 | Extracellular Space | enzyme |
| FNIP2_HUMAN | FNIP2 | folliculin interacting protein 2 | Cytoplasm | other |
| VTDB_HUMAN | GC | group-specific component (vitamin D binding protein) | Extracellular Space | transporter |
| GSCR1_HUMAN | GLTSCR1 | glioma tumor suppressor candidate region gene 1 | Extracellular Space | other |
| GOG8A_HUMAN | GOLGA8A/GOLGA8B | golgin A8 family, member B | Cytoplasm | other |
| GRIN1_HUMAN | GPRIN1 | G protein regulated inducer of neurite outgrowth 1 | Plasma Membrane | other |
| HBB_HUMAN | HBB | hemoglobin, beta | Cytoplasm | transporter |
| HELZ_HUMAN | HELZ | helicase with zinc finger | Nucleus | enzyme |
| HJURP_HUMAN | HJURP | Holliday junction recognition protein | Nucleus | other |
| 1B39_HUMAN | HLA-B | major histocompatibility complex, class I, B | Plasma Membrane | transmembrane receptor |
| H90B3_HUMAN | HSP90AB3P | heat shock protein 90 kDa alpha (cytosolic), class B member 3, pseudogene | unknown | other |
| I22R1_HUMAN | IL22RA1 | interleukin 22 receptor, alpha 1 | Plasma Membrane | transmembrane receptor |
| ITIH2_HUMAN | ITIH2 | inter-alpha-trypsin inhibitor heavy chain 2 | Extracellular Space | other |
| K2C1_HUMAN | KRT1 | keratin 1 | Cytoplasm | other |
| K2C5_HUMAN | KRT5 | keratin 5 | Cytoplasm | other |
| AMPL_HUMAN | LAP3 | leucine aminopeptidase 3 | Cytoplasm | peptidase |
| TRFL_HUMAN | LTF | lactotransferrin | Extracellular Space | peptidase |
| MLAS1_HUMAN | MLLT4-AS1 | MLLT4 antisense RNA 1 | unknown | other |
| MYH4_HUMAN | MYH4 | myosin, heavy chain 4, skeletal muscle | Cytoplasm | enzyme |
| ULA1_HUMAN | NAE1 | NEDD8 activating enzyme E1 subunit 1 | Cytoplasm | enzyme |
| NGEF_HUMAN | NGEF | neuronal guanine nucleotide exchange factor | Cytoplasm | other |
| NAL13_HUMAN | NLRP13 | NLR family, pyrin domain containing 13 | unknown | other |
| NOL4_HUMAN | NOL4 | nucleolar protein 4 | Nucleus | other |
| NTM1A_HUMAN | NTMT1 | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | Nucleus | enzyme |
| TEN3_HUMAN | ODZ3 | odz, odd Oz/ten-m homolog 3 (Drosophila) | Plasma Membrane | other |
| O10A7_HUMAN | OR10A7 | olfactory receptor, family 10, subfamily A, member 7 | Plasma Membrane | other |
| OSGEP_HUMAN | OSGEP | O-sialoglycoprotein endopeptidase | unknown | peptidase |
| PGK1_HUMAN | PGK1 | phosphoglycerate kinase 1 | Cytoplasm | kinase |
| PHF10_HUMAN | PHF10 | PHD finger protein 10 | Nucleus | other |
| KCC1B_HUMAN | PNCK | pregnancy up-regulated non-ubiquitously expressed CaM kinase | unknown | kinase |
| P2R3C_HUMAN | PPP2R3C | protein phosphatase 2, regulatory subunit B”, gamma | Cytoplasm | other |
| PR38A_HUMAN | PRPF38A | PRP38 pre-mRNA processing factor 38 (yeast) domain containing A | Nucleus | other |
| PTPRK_HUMAN | PTPRK | protein tyrosine phosphatase, receptor type, K | Plasma Membrane | phosphatase |
| PUSL1_HUMAN | PUSL1 | pseudouridylate synthase-like 1 | unknown | enzyme |
| PXDN_HUMAN | PXDN | peroxidasin homolog (Drosophila) | Extracellular Space | enzyme |
| PXK_HUMAN | PXK | PX domain containing serine/threonine kinase | Cytoplasm | kinase |
| PZP_HUMAN | PZP | pregnancy-zone protein | Extracellular Space | other |
| RAB10_HUMAN | RAB10 | RAB10, member RAS oncogene family | Cytoplasm | enzyme |
| REST_HUMAN | REST | RE1-silencing transcription factor | Nucleus | transcription regulator |
| RFX8_HUMAN | RFX8 | regulatory factor X, 8 | unknown | other |
| SALL4_HUMAN | SALL4 | sal-like 4 (Drosophila) | Nucleus | other |
| A2AP_HUMAN | SERPINF2 | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 | Extracellular Space | other |
| SHSA7_HUMAN | SHISA7 | shisa homolog 7 (Xenopus laevis) | unknown | other |
| S12A7_HUMAN | SLC12A7 | solute carrier family 12 (potassium/chloride transporters), member 7 | Plasma Membrane | transporter |
| S35A1_HUMAN | SLC35A1 | solute carrier family 35 (CMP-sialic acid transporter), member A1 | Cytoplasm | transporter |
| SMTN_HUMAN | SMTN | smoothelin | Extracellular Space | other |
| SPC25_HUMAN | SPC25 | SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) | Cytoplasm | other |
| SPP24_HUMAN | SPP2 | secreted phosphoprotein 2, 24 kDa | Extracellular Space | other |
| SYNJ1_HUMAN | SYNJ1 | synaptojanin 1 | Cytoplasm | phosphatase |
| TANC1_HUMAN | TANC1 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | Plasma Membrane | other |
| TCEA3_HUMAN | TCEA3 | transcription elongation factor A (SII), 3 | Nucleus | transcription regulator |
| TET1_HUMAN | TET1 | tet methylcytosine dioxygenase 1 | Nucleus | other |
| TEX2_HUMAN | TEX2 | testis expressed 2 | unknown | other |
| TRFE_HUMAN | TF | transferrin | Extracellular Space | transporter |
| TGFR1_HUMAN | TGFBR1 | transforming growth factor, beta receptor 1 | Plasma Membrane | kinase |
| TSP1_HUMAN | THBS1 | thrombospondin 1 | Extracellular Space | other |
| TITIN_HUMAN | TTN | titin | unknown | kinase |
| VAT1_HUMAN | VAT1 | vesicle amine transport protein 1 homolog (T. californica) | Plasma Membrane | transporter |
| MELT_HUMAN | VEPH1 | ventricular zone expressed PH domain homolog 1 (zebrafish) | Nucleus | other |
| VTNC_HUMAN | VTN | vitronectin | Extracellular Space | other |
| XKR3_HUMAN | XKR3 | XK, Kell blood group complex subunit-related family, member 3 | unknown | other |
| XPO5_HUMAN | XPO5 | exportin 5 | Nucleus | transporter |
| ZDH19_HUMAN | ZDHHC19 | zinc finger, DHHC-type containing 19 | unknown | other |
| ZMYM4_HUMAN | ZMYM4 | zinc finger, MYM-type 4 | unknown | other |
| ZN143_HUMAN | ZNF143 | zinc finger protein 143 | Nucleus | transcription regulator |
| ZN333_HUMAN | ZNF333 | zinc finger protein 333 | Nucleus | other |
| ZN486_HUMAN | ZNF486 | zinc finger protein 486 | Nucleus | other |
| ZN516_HUMAN | ZNF516 | zinc finger protein 516 | Nucleus | other |
| ZN607_HUMAN | ZNF607 | zinc finger protein 607 | Nucleus | other |
| ZN645_HUMAN | ZNF645 | zinc finger protein 645 | Extracellular Space | other |
| ZN646_HUMAN | ZNF646 | zinc finger protein 646 | Nucleus | other |
| ZN770_HUMAN | ZNF770 | zinc finger protein 770 | unknown | other |
| ZN808_HUMAN | ZNF808 | zinc finger protein 808 | unknown | other |
| ZN865_HUMAN | ZNF865 | zinc finger protein 865 | unknown | other |
| ZNF98_HUMAN | ZNF98 | zinc finger protein 98 | unknown | other |
| Exo-pMSC-8%O2 | ||||
| ID | Symbol | Entrez Gene Name | Location | Type(s) |
| ACTS_HUMAN | ACTA1 | actin, alpha 1, skeletal muscle | Cytoplasm | other |
| ACTB_HUMAN | ACTB | actin, beta | Cytoplasm | other |
| PACA_HUMAN | ADCYAP1 | adenylate cyclase activating polypeptide 1 (pituitary) | Extracellular Space | other |
| FETA_HUMAN | AFP | alpha-fetoprotein | Extracellular Space | transporter |
| FETUA_HUMAN | AHSG | alpha-2-HS-glycoprotein | Extracellular Space | other |
| ALBU_HUMAN | ALB | albumin | Extracellular Space | transporter |
| ANKH1_HUMAN | ANKHD1 | ankyrin repeat and KH domain containing 1 | unknown | other |
| ARMX1_HUMAN | ARMCX1 | armadillo repeat containing, X-linked 1 | unknown | other |
| ASXL3_HUMAN | ASXL3 | additional sex combs like 3 (Drosophila) | unknown | other |
| ATG2B_HUMAN | ATG2B | autophagy related 2B | unknown | other |
| BAI1_HUMAN | BAI1 | brain-specific angiogenesis inhibitor 1 | Plasma Membrane | G-protein coupled receptor |
| BCL3_HUMAN | BCL3 | B-cell CLL/lymphoma 3 | Nucleus | transcription regulator |
| CL043_HUMAN | C12orf43 | chromosome 12 open reading frame 43 | unknown | other |
| CO3_HUMAN | C3 | complement component 3 | Extracellular Space | peptidase |
| CALB1_HUMAN | CALB1 | calbindin 1, 28 kDa | Cytoplasm | other |
| CALR_HUMAN | CALR | calreticulin | Cytoplasm | transcription regulator |
| CAND1_HUMAN | CAND1 | cullin-associated and neddylation-dissociated 1 | Cytoplasm | transcription regulator |
| CAD20_HUMAN | CDH20 | cadherin 20, type 2 | Plasma Membrane | other |
| CDK4_HUMAN | CDK4 | cyclin-dependent kinase 4 | Nucleus | kinase |
| CEP97_HUMAN | CEP97 | centrosomal protein 97 kDa | Cytoplasm | other |
| CIZ1_HUMAN | CIZ1 | CDKN1A interacting zinc finger protein 1 | Nucleus | transporter |
| CMBL_HUMAN | CMBL | carboxymethylenebutenolidase homolog (Pseudomonas) | unknown | enzyme |
| CO1A1_HUMAN | COL1A1 | collagen, type I, alpha 1 | Extracellular Space | other |
| CO1A2_HUMAN | COL1A2 | collagen, type I, alpha 2 | Extracellular Space | other |
| CSF2R_HUMAN | CSF2RA | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | Plasma Membrane | transmembrane receptor |
| CSTF3_HUMAN | CSTF3 | cleavage stimulation factor, 3′ pre-RNA, subunit 3, 77 kDa | Nucleus | other |
| DIAC_HUMAN | CTBS | chitobiase, di-N-acetyl- | Cytoplasm | enzyme |
| DG2L6_HUMAN | DGAT2L6 | diacylglycerol O-acyltransferase 2-like 6 | unknown | other |
| DYH9_HUMAN | DNAH9 | dynein, axonemal, heavy chain 9 | Cytoplasm | other |
| EMAL5_HUMAN | EML5 | echinoderm microtubule associated protein like 5 | unknown | other |
| ENPP3_HUMAN | ENPP3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 | Plasma Membrane | enzyme |
| FA10_HUMAN | F10 | coagulation factor X | Extracellular Space | peptidase |
| THRB_HUMAN | F2 | coagulation factor II (thrombin) | Extracellular Space | peptidase |
| FA73B_HUMAN | FAM73B | family with sequence similarity 73, member B | unknown | other |
| FBN1_HUMAN | FBN1 | fibrillin 1 | Extracellular Space | other |
| FLT3_HUMAN | FLT3 | fms-related tyrosine kinase 3 | Plasma Membrane | kinase |
| FINC_HUMAN | FN1 | fibronectin 1 | Extracellular Space | enzyme |
| GLBL3_HUMAN | GLB1L3 | galactosidase, beta 1-like 3 | unknown | enzyme |
| GP126_HUMAN | GPR126 | G protein-coupled receptor 126 | Plasma Membrane | G-protein coupled receptor |
| GRIN1_HUMAN | GPRIN1 | G protein regulated inducer of neurite outgrowth 1 | Plasma Membrane | other |
| HBD_HUMAN | HBD | hemoglobin, delta | Cytoplasm | transporter |
| HCFC2_HUMAN | HCFC2 | host cell factor C2 | Nucleus | transcription regulator |
| IL25_HUMAN | IL25 | interleukin 25 | Extracellular Space | cytokine |
| INT4_HUMAN | INTS4 | integrator complex subunit 4 | Nucleus | other |
| IQGA1_HUMAN | IQGAP1 | IQ motif containing GTPase activating protein 1 | Cytoplasm | other |
| ITA4_HUMAN | ITGA4 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | Plasma Membrane | other |
| ITIH2_HUMAN | ITIH2 | inter-alpha-trypsin inhibitor heavy chain 2 | Extracellular Space | other |
| K0232_HUMAN | KIAA0232 | KIAA0232 | Extracellular Space | other |
| SKT_HUMAN | KIAA1217 | KIAA1217 | Cytoplasm | other |
| KNG1_HUMAN | KNG1 | kininogen 1 | Extracellular Space | other |
| K2C1_HUMAN | KRT1 | keratin 1 | Cytoplasm | other |
| K1C10_HUMAN | KRT10 | keratin 10 | Cytoplasm | other |
| LMBL3_HUMAN | L3MBTL3 | l(3)mbt-like 3 (Drosophila) | Nucleus | other |
| LPHN2_HUMAN | LPHN2 | latrophilin 2 | Plasma Membrane | G-protein coupled receptor |
| LRRC9_HUMAN | LRRC9 | leucine rich repeat containing 9 | unknown | other |
| TRFL_HUMAN | LTF | lactotransferrin | Extracellular Space | peptidase |
| MACF1_HUMAN | MACF1 | microtubule-actin crosslinking factor 1 | Cytoplasm | enzyme |
| MCLN2_HUMAN | MCOLN2 | mucolipin 2 | Plasma Membrane | ion channel |
| M4A10_HUMAN | MS4A10 | membrane-spanning 4-domains, subfamily A, member 10 | unknown | other |
| MYB_HUMAN | MYB | v-myb myeloblastosis viral oncogene homolog (avian) | Nucleus | transcription regulator |
| ULA1_HUMAN | NAE1 | NEDD8 activating enzyme E1 subunit 1 | Cytoplasm | enzyme |
| NFL_HUMAN | NEFL | neurofilament, light polypeptide | Cytoplasm | other |
| NOL4_HUMAN | NOL4 | nucleolar protein 4 | Nucleus | other |
| NTF3_HUMAN | NTF3 | neurotrophin 3 | Extracellular Space | growth factor |
| O10A7_HUMAN | OR10A7 | olfactory receptor, family 10, subfamily A, member 7 | Plasma Membrane | other |
| ORC1_HUMAN | ORC1 | origin recognition complex, subunit 1 | Nucleus | other |
| OSBL7_HUMAN | OSBPL7 | oxysterol binding protein-like 7 | Cytoplasm | other |
| PARP8_HUMAN | PARP8 | poly (ADP-ribose) polymerase family, member 8 | unknown | other |
| PCOC1_HUMAN | PCOLCE | procollagen C-endopeptidase enhancer | Extracellular Space | other |
| PENK_HUMAN | PENK | proenkephalin | Extracellular Space | other |
| PHIP_HUMAN | PHIP | pleckstrin homology domain interacting protein | Nucleus | other |
| PI3R4_HUMAN | PIK3R4 | phosphoinositide-3-kinase, regulatory subunit 4 | Cytoplasm | kinase |
| PIWL1_HUMAN | PIWIL1 | piwi-like 1 (Drosophila) | Cytoplasm | other |
| PRDM9_HUMAN | PRDM9 | PR domain containing 9 | Nucleus | enzyme |
| PYRD1_HUMAN | PYROXD1 | pyridine nucleotide-disulphide oxidoreductase domain 1 | unknown | other |
| PZP_HUMAN | PZP | pregnancy-zone protein | Extracellular Space | other |
| RBL1_HUMAN | RBL1 | retinoblastoma-like 1 (p107) | Nucleus | other |
| THBG_HUMAN | SERPINA7 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 | Extracellular Space | transporter |
| ANT3_HUMAN | SERPINC1 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | Extracellular Space | other |
| SHSA7_HUMAN | SHISA7 | shisa homolog 7 (Xenopus laevis) | unknown | other |
| S12A7_HUMAN | SLC12A7 | solute carrier family 12 (potassium/chloride transporters), member 7 | Plasma Membrane | transporter |
| SMC4_HUMAN | SMC4 | structural maintenance of chromosomes 4 | Nucleus | transporter |
| RU2B_HUMAN | SNRPB2 | small nuclear ribonucleoprotein polypeptide B | Nucleus | other |
| OSTP_HUMAN | SPP1 | secreted phosphoprotein 1 | Extracellular Space | cytokine |
| SPP24_HUMAN | SPP2 | secreted phosphoprotein 2, 24 kDa | Extracellular Space | other |
| F10A1_HUMAN | ST13 | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | Cytoplasm | other |
| SPT6H_HUMAN | SUPT6H | suppressor of Ty 6 homolog (S. cerevisiae) | Nucleus | transcription regulator |
| TRBP2_HUMAN | TARBP2 | TAR (HIV-1) RNA binding protein 2 | Nucleus | other |
| TBL3_HUMAN | TBL3 | transducin (beta)-like 3 | Cytoplasm | peptidase |
| TET1_HUMAN | TET1 | tet methylcytosine dioxygenase 1 | Nucleus | other |
| TRFE_HUMAN | TF | transferrin | Extracellular Space | transporter |
| TSP1_HUMAN | THBS1 | thrombospondin 1 | Extracellular Space | other |
| TM117_HUMAN | TMEM117 | transmembrane protein 117 | Cytoplasm | other |
| TMTC3_HUMAN | TMTC3 | transmembrane and tetratricopeptide repeat containing 3 | unknown | other |
| TPD53_HUMAN | TPD52L1 | tumor protein D52-like 1 | Cytoplasm | other |
| TPM3_HUMAN | TPM3 | tropomyosin 3 | Cytoplasm | other |
| TRAF3_HUMAN | TRAF3 | TNF receptor-associated factor 3 | Cytoplasm | other |
| UB2V2_HUMAN | UBE2V2 | ubiquitin-conjugating enzyme E2 variant 2 | Cytoplasm | enzyme |
| UHRF2_HUMAN | UHRF2 | ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase | Nucleus | enzyme |
| UN13C_HUMAN | UNC13C | unc-13 homolog C (C. elegans) | Cytoplasm | other |
| VAMP5_HUMAN | VAMP5 | vesicle-associated membrane protein 5 (myobrevin) | Plasma Membrane | transporter |
| VAT1_HUMAN | VAT1 | vesicle amine transport protein 1 homolog (T. californica) | Plasma Membrane | transporter |
| MELT_HUMAN | VEPH1 | ventricular zone expressed PH domain homolog 1 (zebrafish) | Nucleus | other |
| VTNC_HUMAN | VTN | vitronectin | Extracellular Space | other |
| 1433B_HUMAN | YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Cytoplasm | transcription regulator |
| ZDH23_HUMAN | ZDHHC23 | zinc finger, DHHC-type containing 23 | unknown | other |
| ZMYM3_HUMAN | ZMYM3 | zinc finger, MYM-type 3 | Nucleus | other |
| ZN416_HUMAN | ZNF416 | zinc finger protein 416 | Nucleus | other |
| ZN671_HUMAN | ZNF671 | zinc finger protein 671 | Nucleus | other |
| ZNF74_HUMAN | ZNF74 | zinc finger protein 74 | Nucleus | other |
| ZN778_HUMAN | ZNF778 | zinc finger protein 778 | unknown | other |
| ZN841_HUMAN | ZNF841 | zinc finger protein 841 | unknown | other |
Figure 6. Analysis of pMSC derived-exosomes proteins identified by mass spectrometry using PANTHER software.
Exosomal proteins isolated from pMSC exposed to 1%, 3% or 8% O2 were classified using PANTHER program based on their (A) Biological process and (B) Molecular function.
Figure 7. Ingenuity pathway analysis of pMSC derived-exosomes proteins.
(A) The Venn diagram depicts the distribution of common and unique proteins identified by nanospray LC-MS/MS (ABSciex 5600) in exosomes released from pMSC exposed to 1%, 3% and 8% oxygen. Comparison of canonical pathways: (B) actin cytoskeleton signaling, (C) growth hormone signaling, (D) VEGF signaling, and (E) clathrin-mediated endocytosis signaling identified by IPA core analysis. Values are mean ± SEM. In B, C, D and E, *p<0.005 versus all condition.
Discussion
Mesenchymal stem cells are present in the human placenta during early pregnancy. During early pregnancy, placental vasculogenesis and angiogenesis proceed under low oxygen conditions prior to the establishment of a materno-placental perfusion. The role of MSC in directing and promoting placental vascular development remains to be clearly elucidated. The aim of this study was to establish the effects of oxygen tension on the release of exosomes from pMSC and to determine the effects of pMSC exosomes on endothelial cell migration and tube formation. The data obtained in the study are consistent with the hypothesis that the release of exosomes from pMSC is increased in hypoxic conditions and that pMSC exosomes promote endothelial cell migration and tube formation. Based on the data obtained, we suggest that pMSC exosomes contribute to the development of new vessels and promote angiogenesis within the placenta under low oxygen conditions. During early pregnancy this occurs as a physiological and developmental process. In pathological pregnancies characterized by compromized placental perfusion and ischaemia, such as preeclampsia and intrauterine growth restriction, we propose that pMSC may also increase exosome release as an adaptive response.
Germane to any study seeking to elucidate the physiological or pathophysiological role of exosomes is their specific isolation. Several methods for isolating exosomes have been developed and partially characterized. These methods are primarily based on particle size and density. By definition, exosomes are nanovesicles with a diameter of 30–100 nm, a buoyant density of 1.12 to 1.19 g/ml and express characteristic cell-surface markers. In this study, pMSC exosomes were isolated by differential centrifuge and sucrose gradient purification and were characterized by a diameter of 50 nm, a buoyant density of 1.1270 g/ml, and expressed exosome-specific cell surface markers.
Under hypoxic conditions (1% or 3% O2), pMSC exosome release increased by up to 7-fold compared to cells incubated under normoxic conditions (8% O2). These data are consistent with the effects of hypoxia on the release of exosomes from umbilical cord (UC)-derived MSCs, where low oxygen tension increases exosome release by ∼ 5.6-fold [37]. Hypoxia also has been reported to increase the release of exosomes from breast cancer cell lines (MCF7, SKBR3, and MDA- MB 231), squamous carcinoma cells (A431 cells) [26] and cardiac myocytes [38]. The mechanism by which hypoxia induces exosome release remains to be clearly established.
Recent evidence suggests that increased release of exosomes from breast cancer cells under hypoxic condition may be mediated by transcriptional factor HIF-1α[39]. In this study, the authors also observed higher expression of miR-210 in exosomes isolated from cancer cells exposed to hypoxia compared to normaxia cell-derived exosomes. Exosomal miR-210 from metastatic cancer cells enhances endothelial cell angiogenesis [40].
In MSCs, HIFs have been reported to promote MSC-mediated angiogenic effect on endothelial cells through the release of interleukin 8, VEGF and other growth factors [41]. It has been demonstrated that the secretion of soluble VEGF requires functional ADP-ribosylation factor 6 (Arf6) [42]. Interestingly, Arf6 is expressed on the membrane of exosomes and may promote exosome release [43]. An association between VEGF and Arf6 within exosomes, however, has not yet been demonstrated. Similarly, HIFs may contribute to the hypoxia-induced release of exosomes from pMSC observed in this study.
Previous studies have established that MSC promote angiogenesis via paracrine mechanisms [44]. The possible contribution of exosomes in mediating such paracrine actions has not been established. It is likely that exosomes were present (and not accounted for) in all conditioned media previously used to establish such paracrine effects. In this study, exosomes were isolated from pMSC, promoting hPMEC cell migration and tube formation. This effect was enhanced when pMSC were cultured under hypoxic conditions. Previously, Zhang et al., 2012, demonstrated that exosomes released from UC-MSC are internalized into umbilical cord endothelial cells and enhance in vitro the proliferation and network formation in a dose-dependent manner [37]. Interestingly, pMSC have ∼3.2-fold higher than that UC-MSC migration capacity [20]. Recently, Mineo et al., reported that the effect of exosomes on angiogenesis involves the Src family of kinases [45]. In addition, the role of Src family members in angiogenesis, promotion of tube formation and prevention of their regression has been reported [46], [47]. Recent commentary, suggests that mesenchymal stem cells-derived exosomes may not only afford therapeutic opportunities in regenerative medicine to repair damaged tissue but also in the cell-specific delivery of anticancer agents [48].
The exosomal content is highly dependent on the cell type and pre-conditioning. One of the first exosomal proteomes characterized was from mesothelioma cells, in which 38 different proteins were identified [49]. Studies in cancer cells show the great variability of proteins expressed in exosomes [50]–[54]. Supporting our results, exosomes isolated from a human first trimester cell line (Sw71) Atay et al., using an ion trap mass spectrometry approach, identified proteins implicated in a wide range of cellular processes including: cytoskeleton structure, ion channels, lysosomal degradation, molecular chaperones, amino-acid metabolism, carbohydrate metabolism, lipid metabolism, regulatory proteins, mRNA splicing, immune function and others [55]. Our study provides the first extensive analysis of the proteome of the exosomes derived-MSC primary culture, highlighting the extent of putative functional interactions that may be mediated by exosomes.
Endothelial cell migration requires the initiation of numerous signaling pathways that remodels cytoskeleton. Also, actin and related proteins of cytoskeletal organization are critical for cell motility and migration. From the canonical pathway analysis, we found significantly more proteins associated with actin cytoskeleton, growth hormone, and VEGF signaling in exosomes isolated from pMSC exposed to 1% O2 compare to 3% or 8% O2. Likewise, clathrin-mediated endocytosis signaling was enhanced, possibly increasing the exosome uptake of target cells., Cell migration, however, is the final functional outcome of multiple pathways and the involvement of other regulatory moieties (e.g. miRNA) cannot be negated.
In summary, pMSC isolated from first trimester placenta release exosomes in response to decreased oxygen tension. pMSC exosomes stimulate microvascular endothelial cells migration in a concentration and oxygen-dependent manner, and promote vascular network formation. The data obtained in this study are consistent with the hypothesis that under normal developmental conditions, pMSC promote vasculogenesis and angiogenesis within the early pregnancy placenta via a mechanism(s) involving exosomal trafficking to endothelial cells. We further suggest that in pathological pregnancies associated with under perfusion of the placenta, such as those complicated by pre-eclampsia and intra-uterine growth restriction, increased release of exosomes from pMSC may occur as an adaptive response.
Funding Statement
This research was supported by CONICYT (ACT-73 PIA, Pasantía Doctoral en el Extranjero BECAS Chile) and FONDECYT (1110977). CS holds CONICYT-PhD fellowships and Faculty of Medicine/PUC-PhD fellowships. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bang C, Thum T (2012) Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol 44: 2060–2064. [DOI] [PubMed] [Google Scholar]
- 2.Thebaud B, Stewart DJ (2012) Exosomes: cell garbage can, therapeutic carrier, or trojan horse? Circulation 126: 2553–2555. [DOI] [PubMed] [Google Scholar]
- 3.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, et al.. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Communications 2. [DOI] [PMC free article] [PubMed]
- 4.Bullerdiek J, Flor I (2012) Exosome-delivered microRNAs of “chromosome 19 microRNA cluster” as immunomodulators in pregnancy and tumorigenesis. Molecular Cytogenetics 5. [DOI] [PMC free article] [PubMed]
- 5.Kshirsagar SK, Alam SM, Jasti S, Hodes H, Nauser T, et al.. (2012) Immunomodulatory molecules are released from the first trimester and term placenta via exosomes. Placenta. [DOI] [PMC free article] [PubMed]
- 6.Tolosa JM, Schjenken JE, Clifton VL, Vargas A, Barbeau B, et al. (2012) The endogenous retroviral envelope protein syncytin-1 inhibits LPS/PHA-stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta 33: 933–941. [DOI] [PubMed] [Google Scholar]
- 7.Zhang HG, Zhuang X, Sun D, Liu Y, Xiang X, et al. (2012) Exosomes and immune surveillance of neoplastic lesions: a review. Biotech Histochem 87: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mincheva-Nilsson L, Nagaeva O, Chen T, Stendahl U, Antsiferova J, et al. (2006) Placenta-derived soluble MHC class I chain-related molecules down-regulate NKG2D receptor on peripheral blood mononuclear cells during human pregnancy: a possible novel immune escape mechanism for fetal survival. J Immunol 176: 3585–3592. [DOI] [PubMed] [Google Scholar]
- 9.Alderton GK (2012) METASTASIS Exosomes drive premetastatic niche formation. Nature Reviews Cancer 12: 447–447. [DOI] [PubMed] [Google Scholar]
- 10.Higginbotham JN, Beckler MD, Gephart JD, Franklin JL, Bogatcheva G, et al. (2011) Amphiregulin Exosomes Increase Cancer Cell Invasion. Current Biology 21: 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross JC, Chaudhary V, Bartscherer K, Boutros M (2012) Active Wnt proteins are secreted on exosomes. Nature Cell Biology 14: 1036−+. [DOI] [PubMed]
- 12.Meckes DG, Shair KHY, Marquitz AR, Kung CP, Edwards RH, et al. (2010) Human tumor virus utilizes exosomes for intercellular communication. Proceedings of the National Academy of Sciences of the United States of America 107: 20370–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Li JJ, Yang JY, Wang DS, Zhao W, et al.. (2012) Tolerance Induction by Exosomes from Immature Dendritic Cells and Rapamycin in a Mouse Cardiac Allograft Model. PLoS ONE 7. [DOI] [PMC free article] [PubMed]
- 14.Vrijsen KR, Sluijter JPG, Schuchardt MWL, van Balkom BWM, Noort WA, et al. (2010) Cardiomyocyte progenitor cell-derived exosomes stimulate migration of endothelial cells. Journal of Cellular and Molecular Medicine 14: 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corrado C, Flugy AM, Taverna S, Raimondo S, Guggino G, et al.. (2012) Carboxyamidotriazole-Orotate Inhibits the Growth of Imatinib-Resistant Chronic Myeloid Leukaemia Cells and Modulates Exosomes-Stimulated Angiogenesis. PLoS ONE 7. [DOI] [PMC free article] [PubMed] [Retracted]
- 16.Cantaluppi V, Biancone L, Figliolini F, Beltramo S, Medica D, et al. (2012) Microvesicles Derived From Endothelial Progenitor Cells Enhance Neoangiogenesis of Human Pancreatic Islets. Cell Transplantation 21: 1305–1320. [DOI] [PubMed] [Google Scholar]
- 17.Zhu W, Huang L, Li YH, Zhang X, Gu JM, et al. (2012) Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Letters 315: 28–37. [DOI] [PubMed] [Google Scholar]
- 18.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, et al. (2010) Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res 4: 214–222. [DOI] [PubMed] [Google Scholar]
- 19.Abumaree MH, Al Jumah MA, Kalionis B, Jawdat D, Al Khaldi A, et al.. (2012) Phenotypic and Functional Characterization of Mesenchymal Stem Cells from Chorionic Villi of Human Term Placenta. Stem Cell Rev. [DOI] [PubMed]
- 20.Li G, Zhang XA, Wang H, Wang X, Meng CL, et al. (2011) Comparative proteomic analysis of mesenchymal stem cells derived from human bone marrow, umbilical cord and placenta: implication in the migration. Adv Exp Med Biol 720: 51–68. [DOI] [PubMed] [Google Scholar]
- 21.Welt FG, Gallegos R, Connell J, Kajstura J, D’Amario D, et al.. (2012) Effect of Cardiac Stem Cells on Left Ventricular Remodeling in a Canine Model of Chronic Myocardial Infarction. Circ Heart Fail. [DOI] [PubMed]
- 22.Zhang ZY, Teoh SH, Hui JH, Fisk NM, Choolani M, et al. (2012) The potential of human fetal mesenchymal stem cells for off-the-shelf bone tissue engineering application. Biomaterials 33: 2656–2672. [DOI] [PubMed] [Google Scholar]
- 23.Burlacu A, Grigorescu G, Rosca AM, Preda MB, Simionescu M (2012) Factors Secreted by Mesenchymal Stem Cells and Endothelial Progenitor Cells Have Complementary Effects on Angiogenesis In Vitro. Stem Cells Dev. [DOI] [PMC free article] [PubMed]
- 24.Boomsma RA, Geenen DL (2012) Mesenchymal stem cells secrete multiple cytokines that promote angiogenesis and have contrasting effects on chemotaxis and apoptosis. PLoS One 7: e35685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Luca A, Lamura L, Gallo M, Maffia V, Normanno N (2012) Mesenchymal stem cell-derived interleukin-6 and vascular endothelial growth factor promote breast cancer cell migration. J Cell Biochem 113: 3363–3370. [DOI] [PubMed] [Google Scholar]
- 26.Byeon YE, Ryu HH, Park SS, Koyama Y, Kikuchi M, et al. (2010) Paracrine effect of canine allogenic umbilical cord blood-derived mesenchymal stromal cells mixed with beta-tricalcium phosphate on bone regeneration in ectopic implantations. Cytotherapy 12: 626–636. [DOI] [PubMed] [Google Scholar]
- 27.Xu RX, Chen X, Chen JH, Han Y, Han BM (2009) Mesenchymal stem cells promote cardiomyocyte hypertrophy in vitro through hypoxia-induced paracrine mechanisms. Clin Exp Pharmacol Physiol 36: 176–180. [DOI] [PubMed] [Google Scholar]
- 28.Lee C, Mitsialis SA, Aslam M, Vitali SH, Vergadi E, et al.. (2012) Exosomes Mediate the Cytoprotective Action of Mesenchymal Stromal Cells on Hypoxia-Induced Pulmonary Hypertension. Circulation. [DOI] [PMC free article] [PubMed]
- 29.Rodesch F, Simon P, Donner C, Jauniaux E (1992) Oxygen measurements in endometrial and trophoblastic tissues during early pregnancy. Obstet Gynecol 80: 283–285. [PubMed] [Google Scholar]
- 30.Jauniaux E, Gulbis B, Burton GJ (2003) Physiological implications of the materno-fetal oxygen gradient in human early pregnancy. Reprod Biomed Online 7: 250–253. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann NA, Ortner A, Jacamo RO, Reinisch A, Schallmoser K, et al. (2012) Oxygen sensing mesenchymal progenitors promote neo-vasculogenesis in a humanized mouse model in vivo. PLoS One 7: e44468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steigman SA, Fauza DO (2007) Isolation of mesenchymal stem cells from amniotic fluid and placenta. Curr Protoc Stem Cell Biol Chapter 1: Unit 1E 2. [DOI] [PubMed]
- 33.Chen YS, Pelekanos RA, Ellis RL, Horne R, Wolvetang EJ, et al. (2012) Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. Stem Cells Transl Med 1: 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomon C, Westermeier F, Puebla C, Arroyo P, Guzman-Gutierrez E, et al. (2012) Gestational diabetes reduces adenosine transport in human placental microvascular endothelium, an effect reversed by insulin. PLoS One 7: e40578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thery C, Amigorena S, Raposo G, Clayton A (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3: Unit 3 22. [DOI] [PubMed]
- 36.Thon JN, Devine MT, Jurak Begonja A, Tibbitts J, Italiano JE Jr (2012) High-content live-cell imaging assay used to establish mechanism of trastuzumab emtansine (T-DM1)–mediated inhibition of platelet production. Blood 120: 1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang HC, Liu XB, Huang S, Bi XY, Wang HX, et al.. (2012) Microvesicles Derived from Human Umbilical Cord Mesenchymal Stem Cells Stimulated by Hypoxia Promote Angiogenesis Both In Vitro and In Vivo. Stem Cells Dev. [DOI] [PMC free article] [PubMed]
- 38.Gupta N, Su X, Popov B, Lee JW, Serikov V, et al. (2007) Intrapulmonary delivery of bone marrow-derived mesenchymal stem cells improves survival and attenuates endotoxin-induced acute lung injury in mice. J Immunol 179: 1855–1863. [DOI] [PubMed] [Google Scholar]
- 39.King HW, Michael MZ, Gleadle JM (2012) Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, et al. (2013) Neutral Sphingomyelinase 2 (nSMase2)-dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis. J Biol Chem 288: 10849–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang J, Huang W, Yu X, Ashraf A, Wary KK, et al. (2012) Suicide gene reveals the myocardial neovascularization role of mesenchymal stem cells overexpressing CXCR4 (MSC(CXCR4)). PLoS One 7: e46158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung JJ, Tiwari A, Inamdar SM, Thomas CP, Goel A, et al. (2012) Secretion of soluble vascular endothelial growth factor receptor 1 (sVEGFR1/sFlt1) requires Arf1, Arf6, and Rab11 GTPases. PLoS One 7: e44572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afroze SH, Uddin MN, Cao X, Asea A, Gizachew D (2011) Internalization of exogenous ADP-ribosylation factor 6 (Arf6) proteins into cells. Mol Cell Biochem 354: 291–299. [DOI] [PubMed] [Google Scholar]
- 44.Biancone L, Bruno S, Deregibus MC, Tetta C, Camussi G (2012) Therapeutic potential of mesenchymal stem cell-derived microvesicles. Nephrol Dial Transplant 27: 3037–3042. [DOI] [PubMed] [Google Scholar]
- 45.Mineo M, Garfield SH, Taverna S, Flugy A, De Leo G, et al. (2012) Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a src-dependent fashion. Angiogenesis 15: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Im E, Kazlauskas A (2007) Src family kinases promote vessel stability by antagonizing the Rho/ROCK pathway. J Biol Chem 282: 29122–29129. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Sainz IM, Wu Y, Pixley R, Espinola RG, et al. (2008) The inhibition of tube formation in a collagen-fibrinogen, three-dimensional gel by cleaved kininogen (HKa) and HK domain 5 (D5) is dependent on Src family kinases. Exp Cell Res 314: 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai S, Wei D, Wu Z, Zhou X, Wei X, et al. (2008) Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther 16: 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, et al. (2004) Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol 164: 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, et al. (2010) Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics 9: 1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pisitkun T, Gandolfo MT, Das S, Knepper MA, Bagnasco SM (2012) Application of systems biology principles to protein biomarker discovery: Urinary exosomal proteome in renal transplantation. Proteomics Clin Appl 6: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, et al. (2009) Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20: 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Zhang Y, Qiu F, Qiu Z (2011) Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 32: 1976–1983. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Li Y, Qiu F, Qiu Z (2010) Comprehensive analysis of low-abundance proteins in human urinary exosomes using peptide ligand library technology, peptide OFFGEL fractionation and nanoHPLC-chip-MS/MS. Electrophoresis 31: 3797–3807. [DOI] [PubMed] [Google Scholar]
- 55.Atay S, Gercel-Taylor C, Kesimer M, Taylor DD (2011) Morphologic and proteomic characterization of exosomes released by cultured extravillous trophoblast cells. Exp Cell Res 317: 1192–1202. [DOI] [PubMed] [Google Scholar]