Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean (original) (raw)
Abstract
Planktonic bacteria dominate surface ocean biomass and influence global biogeochemical processes, but remain poorly characterized owing to difficulties in cultivation. Using large-scale single cell genomics, we obtained insight into the genome content and biogeography of many bacterial lineages inhabiting the surface ocean. We found that, compared with existing cultures, natural bacterioplankton have smaller genomes, fewer gene duplications, and are depleted in guanine and cytosine, noncoding nucleotides, and genes encoding transcription, signal transduction, and noncytoplasmic proteins. These findings provide strong evidence that genome streamlining and oligotrophy are prevalent features among diverse, free-living bacterioplankton, whereas existing laboratory cultures consist primarily of copiotrophs. The apparent ubiquity of metabolic specialization and mixotrophy, as predicted from single cell genomes, also may contribute to the difficulty in bacterioplankton cultivation. Using metagenome fragment recruitment against single cell genomes, we show that the global distribution of surface ocean bacterioplankton correlates with temperature and latitude and is not limited by dispersal at the time scales required for nucleotide substitution to exceed the current operational definition of bacterial species. Single cell genomes with highly similar small subunit rRNA gene sequences exhibited significant genomic and biogeographic variability, highlighting challenges in the interpretation of individual gene surveys and metagenome assemblies in environmental microbiology. Our study demonstrates the utility of single cell genomics for gaining an improved understanding of the composition and dynamics of natural microbial assemblages.
Keywords: comparative genomics, marine microbiology, microbial ecology, microbial microevolution, operational taxonomic unit
Planktonic bacteria dominate surface ocean biomass and have a major impact on the global cycling of carbon, nitrogen, and other elements (1). Among the available pure cultures of marine bacterioplankton, only a limited number represent bacterioplankton that are abundant in the ocean, such as the cyanobacteria Prochlorococcus and Synechococcus and the Alphaproteobacteria Pelagibacter (collectively termed PSP cultures). This limits the scope of studies of the microbial metabolic processes and evolutionary changes that impact marine ecosystems and their geochemical cycles (2–6). Unusual nutritional requirements resulting from genome reduction may contribute to cultivation difficulties, as suggested by studies of the chemoheterotroph Pelagibacter (7, 8) and the methylotroph OM43 (9).
Although prevailing culture-independent tools, including microbial community shotgun sequencing, targeted gene surveys, and fluorescent in situ hybridization, have revealed the extent and significance of microbial diversity, they have not been able to provide the genome context information required for accurate metabolic reconstruction spanning organismal, population, and community levels of organization (10). As a result, the genomic repertoires, natural histories, and geographic distribution of even the most abundant taxonomic groups of marine bacterioplankton remain largely unknown (1, 11). Microbial studies in other environments, such as the human body and soils, face similar challenges (10). The recent development of robust protocols for single cell genomics provides a versatile, cultivation-independent approach for assessing natural microbial diversity with corresponding genome context information (12).
To determine whether genome streamlining is a prevalent feature among free-living marine bacterioplankton, and to analyze global patterns of surface ocean bacterioplankton distribution, we obtained draft genomes of 56 single amplified genomes (SAGs) (5, 13–15) and compared them with existing bacterioplankton cultures and metagenomes. The sequenced SAGs represent many ubiquitous surface ocean bacteria lineages, including Marine Group A, Verrucomicrobia, Actinobacteria, Bacteroidetes, and Proteobacteria lineages SAR86, ARCTIC96BD-19, SAR92, SAR116, and Roseobacter (SI Appendix, Fig. S1). The majority of these groups have few or no cultured representatives. Members of the PSP group were excluded from SAG selection, because their genome streamlining and environmental abundance have been demonstrated previously (1, 2, 4, 11). Samples for SAG generation were collected from the Gulf of Maine, the Mediterranean Sea, and the subtropical gyres of the North Pacific and South Atlantic Oceans (SI Appendix, Table S1). On average, 55% (range, 0.3–97.8%) of the genome was recovered from each analyzed cell (SI Appendix, Table S2). A subset of 41 SAGs, each >0.75 Mbp in size and with >30% estimated genome recovery, was used for our comparative genomics and biogeographic analyses. Our results demonstrate that genome streamlining is a prevalent evolutionary strategy among free-living bacterioplankton in the surface ocean. They also suggest that the global distribution of the majority of surface ocean bacterioplankton might not be limited by dispersal and is correlated with temperature and latitude.
Results and Discussion
Genomic Signatures of Streamlining and Oligotrophy Among Uncultured Marine Bacteria.
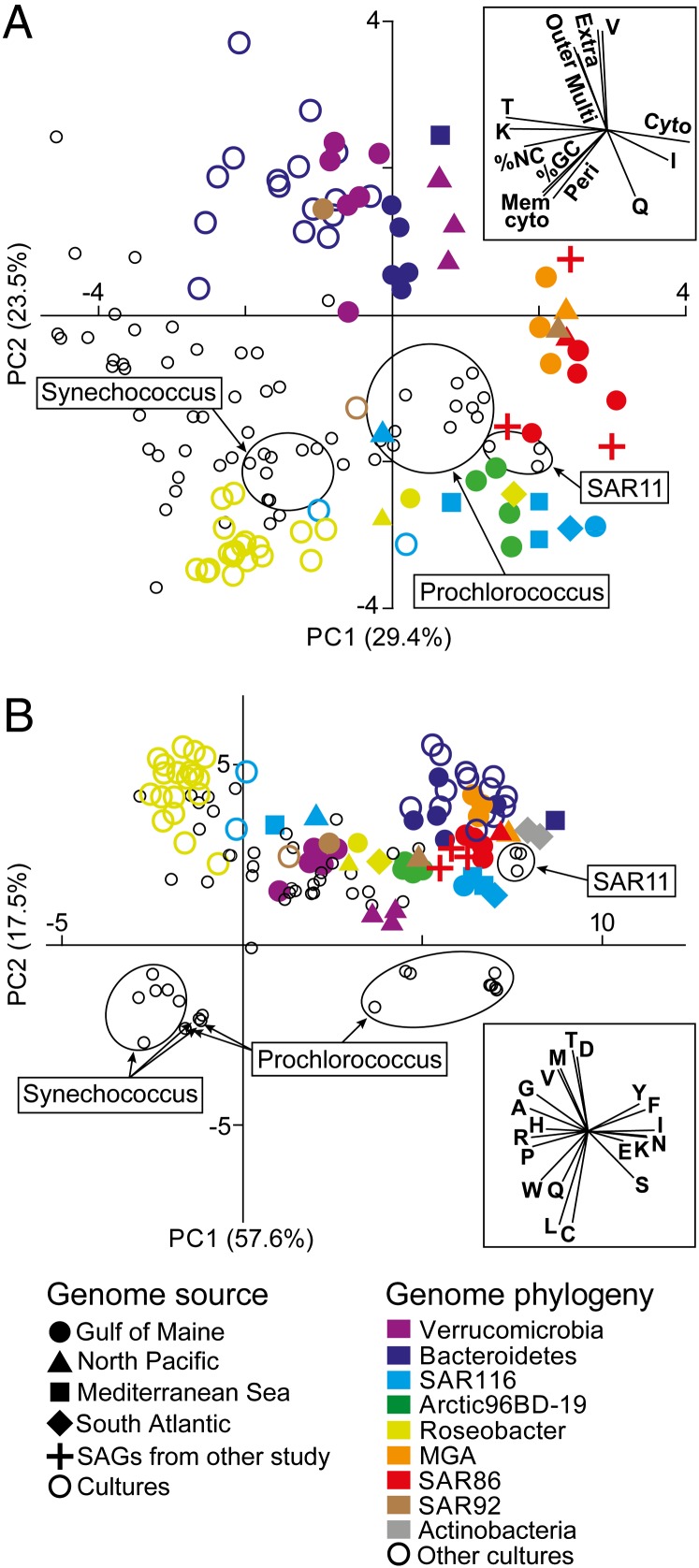
A comparison of general genome features among marine bacterioplankton revealed that the majority of our SAGs clustered with cultures of Prochlorococcus and Pelagibacter, as well as with the SAR86 SAGs sequenced by Dupont et al. (6) (Fig. 1_A_ and SI Appendix, Table S3). SAGs segregated from cultures along a principal component axis associated with low guanine and cytosine (GC) content, low percentage of noncoding nucleotides, low fraction of genes encoding periplasm and cytoplasm membrane proteins, and Clusters of Orthologous Groups (COG) categories K (transcription) and T (signal transduction). These genomic signatures have been identified as indicators of genome streamlining and oligotrophy (16, 17). All Verrucomicrobia and Bacteroidetes SAGs, one SAR92 SAG, and all Bacteroidetes cultures clustered separately from other SAGs and cultures (Fig. 1_A_). These genomes are associated with elevated frequency of genes encoding extracellular, outer membrane and multilocation proteins, and COG category V (defense mechanisms), corroborating the previously proposed role of Bacteroidetes (18, 19) and the recently suggested importance of Verrucomicrobia (14) in macromolecule degradation, a process requiring cell surface-associated or extracellular hydrolases. SAGs of the same taxonomic group but retrieved from different geographic locations had similar genomic signatures, indicating that the selection for these signatures operates in both the open ocean and coastal waters, and in diverse climate zones. In contrast, large differences in genomic signatures were found between SAGs and their cultured relatives within each taxonomic group that contains multiple SAGs and cultures, such as Roseobacter, SAR116, and Bacteroidetes (SI Appendix, Table S4).
Fig. 1.
Genomic differences between SAGs and cultured bacterioplankton. PCA of general genome characteristics (A) and encoded amino acid frequency (B) of SAGs (solid colored symbols) and cultures of marine bacterioplankton (open circles) are shown. Cultures belonging to the same taxonomic group as SAGs have the same color. The two Actinobacteria SAGs were excluded from the genome characteristics analysis because they are Gram-positive bacteria, which have a different cell wall architecture, and were not included in the development of the trophic strategy model of Lauro et al. (17). (Insets) Variable vectors corresponding to each PCA plot. The following input variables were used for the genome characteristic analysis: abundance of genes encoding proteins localized in the cytoplasm; cytoplasmic membrane, periplasm, outer membrane, extracellular, and multiple locations; COG categories I, K, Q, T, and V; %NC, % noncoding DNA.
Obligate oligotrophy has been proposed as a key factor leading to poor recovery of environmental microorganisms in pure cultures (19–21), and our study provides clear evidence for the predominance of a copiotroph lifestyle among existing marine cultures across taxonomic groups. Our data also suggest that oligotroph characteristics in surface ocean bacteria are not limited to members of Prochlorococcus and Pelagibacter in tropical regions, as previously thought (16, 22), but rather is a common trophic strategy among many bacterioplankton lineages around the globe.
As one of the variables contributing to genomic differences between SAGs and cultures (Fig. 1_A_), the average GC content of SAGs (37.9%) was significantly lower than that of the 101 marine bacterioplankton cultures (48.5%; SI Appendix, Fig. S2_A_). Although multiple displacement amplification (MDA) of mixed templates may introduce GC biases, here such biases were eliminated by performing MDA on individual cells, followed by high-coverage sequencing and de novo assembly, which have been demonstrated to accurately reconstruct GC of the analyzed genomes (23–25). The high similarity of the average GC content of SAGs (37.9%) and available surface ocean metagenomes (39.6%) provides further support for the representativeness of our SAG data (SI Appendix, Table S5). The difference in %GC between SAGs and cultures was significant in both coding and noncoding genome regions, suggesting GC content rather than protein composition as the primary adaptive trait (SI Appendix, Fig. S2_B_).
SAGs differed from cultures in the frequency of encoded amino acids (Fig. 1_B_ and SI Appendix, Table S6), with SAGs being enriched in tyrosine, phenylalanine, isoleucine, glutamic acid, asparagine, lysine, and serine and depleted in valine, glycine, alanine, arginine, proline, histidine, and tryptophan. These two groups of amino acids were similar in terms of chemical properties, synthesis costs, and numbers of C and N atoms (SI Appendix, Table S7), but diverged in average GC content of the first two nucleotides of their codons (14% and 79%, respectively). This finding provides further evidence that differences in amino acid utilization between SAGs and cultures are driven primarily by differences in %GC. Recent experimental work suggests that high GC content may enhance bacterial growth in laboratory conditions (26). In contrast, low genomic GC content may be an adaptation to nitrogen limitation (27) or a result of mutational biases in the absence of effective DNA repair systems (16). It remains to be understood how the observed GC depletion in bacterioplankton and the resulting shifts in amino acid use impact surface ocean processes.
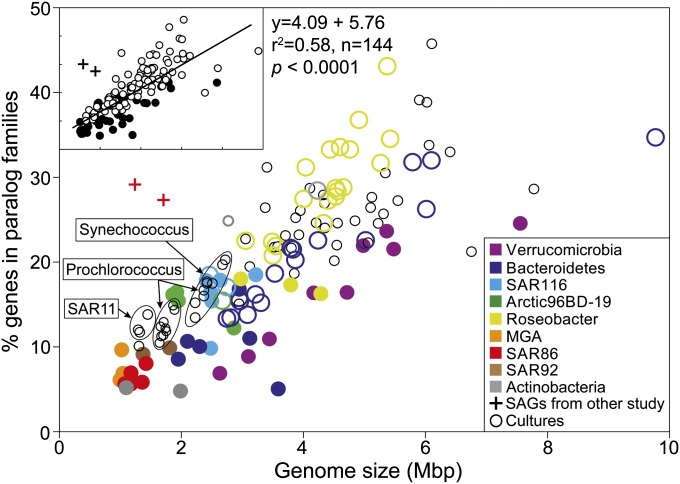
One predicted cost of genome streamlining in free-living bacteria is a reduction in physiological flexibility, leading to specialization in resource utilization. Accordingly, SAGs had fewer paralogs and smaller genomes compared with cultures from the same taxonomic groups, with the exception of SAR116 (Fig. 2). The low paralog frequency is not likely the result of incomplete genome recovery from SAGs, given that partial genes at the ends of contigs may be incorrectly assigned as paralogs, leading to overestimation of paralogs. This effect is evident in the substantially higher fraction of paralogs identified from highly fragmented SAR86 SAG assemblies sequenced by Dupont et al. (6) compared with the SAR86 SAGs reported here. This overall trend suggests that the small genome size and fewer gene duplications may provide an adaptive advantage to life in the oligotrophic ocean.
Fig. 2.
Genome size and paralogous gene frequency of SAGs and bacterioplankton cultures. The percentages of genes belonging to paralog families in SAGs (solid colored circles) and cultures (open circles) were estimated using BLASTCLUST. Cultures belonging to the same taxonomic group as SAGs have the same color. (Inset) Results of least squares linear regression between genome size and paralog frequency.
Comparisons of metabolic potential among taxonomic groups represented by multiple SAGs provide strong evidence for specialized resource utilization despite incomplete genome recovery from individual SAGs (SI Appendix, Figs. S3–S8 and Tables S8–S10). For example, Gammaproteobacteria lineages SAR86, SAR92, and ARCTIC96BD-19 encode a heterotrophic central metabolism but differ in terms of pathway completeness and variation. Moreover, genes encoding the oxidative component of the pentose phosphate metabolism are absent in most SAR86 SAGs, but this pathway was found to be complete in most ARCTIC96BD-19 SAGs (SI Appendix, Table S9). Evidence of autotrophic carbon fixation was found only in ARCTIC96BD-19 SAGs, which harbor the RuBisCO operon, as previously reported for SAGs of this lineage from the mesopelagic zone (15). Only the SAR116 SAGs encoded form I coxL, indicating a functional carbon monoxide dehydrogenase (SI Appendix, Fig. S7). Genes supporting various inorganic sulfur utilization pathways were common and lineage-specific, including polysulfide reductase (psr) in Marine Group A, the sox (sulfur oxidation) operon in SAR116, and adenylylsulfate reductase (aprA) among members of ARCTIC96BD-19. Proteorhodopsin genes were found consistently in Marine Group A and ARCTIC96BD-19 SAGs, expanding the taxonomic groups known to encode these photometabolic systems (SI Appendix, Fig. S4 and Table S10).
The ubiquity of metabolic specialization and mixotrophy, as suggested by these data, may contribute to difficulties in cultivating marine bacterioplankton. Accordingly, a member of the ARCTIC96BD-19 lineage was recently cultured from the surface ocean and found to oxidize thiosulfate (28), as was suggested by genome information obtained from SAGs in our previous study (15). Thus, single cell genomics provides a means for the discovery of genes that can be unequivocally assigned to uncultured taxonomic groups, thereby providing critical knowledge about their biology, including clues for cultivation strategies.
Biogeography of Marine Bacterioplankton.
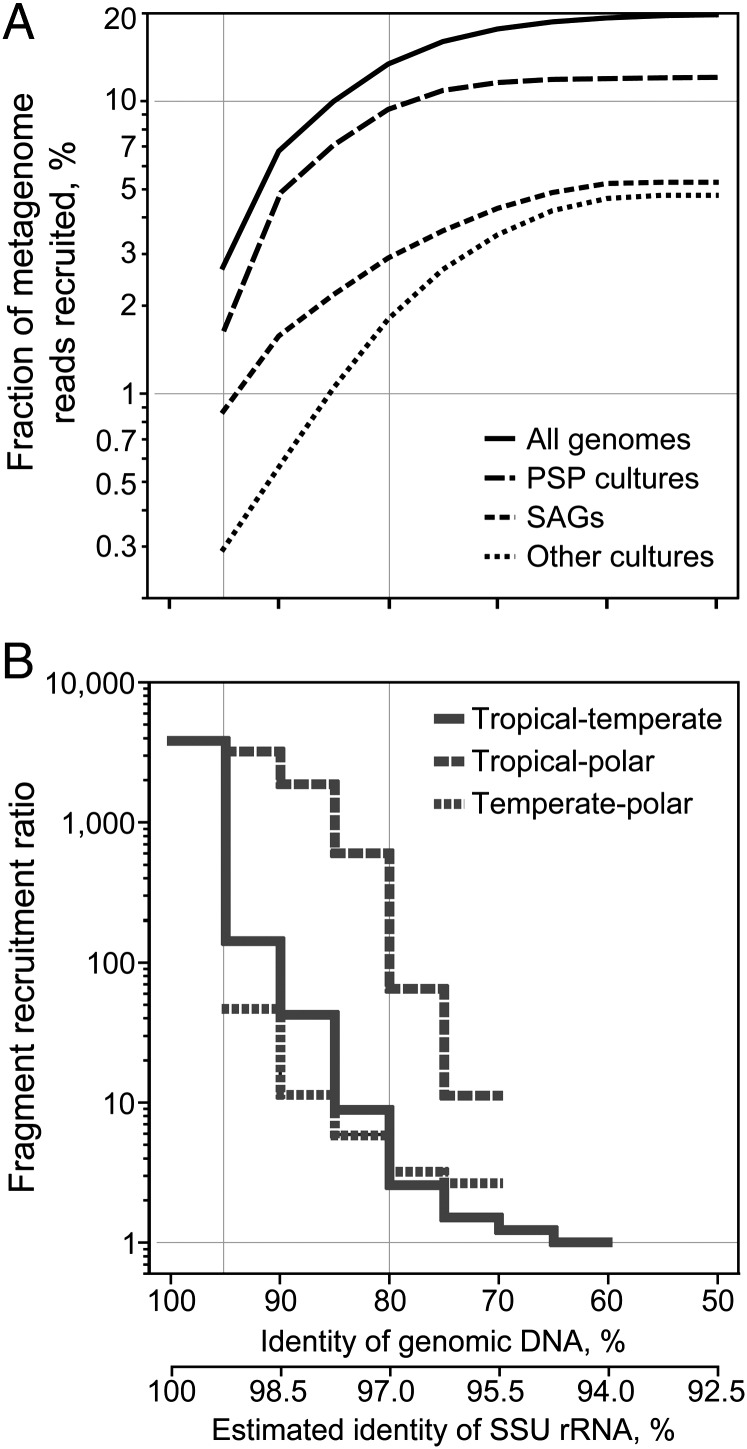
We analyzed the global distribution of surface ocean bacterioplankton using SAGs as references in fragment recruitment (4–6) of publicly available metagenomes, which span diverse geographic regions and climate zones and contain 45 million sequence reads totaling 23 Gbp (SI Appendix, Table S5 and Fig. S9). Using the 95% genomic DNA identity threshold, an operational delineation of taxonomically defined microbial species (29), the combined set of our 41 SAGs recruited an average of 0.9% reads from each surface ocean metagenome (Figs. 3 and 4_A_). The available PSP genomes (Prochlorococcus, Synechococcus, and Pelagibacter; a total of 24) recruited 1.6%, whereas the remaining 82 genomes of marine bacterioplankton cultures recruited only 0.3% (Fig. 4_A_). Lowering the DNA identity threshold in fragment recruitment resulted in a linear increase in the fraction of recruited reads until BLAST effectiveness diminished at nucleotide identities <60%. At this relaxed threshold, which corresponds to ∼94% identity of the small subunit (SSU) rRNA gene (30) and an approximate, operational delineation of taxonomic order (31), 5.2%, 12.0%, 4.7%, and 19.3% of marine metagenome reads were recruited by SAGs, PSP genomes, 82 other bacterioplankton cultures, and a combined set of all genomes, respectively. Although the majority of marine bacterioplankton remains genomically unexplored, single cell sequencing offers a practical solution for genome recovery of uncultivated environmental microorganisms.
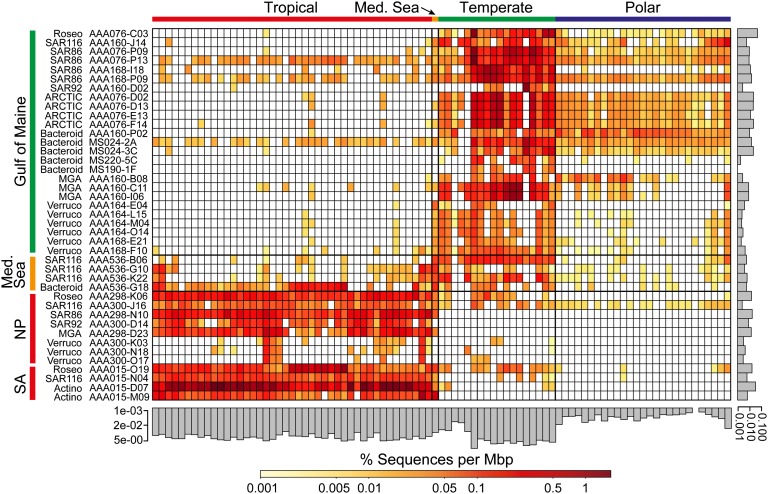
Fig. 3.
Global distribution of SAG-related microorganisms, as determined by metagenomic fragment recruitment. SAGs are listed along the _y_-axis, where color bars indicate source locations. Color bars along the _x_-axis indicate the surface ocean climate zone (SI Appendix, Table S5 provides locations). Metagenomes are in the same order as presented in SI Appendix, Fig. S10 along the top _x_-axis. The scale bar indicates the percentage of aligned metagenome sequences with alignments ≥200 bp long and ≥95% identity, normalized by the length of each SAG assembly. Percentages of aligned sequences from each metagenome to all SAGs, and from all metagenomes to individual SAGs, are presented as gray bars on the _y_-axis and _x_-axis, respectively. Med. Sea, Mediterranean Sea; NP, North Pacific; SA, South Atlantic; Roseo, Roseobacter; ARCTIC, ARCTIC96-BD19 cluster; Bacteroid, Bacteroidetes; MGA, Marine Group A; Verruco, Verrucomicrobia; Actino, Actinobacteria. A threshold of ≥95% nucleotide sequence identity of alignments ≥200 bp was applied for the BLASTN-based recruitment.
Fig. 4.
Capacity of available genomes to represent surface ocean bacterioplankton assemblages, as related to genetic divergence and geographic differences. (A) Fraction of marine metagenome reads recruited by SAGs, genomes of bacterioplankton cultures, and the combined set of genomes using a range of genomic DNA identity thresholds. (B) Ratio of recruitment in the SAGs’ native versus nonnative environment as a function of genomic DNA identity. Averages of values calculated for each metagenome (A) or genome (B) are provided. The scale of the SSU rRNA gene divergence was estimated using a Bacteria domain-wide correlation between SSU rRNA gene identity and the average nucleotide identity of available genomes (31). A threshold of ≥200-bp alignment was applied for the BLASTN-based recruitment.
Using the 95% genomic DNA identity threshold, all SAGs obtained from the Gulf of Maine recruited the highest fraction of metagenomes from temperate regions (average temperature, 11.7 °C; range, 4.0–18.2 °C), which are represented by the northeast and northwest coasts of North America, the Atlantic coast of Europe, and the Indian Ocean off New Zealand in available datasets (Fig. 3). In contrast, SAGs obtained from the two subtropical gyres recruited primarily from warm-water metagenomes in the Atlantic, Pacific, and Indian Oceans (average temperature, 25.7 °C; range, 18.6–29.3 °C; designated “tropical”). SAGs recovered from the Mediterranean Sea, which has an intermediate climate, recruited relatively evenly across temperate and tropical metagenomes. Metagenomes from the Southern Ocean (average temperature, −0.1 °C; range, −2.0 to 4.2 °C; designated “polar”) recruited primarily to SAGs from the Gulf of Maine, although significantly less compared with temperate metagenomes. In contrast to recruitment to SAGs, metagenome fragment recruitment to the majority of marine cultures was limited, and fewer clear biogeographic patterns were apparent (SI Appendix, Figs. S10 and S11), in agreement with previous observations (4, 32).
The abundance of specific genotypes, determined by metagenome fragment recruitment, was most strongly correlated with surface water temperature and latitude (SI Appendix, Fig. S12). Chlorophyll a concentration, water column depth, and longitude were minor factors in the ordination, suggesting that phytoplankton abundance, proximity to the coast, and geographic distance among sampling stations are less important than latitude in determining the abundance of most analyzed genotypes. These findings corroborate recent reports of temperature as a major driver of the global distribution of marine algae (33, 34) and Pelagibacter (35). Temperature and latitude also have been identified as key determinants of less-specific descriptors of marine bacterioplankton biogeography, such as community richness (36) and the frequency of functionally related genes (37–39), for which our study provides extensive genomic context.
We estimated the ratio of metagenomic fragment recruitment from native versus nonnative climate zones, relative to SAG collection site, at various DNA identity intervals as proxies for evolutionary distance (Fig. 4_B_). In the case of temperate versus tropical zones, the ratio was highest (3,827) at 95–100% DNA identity, decreased to 154 at 90–95% identity, and declined to <10 at 80–85% identity. This pattern was similar for all taxonomic groups analyzed. The corresponding ratios were similar when comparing recruitment by temperate SAGs in temperate versus polar environments, but were higher when comparing recruitment by tropical SAGs in tropical versus polar environments. Thus, operationally defined species (>95% genomic DNA identity) were highly specific to their climate zones, but little geographic specificity was observed within phylogenetic groups that shared <80% genomic DNA identity, which corresponds to ∼97% identity of the SSU rRNA gene (31). Accordingly, several bacterioplankton cells analyzed in this study shared >97% identity of their SSU rRNA genes even though they originated from divergent climate zones and demonstrated contrasting geography in metagenome fragment recruitment; examples include SAR116 SAGs AAA158-M15 versus AAA015-N04 and SAR86 SAGs AAA298-N10 versus AAA076-P09 (SI Appendix, Fig. S1).
Whereas the SSU rRNA gene identities were high in these pairs of SAGs, the average nucleotide identity (29) was only 75% and 71%, respectively. The >97% identity of the SSU rRNA gene is the most widely used delineator of operational taxonomic units (OTUs) in microbial ecology. However, it is often overlooked that such OTUs encompass much broader phylogenetic groups than the currently accepted, operationally defined bacterial species, and may contain organisms with divergent adaptations. Thus, insufficient phylogenetic resolution might explain the difficulties encountered in earlier studies in detecting consistent differentiation of bacterioplankton along longitudinal gradients when using SSU rRNA gene surveys (35, 39, 40) or metagenome fragment recruitment with relaxed settings (32), although the more pronounced differences between polar and tropical bacterioplankton have been reported from such studies (35, 39, 40). Here, metagenome fragment recruitment using stringent settings and environmentally relevant, single cell genomes as references enabled us to identify previously undetected, community-wide genetic divergence among tropical, temperate, and polar marine bacterioplankton.
Assuming 1% divergence of the SSU rRNA gene every 50 Ma (41), we estimate that bacterioplankton genetic differences among the three climate zones might have accumulated over tens to hundreds of millions of years. Although such estimates contain significant uncertainties (42, 43), it is clear that the required evolutionary timeframe encompasses numerous overturns of the global ocean by surface currents and thermohaline circulation, which take 1,000–2,000 y each (44). These estimates corroborate the absence of longitudinal effects on fragment recruitment (Figs. 3 and 4) and suggest that the observed differences in bacterioplankton composition between nonpolar climate zones are not driven by dispersal limitations, but are defined by evolutionary innovation enabling certain genotypes to thrive in a specific climate zone. Given our lack of direct evidence for the genomic context of recruited metagenome fragments, how local populations of surface ocean bacterioplankton vary by their genome organization remains to be determined. Nevertheless, our data suggest that the global distribution of surface ocean bacterioplankton genes is not limited by dispersal at the time scales required for nucleotide substitution to exceed the current operational definition of bacteria species, thus adding some evolutionary constraints to the famous statement that “everything is everywhere, but the environment selects” (45).
Summary
Using large-scale single cell genomic sequencing and metagenome fragment recruitment, we have provided extensive, cultivation-independent insight into the genome-level diversity, metabolic potential, and biogeography of many abundant bacterial lineages inhabiting the surface ocean. Our data provide clear evidence that existing laboratory cultures consist mostly of copiotrophic genotypes, compared with free-living bacterioplankton that are streamlined for growth under resource-poor conditions. We also show that the global distribution of the majority of surface ocean bacterioplankton is correlated with temperature and latitude and is not likely limited by dispersal. Individual cells with highly similar SSU rRNA gene sequences exhibited significant genomic and biogeographic variability, highlighting challenges in the interpretation of individual gene surveys and metagenome assemblies in environmental microbiology. Our study demonstrates the utility of single cell genomics in providing a significantly improved understanding of the composition and dynamics of natural microbial assemblages in the ocean and other environments, which will be critical in predicting how ecosystems respond to large-scale environmental shifts, such as global warming and ocean acidification.
Materials and Methods
Collection and Construction of SAGs.
Replicate, 1-mL aliquots of water collected for single cell analyses were cryopreserved with 6% glycine betaine (Sigma-Aldrich) and stored at −80 °C or in liquid nitrogen (46). Single cell sorting, whole-genome amplification, real-time PCR screens, and PCR product sequence analyses were performed at the Bigelow Laboratory Single Cell Genomics Center (www.bigelow.org/scgc), as described by Stepanauskas and Sieracki (13) for SAGs MS024-2A, MS024-3C, MS190-1F, and MS220-5C and by Swan et al. (15) and Martinez-Garcia et al. (14) for the remaining SAGs.
SSU rRNA gene sequences were edited using Sequencher v4.7 (Gene Codes) and compared with previously deposited sequences using the RDP v10 Classifier (SSU rRNA) and National Center for Biotechnology Information BLAST. SAG SSU rRNA sequences were aligned with selected database sequences using ClustalW. Alignment columns with > 90% gaps were removed, and a maximum likelihood tree (100 bootstrap replicates) was constructed using PhyML implemented in Geneious v6.0.5 (47). Details of SAG sequencing, assembly, and annotation are provided in the SI Appendix.
Genome Recovery Estimation of SAGs and Determination of Paralogs.
To estimate the completeness of each assembled SAG genome, we analyzed all finished genome sequences of the taxonomic phyla Alphaproteobacteria (n = 145), Gammaproteobacteria (n = 317), Bacteroidetes (n = 22), and Actinobacteria (n = 131); the taxonomic phylum Verrucomicrobia (n = 4); and the taxonomic domain Bacteria (n = 1,023) available from the Integrated Microbial Genomes (IMG) database (48). Based on COG gene classifications, a set of conserved single copy genes (CSCGs) was extracted for each group of finished genomes from the IMG database. A CSCG was defined as a gene that occurs only once in each of 99% (95% in the case of the domain Bacteria) of the genomes contributing to the taxonomic group. The number of CSCGs for each group was as follows: Alphaproteobacteria, n = 58; Gammaproteobacteria, n = 47; Bacteroidetes, n = 86; Actinobacteria, n = 60; Verrucomicrobia, n = 330; Bacteria, n = 45. The ratio of the number of CSCGs observed for each SAG assembly and for the corresponding taxonomic group of finished genomes was used as a measure of genome recovery (SI Appendix, Table S2).
The frequency of paralog gene families within SAGs and marine cultures was determined using BLASTCLUST with the following settings: −L 0.5 −S 30.0 −e 1e-6. The number of paralogs out of the total number of protein coding genes was calculated for each genome.
Multivariate Analysis of SAG and Marine Culture Genome Signatures.
The amino acid frequencies of 41 SAGs and bacterioplankton genomes were determined using Geneious v. 6.0.5, arcsin square root-transformed, and analyzed using principal components analysis (PCA) after standardization of values. Several genome characteristics found to separate marine prokaryotes by lifestyle (i.e., frequency of protein localizations and several COG categories) were calculated for SAGs and marine culture genomes as described previously (17), as was %GC and noncoding DNA, and these values were used as input for a second PCA analysis as described above. For this second PCA, the two Actinobacteria SAGs AAA015-D07 and AAA015-M09 were excluded. All PCAs were conducted using PRIMER v6.0.
Fragment Recruitment Analysis.
The basic approach of Rusch et al. (4) was used to estimate the abundances of relatives of SAGs and bacterioplankton cultures within each metagenome. BLAST+ v2.2.25 was used to recruit metagenome sequences to each SAG assembly using default parameter values, except for the following: -evalue 0.0001 -reward 1 -penalty -1 -soft_masking true -lcase_masking -xdrop_gap 150. Genome contigs ≥2,000 kbp from each SAG were used in the fragment recruitment analysis. The 23S, 16S, 5S, and ITS regions were masked in each genome before recruitment. The percentage of unique recruits (≥200 bp long and matching at ≥95% identity) from each metagenome matching to each SAG was normalized by genome length. The percentage of unique reads for each metagenome–genome pair was also determined at 90%, 85%, 80%, 75%, 70%, 65%, 60%, 55%, and 50% identity thresholds. SAG abundances from each metagenome were calculated from BLAST output and plotted using custom R scripts. Metagenomes used in fragment recruitment analysis were quality processed using PRINSEQ (49), and all sequences with the following characteristics were removed from further analysis: sequences <100 bp, sequences containing any ambiguities (Ns), all forms of replicate and duplicate sequences, and sequences with a minimum entropy value of 70 (applied to pyrosequencing datasets only).
Environmental and Sample Location Correlations with Fragment Recruitment Abundances.
The influence of environmental factors on fragment recruitment-derived community composition was determined using nonmetric multidimensional scaling (MDS). MDS is an ordination technique that plots samples as points in low-dimensional space while attempting to maintain the relative distances between points as close as possible to the actual rank order of similarities between samples (50). Thus, metagenomes with similar community composition are plotted closer together in ordination space. A stress factor calculated for each MDS ordination indicates how well plotted configurations of sample distances agree with original rank orders calculated from the similarity matrices. SAG recruitment abundances were arcsin square root-transformed, and the Bray–Curtis distance was calculated for the MDS analysis. Sampling and environmental factors used for axis correlations were temperature, chlorophyll a concentration, water column depth at the sampling location (log-transformed), and latitude and longitude of the sampling location. All MDS calculations were performed using PC-ORD v6.08.
Calculation of Average Nucleotide Identity Between Genomes.
Average nucleotide identity (ANI) values between the pairs of SAR116 SAGs AAA158-M15 and AAA015-N04 and SAR86 SAGs AAA298-N10 and AAA076-P09 were calculated following the method described by Goris et al. (29), using a custom Perl script. Each SAG served as a reference genome, and resulting ANI values were averaged.
Supplementary Material
Supporting Information
Acknowledgments
We thank David Emerson, William Hanage, Siv Anderson, and Jessica Labonte for valuable comments; Edward F. DeLong for metagenomes from Hawaii ocean time (HOT) station ALOHA; the officers and crew of the RV Ka'imikai-O-Kanaloa and the HOT team for sample collection at station ALOHA; and Jane Heywood and Michael Sieracki for South Atlantic field sample collection. This work was supported by National Science Foundation Grants EF-826924 (to R.S.), OCE-821374 (to R.S.), and OCE-1232982 (to R.S. and B.K.S.); US Department of Energy (DOE) JGI 2011 Microbes Program Grant CSP 387 (to R.S., B.K.S., S.G., M.A.M., F.M.L., R.C. and S.G.A.); the Gordon and Betty Moore Foundation (M.A.M.); Spanish Ministry of Science and Innovation Grant CGL2011-26848/BOS (to S.G.A) and CONSOLIDER-INGENIO2010 Program Grant CSD2008-00077 (to S.G.A. and J.M.G.); the Natural Sciences and Engineering Research Council of Canada (NSERC); the Canada Foundation for Innovation, and the Canadian Institute for Advanced Research (CIFAR; S.J.H.). J.J.W. was supported by NSERC. Research activities of R.C. and F.M.L. are supported by the Australian Research Council, and research activities of R.C. are supported by the Australian Antarctic Science program. Work conducted by the DOE Joint Genome Institute is supported by the DOE’s Office of Science under Contract DE-AC02-05CH11231. This is contribution no. 006 of the Tara Oceans Expedition 2009–2012.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Whole-genome sequence data for single amplified genomes used for our analyses are available in the Joint Genome Institute's Integrated Microbial Genome database, http://img.jgi.doe.gov/cgi-bin/w/main.cgi (accession nos. 643886079, 643886118, 2228664025-26, 2228664028-29, 2228664032, 2228664034, 2228664052-53, 2228664055-56, 2236347001, 2236347003, 2236347013, 2236347015, 2236347017-19, 2236347021-24, 2236347026-27, 2236347030-33, 2236347035-36, 2236347039, 2236347041, 2236347043, 2236661010, 2236661014, 2236661017-18, 2507262045, 2507262047, and 2517572139).
References
- Glockner FO, et al. (2012) Marine Microbial Diversity and Its Role in Ecosystem Functioning and Environmental Change. Marine Board Position Paper 17. eds Calewaert JB, McDonough N (Marine Board, European Science Foundation, Ostend, Belgium)
- 2.Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345(6270):60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 3.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5(3):e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woyke T, et al. Assembling the marine metagenome, one cell at a time. PLoS ONE. 2009;4(4):e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6(6):1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tripp HJ, et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452(7188):741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 8.Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph “Candidatus Pelagibacter ubique” HTCC1062 on a defined medium. ISME J. 2013;7(3):592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halsey KH, Carter AE, Giovannoni SJ. Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ Microbiol. 2012;14(3):630–640. doi: 10.1111/j.1462-2920.2011.02605.x. [DOI] [PubMed] [Google Scholar]
- 10.Temperton B, Giovannoni SJ. Metagenomics: Microbial diversity through a scratched lens. Curr Opin Microbiol. 2012;15(5):605–612. doi: 10.1016/j.mib.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 12.Stepanauskas R. Single cell genomics: An individual look at microbes. Curr Opin Microbiol. 2012;15(5):613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA. 2007;104(21):9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Garcia M, et al. Capturing single cell genomes of active polysaccharide degraders: An unexpected contribution of Verrucomicrobia. PLoS ONE. 2012;7(4):e35314. doi: 10.1371/journal.pone.0035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swan BK, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333(6047):1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 16.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309(5738):1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 17.Lauro FM, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106(37):15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cottrell MT, Kirchman DL. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66(4):1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo H. Predicted protein subcellular localization bacterioplankton in dominant surface ocean. Appl Environ Microbiol. 2012;78(18):6550–6557. doi: 10.1128/AEM.01406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannoni S, Stingl U. The importance of culturing bacterioplankton in the “omics” age. Nat Rev Microbiol. 2007;5(10):820–826. doi: 10.1038/nrmicro1752. [DOI] [PubMed] [Google Scholar]
- 21.Schut F, Prins RA, Gottschal JC. Oligotrophy and pelagic marine bacteria: Facts and fiction. Aquat Microb Ecol. 1997;12:177–202. [Google Scholar]
- 22.Coleman ML, Chisholm SW. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 2007;15(9):398–407. doi: 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigue S, et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE. 2009;4(9):e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71(6):3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104(29):11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci USA. 2012;109(36):14504–14507. doi: 10.1073/pnas.1205683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grzymski JJ, Dussaq AM. The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 2012;6(1):71–80. doi: 10.1038/ismej.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall KT, Morris RM. Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J. 2013;7(2):452–455. doi: 10.1038/ismej.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 30.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102(7):2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187(18):6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yooseph S, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468(7320):60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 33.Barton AD, Dutkiewicz S, Flierl G, Bragg J, Follows MJ. Patterns of diversity in marine phytoplankton. Science. 2010;327(5972):1509–1511. doi: 10.1126/science.1184961. [DOI] [PubMed] [Google Scholar]
- 34.Thomas MK, Kremer CT, Klausmeier CA, Litchman E. A global pattern of thermal adaptation in marine phytoplankton. Science. 2012;338(6110):1085–1088. doi: 10.1126/science.1224836. [DOI] [PubMed] [Google Scholar]
- 35.Brown MV, et al. Global biogeography of SAR11 marine bacteria. Mol Syst Biol. 2012;8:595. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuhrman JA, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105(22):7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gianoulis TA, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci USA. 2009;106(5):1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X, et al. Functional biogeography of ocean microbes revealed through non-negative matrix factorization. PLoS ONE. 2012;7(9):e43866. doi: 10.1371/journal.pone.0043866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghiglione JF, et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA. 2012;109(43):17633–17638. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pommier T, Pinhassi J, Hagstrom A. Biogeographic analysis of ribosomal RNA clusters from marine bacterioplankton. Aquat Microb Ecol. 2005;41(1):79–89. [Google Scholar]
- 41.Ochman H, Wilson AC. Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26(1-2):74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 42.Kuo CH, Ochman H. Inferring clocks when lacking rocks: The variable rates of molecular evolution in bacteria. Biol Direct. 2009;4:35. doi: 10.1186/1745-6150-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho SYW, et al. Time-dependent rates of molecular evolution. Mol Ecol. 2011;20(15):3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- 44.Doos K, Nilsson J, Nycander J, Brodeau L, Ballarotta M. The world ocean thermohaline circulation. J Phys Oceanogr. 2012;42:1445–1460. [Google Scholar]
- 45.Baas Becking LGM. Geobiologie of Inleiding tot de Milieukunde. The Hague, The Netherlands: W.P. Van Stockum & Zoon; 1934. [Google Scholar]
- 46.Cleland D, Krader P, McCree C, Tang J, Emerson D. Glycine betaine as a cryoprotectant for prokaryotes. J Microbiol Methods. 2004;58(1):31–38. doi: 10.1016/j.mimet.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 47.Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct. 2007;2:27. doi: 10.1186/1745-6150-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markowitz VM, et al. The integrated microbial genomes system: An expanding comparative analysis resource. Nucleic Acids Res. 2010;38(Suppl. 1):D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information