Fluorescence-Based Sensors to Monitor Localization and Functions of Linear and K63-Linked Ubiquitin Chains in Cells (original) (raw)
. Author manuscript; available in PMC: 2013 Jul 18.
SUMMARY
Ubiquitin chains modify a major subset of the proteome, but detection of ubiquitin signaling dynamics and localization is limited due to a lack of appropriate tools. Here, we employ ubiquitin-binding domain (UBD)-based fluorescent sensors to monitor linear and K63-linked chains in vitro and in vivo. We utilize the UBD in NEMO and ABIN (UBAN) for detection of linear chains, and RAP80 ubiquitin-interacting motif (UIM) and TAB2 Npl4 zinc finger (NZF) domains to detect K63 chains. Linear and K63 sensors decorated the ubiquitin coat surrounding cytosolic Salmonella during bacterial autophagy, whereas K63 sensors selectively monitored Parkin-induced mitophagy and DNA damage responses in fixed and living cells. In addition, linear and K63 sensors could be used to monitor endogenous signaling pathways, as demonstrated by their ability to differentially interfere with TNF- and IL-1-induced NF-κB pathway. We propose that UBD-based biosensors could serve as prototypes to track and trace other chain types and ubiquitin-like signals in vivo.
INTRODUCTION
Eukaryotic cells employ a wide repertoire of ubiquitin (Ub) signals to regulate processes like DNA damage repair, signal transduction pathways, and innate immune responses (Bianchi and Meier, 2009; Haglund and Dikic, 2005; Ravid and Hochstrasser, 2008). Ub signals vary from mono-Ub to extended chains, linked through seven lysine residues present in Ub (K6, K11, K27, K29, K33, K48, and K63) or via the N-terminal methionine residue (linear ubiquitination) (Ikeda and Dikic, 2008; Walczak et al., 2012). Ubiquitin chains are selectively recognized and decoded into downstream signals by specific Ub-binding domains (UBDs), creating the basis of complex signaling networks that are spatiotemporally regulated in vivo (reviewed in Dikic et al., 2009; Grabbe et al., 2011).
K48-linked chains have been studied in the context of protein degradation (reviewed in Finley, 2009), whereas K63-linked chains have been implicated in a variety of nonproteolytic functions (Winget and Mayor, 2010). Other chain types have been less studied, but their specific roles are now starting to emerge. Proteomic analyses revealed that all seven lysine residues as well as the methionine can be used to construct chains with varying linkage abundances between cell types and model organisms (Dammer et al., 2011; Gerlach et al., 2011; Peng et al., 2003; Ziv et al., 2011). The newly identified linear chains are generated by the LUBAC complex, composed of HOIP-1, HOIL-1L, and SHARPIN (Ikeda et al., 2011; Iwai and Tokunaga, 2009; Nakamura et al., 2006; Tokunaga et al., 2009, 2011) and play an important role during tumor necrosis factor α (TNF-α)-mediated nuclear factor κB signaling (reviewed in Verhelst et al., 2011; Walczak et al., 2012). LUBAC has been identified as an integral part of the intracellular TNF-receptor complex 1 (TNFR-C1) upon ligand binding (Haas et al., 2009), and LUBAC-dependent deposition of linear chains onto IKKγ/NEMO as well as RIPK1 is required for efficient downstream signal propagation (Gerlach et al., 2011; Ikeda et al., 2011; Tokunaga et al., 2011). Linear chains adopt open and extended conformations that act as recognition signal for proteins containing Ub-binding in ABIN and NEMO (UBAN) domains, such as NEMO, ABIN1-3, and Optineurin (OPTN) (Wagner et al., 2008). The structure of the NEMO UBAN domain in complex with linear di-Ub reveals a parallel coiled-coil dimer that contacts both the proximal and distal Ub moieties (Rahighi et al., 2009). UBAN-Ub interactions are mediated through residues in the linker region between the linear-linked Ub molecules and explain the high affinity of the UBAN toward linear as compared to K-linked chains (Dynek et al., 2010; Lo et al., 2009; Rahighi et al., 2009). On the other hand, K63-linked poly-Ub chains are also found to adopt open conformations but are selectively recognized by another subset of UBDs, well-studied examples being the Npl4 zinc finger (NZF) of TAK1 binding protein 2 (TAB2) (Komander et al., 2009; Kulathu et al., 2009; Sato et al., 2009b) or the Ub interaction motifs (UIMs) of receptor-associated protein 80 (RAP80/UIMC1) (Sato et al., 2009a). Interestingly, the TAB2 NZF and the UIM of RAP80 do not contact the isopeptide linkage but make specific contacts with the surfaces of two ubiquitin moieties in the K63-linked chains.
To understand the biological significance of differently linked Ub chains, it is essential to study polymeric Ub signals in endogenous settings (reviewed in Ikeda et al., 2010). However, despite years of intense research and recent technological advances in mass spectrometry (Kim et al., 2011; Xu et al., 2010), the development of chain-specific antibodies (Matsumoto et al., 2010, 2011b; Newton et al., 2008), and genetic approaches (Xu et al., 2009), visualizing endogenous ubiquitination events in cells remains problematic. High expression levels, differential abundances of chain types, and the dynamic nature of Ub chains complicate the detailed study of poly-Ub chain behavior. In order to monitor the dynamics of Ub chain formation in vivo, we proposed to establish Ub sensors based on the use of selective UBDs that can recognize endogenous pools of Ub chains coupled to diverse physiological functions (Ikeda et al., 2010).
Here, we describe the technical development of such sensors and demonstrate that they can be used in vitro and in vivo to monitor localization and functions of specific Ub chains. By using GFP as a fluorescent tag, we have established UBD-based biosensors to study linear and K63-linked Ub chains. The ability of linear and K63 chain sensors to label their cognate chains was challenged in different biological processes. Linear and K63 chain sensors were able to decorate cytosolic Salmonella upon infection, depending on recognition and binding to their cognate chains within the Ub coat. Recruitment of linear and K63 sensors to cytosolic Salmonella coincided with costaining by linear and K63-specific poly-Ub antibodies. In addition, K63-specific UIM and NZF-derived sensors were able to monitor DNA double-strand breaks (DSBs) and Parkin-mediated mitochondrial autophagy, respectively. Finally, linear and K63 sensors can act as differential modulators of ubiquitin-dependent signal transduction pathways, such as TNF- and IL-1-induced NF-κB signaling.
RESULTS
Validation of Ubiquitin-Binding Specificity
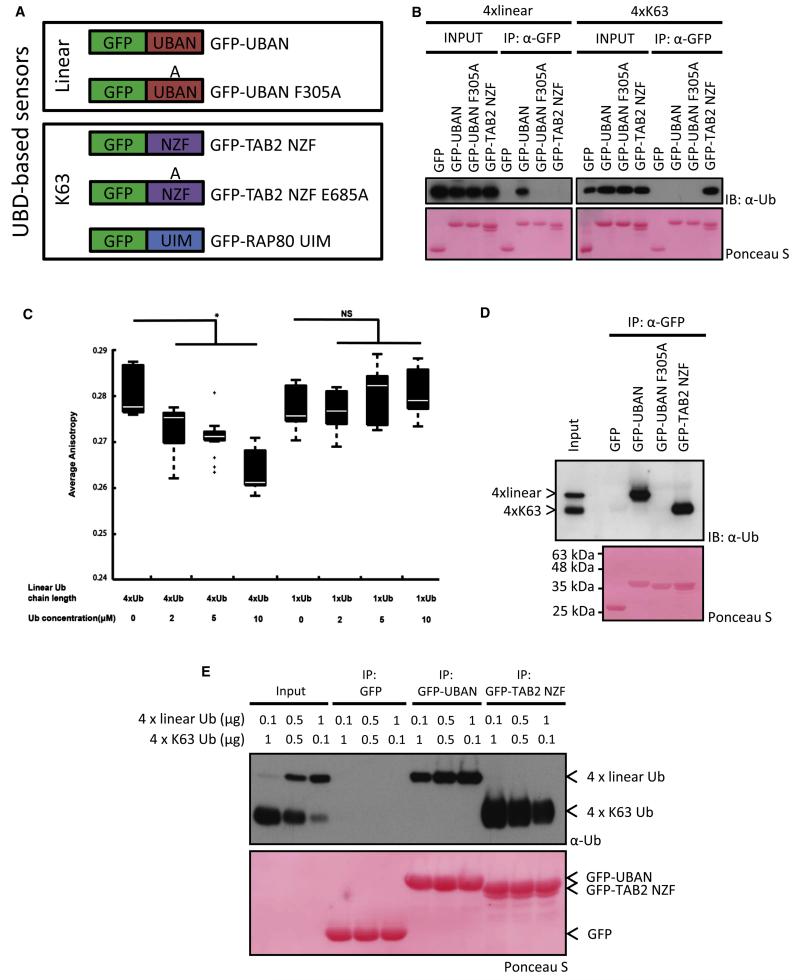
Different polymeric Ub signals adopt distinct structural arrangements, such as the closed K48-linked chains, or more open and elongated conformations, as is the case for linear and K63-linked chains (Fushman and Walker, 2010). From a structural perspective, linear and K63-linked chains appear very similar. Thus, a prerequisite for useful chain-specific sensors is to discriminate between structurally similar chains in order to interact solely with their cognate chain types. The NEMO coiled coil 2 (CC2) and leucine zipper (LZ) region (CoZi domain; amino acids 183–339) encompassing the UBAN domain form a parallel coiled-coiled homodimer that binds linear di-Ub with approximately 100-fold higher affinity than K63- or K48-linked di-Ub (Rahighi et al., 2009). Therefore, we decided to fuse enhanced green fluorescent protein (EGFP) to the CoZi domain of murine NEMO (hereafter referred to as GFP-UBAN) (Figure 1A) and used GFP-UBAN F305A as a Ub-binding-deficient form, as shown previously (Rahighi et al., 2009). Similarly, to monitor K63 chains, a fusion protein of GFP and the NZF domain of human TAB2 was constructed (Figure 1A).
Figure 1. Validation of Sensor Chain Interaction Selectivity.
(A) Schematic representation of GFP-UBD sensors (GFP-UBAN WT and F305A [residues 183–339], GFP-TAB2 NZF WT and E685A [amino acids 628–693], and RAP80 UIM [residues 1–124]).
(B) Pull-down analysis of immobilized GFP-UBDs with linear or K63-linked tetra-Ub. Input is 10%. Ponceau S-stained membrane shows GFP-tagged UBDs.
(C) Steady-state anisotropy of GFP-UBAN with indicated amounts of linear tetra- or mono-Ub as box plots of two independent measurements. A Kolmogorov-Smirnov statistical test was used to determine significance *p < 0.005, NS, nonsignificant.
(D) Ub competition pull-down assay as in (B), in which equal amounts of linear and K63-linked were combined as input. Input is 10%.
(E) Ub competition pull-down analysis of different amounts of linear and K63 Ub. Increasing amounts of linear 4× Ub were mixed with decreasing amounts of K63-linked 4× Ub. GFP immunoprecipitations were performed, followed by SDS-PAGE, immunoblotting, and probing with anti-Ub antibody.
The GFP-tagged UBDs were subsequently expressed and purified from Escherichia coli (see Figure S1A available online). Attaching GFP to the UBAN domains did not interfere with dimer formation, as revealed by gel filtration (data not shown). Selective binding of the GFP-UBDs to differentially linked Ub chains was confirmed in pull-down assays. Incubation of purified linear and K63 tetra-Ub with the sensors showed that GFP-UBAN was able to interact with linear Ub chains, whereas binding to K63 chains was not detected (Figure 1B). In contrast, GFP-TAB2 NZF did not interact with linear chains but specifically pulled down K63 chains (Figure 1B). GFP-UBAN F305A bound neither linear chains nor K63 tetra-Ub chains, confirming its use as a negative control (Figure 1B). The binding affinity of GFP-UBAN for linear tetra-Ub was confirmed by determining a dissociation constant of 4.7 μM, as compared with GFP-UBAN/mono-Ub and GFP-UBAN F305A/linear tetra-Ub, all of which bound below the detection level (Figure S1B). Previous studies and our measurements of the dissociation constants revealed that UBAN domains do not display binding affinities toward mono-Ub (Wagner et al., 2008). Using steady-state GFP anisotropy, we measured in-solution homo-FRET of GFP-UBAN in combination with increasing concentrations of either linear tetra- or mono-Ub. Upon incubation of GFP-UBAN with linear tetra-Ub, a dose-dependent decrease in anisotropy could be observed, whereas equal amounts of mono-Ub did not affect anisotropy (Figure 1C). Additionally, changes in anisotropy were not detected when GFP-TAB2 NZF was combined with increasing concentrations of linear chains or mono-Ub (Figure S1C), emphasizing chain interaction selectivity in solution.
Sensor selectivity was next assessed in in vitro competition assays, in which GFP-UBDs were incubated with equal amounts of linear and K63 tetra-Ub. Despite having identical molecular masses, linear and K63-linked chains display different electrophoretic mobility due to chain-specific structural constraints. Even when both chain types were present, GFP-UBAN exclusively bound to linear chains, efficiently discriminating against K63-linked Ub (Figure 1D). On the other hand, GFP-TAB2 NZF selectively bound K63 chains and did not interact with linear poly-Ub (Figure 1D). As expected, GFP-UBAN F305A did not show any interaction with either linear or K63-linked chains.
Different amounts of K63 and linear chains have been detected in cells at steady state and in cells stimulated with cytokines (Matsumoto et al., 2011b; Phu et al., 2011). To test if the UBD sensors could distinguish these physiologically relevant stoichiometric distributions, GFP-UBD pull-downs were performed with increasing amounts of linear chains combined with decreasing concentrations of K63-linked poly-Ub. Even when low amounts of linear Ub were combined with a 10-fold excess of K63 Ub, GFP-UBAN was able to interact exclusively with linear chains (Figure 1E). Additionally, GFP-TAB2 NZF only interacted with K63-linked chains, indicating that the sensors exhibit selectivity even under conditions where high levels of structurally similar chains are present.
UBD-Based Sensors Label Cytosolic Salmonella typhimurium
The performance of the sensors in vitro prompted us to test their ability to monitor poly-Ub chains in cells. Upon overexpression in HeLa cells, both wild-type and F305A GFP-UBAN sensors showed cytoplasmic localization (Figure S2A), whereas the K63-specific GFP-TAB2 NZF could be observed in the nucleus as well as in the cytoplasm (Figure S2B). Linear and K63-specific sensors were expressed at similar level (Figure S2C).
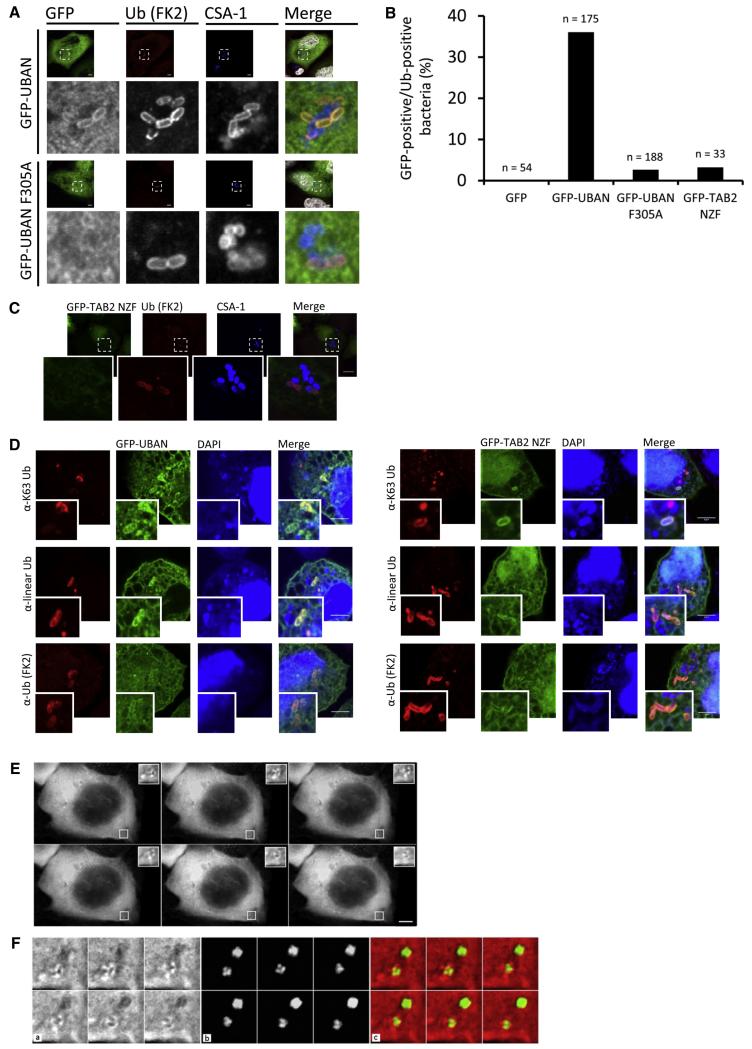
We first tested the sensors in epithelial cells infected with Salmonella typhimurium, which following internalization resides in _Salmonella_-containing vacuoles (SCVs). However, a small fraction evades into the cytosol and becomes covered with a Ub coat that initiates autophagic clearance (Perrin et al., 2004; Thurston et al., 2009; Wild et al., 2011). The identity of substrates as well as the types of Ub chain linkages on the surface of cytosolic Salmonella remains enigmatic. Three autophagy receptors, p62, NDP52, and OPTN, have been shown to link ubiquitinated bacteria with LC3-positive autophagosomal membranes (Perrin et al., 2004; Thurston et al., 2009; Wild et al., 2011) Their UBDs bind to mono-Ub through Ub-binding zinc fingers (NDP52) or K63 and K48 chains through a phosphory-lated UBA domain (p62/SQSTM2) (Matsumoto et al., 2011a). Recruitment of OPTN to bacteria is mediated via recognition of linear chains by the UBAN domain (Wild et al., 2011). This implies that different Ub signals could be part of the Ub coat and recruit distinct autophagy receptors. To evaluate the ability of the sensors to recognize cytosolic bacteria, cells expressing the UBD sensors were infected with Salmonella and processed for immunofluorescence. GFP-UBAN efficiently labeled a subpopulation of cytosolic Salmonella that also stained positive for conjugated Ub (Figure 2A) (using the FK2 anti-Ub antibody). In all cases, GFP-UBAN sensors labeled only Ub-positive Salmonella, while bacteria that stained negative for Ub located in SCVs did not recruit GFP-UBAN (Figure 2A), underscoring that sensor recruitment is not a consequence of nonselective binding. Consistently, the Ub-binding-deficient GFP-UBAN F305A was not enriched on Salmonella, in line with the UBAN-dependent OPTN recruitment described previously (Wild et al., 2011) (Figures 2A and 2C).
Figure 2. UBD-Based Sensors Label Cytosolic Salmonella.
(A) Confocal images of HeLa cells expressing wild-type GFP-UBAN (upper panels) and GFP-UBAN F305A (lower panels), infected with wild-type Salmonella strain SL1344 and processed for immunofluorescence at 2 hr postinfection. Cells were simultaneously stained with antibodies against ubiquitin (FK2) and the Salmonella marker common structural antigen-1 (CSA-1), followed by incubation with Cy3- and Cy5-labeled secondary antibodies, respectively. DNA was counterstained with DAPI (shown false colored in white in merged panels). Scale bars, 10 μm. Note the mutual exclusive FK2/CSA-1 staining on Salmonella.
(B) As (A), but with GFP-TAB2 NZF.
(C) Quantification of UBD sensor recruitment to intracellular wild-type Salmonella. n represents the number of bacteria counted from three independent experiments.
(D) Confocal images of HeLa cells expressing wild-type GFP-UBAN (left panels) and GFP-TAB2 NZF (right panels), infected with Salmonella strain SBM256 (sifA:kan), and processed for immunofluorescence at 1 hr postinfection. Cells were stained with antibodies against ubiquitin (FK2), K63-linked Ub (Apu3.A8), and linear Ub (1F11/3F5/Y102L), followed by incubation with Cy3-labeled secondary antibodies. DNA was counterstained with DAPI. Scale bars, 5 μm.
(E) Time-lapse imaging of HeLa cells expressing mCherry-UBAN were infected with Salmonella strain SFH4 (sifA:kan/puHpF-GFP; green). After infection, cells were grown in medium containing gentamycin, and images were acquired at 45 s intervals for a total period of 15 min. Scale bar, 10 μm. See also Movie S1.
(F) Enlargements of the white-boxed region from (E). Shown are mCherry (a), GFP (b), and merged (c) channels.
Tandem fusion of multiple identical UBDs can affect binding affinities to Ub chains (Hjerpe et al., 2009), and we therefore constructed a GFP fusion with three tandem UBAN domains (GFP-3xUBAN and GFP-3xUBAN F305A) (Figures S2D–S2F). GFP-3xUBAN was recruited to cytosolic Salmonella, while the triple F305A mutant was not detected at the bacteria. In addition, we could detect more GFP-3xUBAN-positive Salmonella compared to GFP-UBAN (Figure 2D and Figure S2F). GFP-TAB2 NZF localization on Ub-coated wild-type Salmonella could be detected at low levels (Figures 2B and 2C).
To further examine the recruitment of sensors to cytosolic bacteria, we performed infection experiments with Salmonella strains lacking the bacterial effector SifA. These bacteria undergo more frequent evasion from SCVs, leading to an enhanced number of ubiquitinated cytosolic Salmonella compared to wild-type strains (Wild et al., 2011). Upon infection with SifA-deficient Salmonella, GFP-TAB2 NZF was recruited to the Ub coat, while enrichment of a Ub-binding deficient NZF E685A could not be detected (Figure 2D and Figure S2G). In agreement with wild-type Salmonella experiments, GFP-UBAN was also detected surrounding SifA-deficient Salmonella (Figure 2D).
Sensor recruitment to the bacterial Ub coat implies the presence of K63 and linear signals, and immunofluorescence staining revealed that antibodies recognizing K63-linked and linear-linked poly-Ub clearly colocalized with bacteria (Figure S2H). Interestingly, GFP-UBAN and GFP-TAB2 NZF were recruited to bacteria that stained positive for linear and K63-linked Ub chains, respectively (Figure 2D), suggesting that UBD sensors detect cytosolic Salmonella through selective recognition of Ub chains. The colocalization of the sensors with K63 and linear chain-specific antibodies on Salmonella suggests that the coat contains multiple types of Ub signals. This also demonstrates that the sensors can be used as genetically encoded reagents to visualize linear and K63 chains in cells. To strengthen this idea, HeLa cells expressing an mCherry-UBAN sensor were infected with constitutively GFP-expressing Salmonella and subjected to live-cell imaging. mCherry-tagged UBAN could be imaged in time lapse on Salmonella during live-cell infection, indicating that UBAN-based sensors are able to track linear chains in living cells (Figures 2E and 2F and Movie S1).
Selective Monitoring of DNA Double-Strand Breaks by RAP80-UIM K63 Sensors
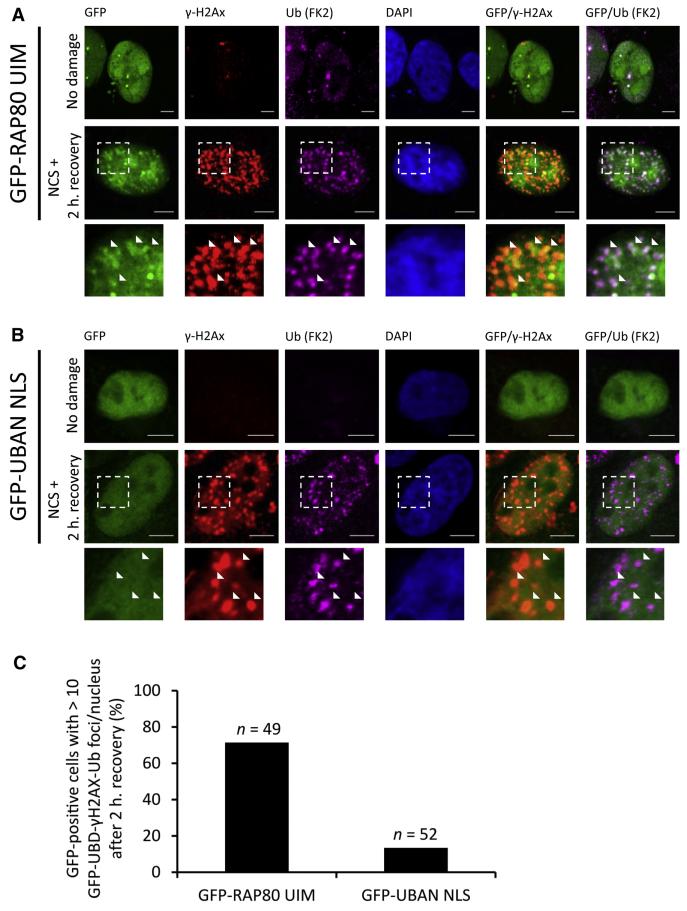
Ub signals have been implicated in the coordinated recruitment and assembly of DNA repair protein complexes (Bergink and Jentsch, 2009; Ulrich and Walden, 2010). DNA double-strand breaks (DSBs) led to the recruitment of the E3 ligases RNF8 and RNF168 that catalyze K63 chain assembly (Doil et al., 2009; Kim et al., 2007; Sobhian et al., 2007; Stewart et al., 2009; Wang et al., 2007). These chains are recognized by RAP80 through its tandem UIMs (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007). Linear and K63 sensors were tested for their ability to monitor DSB damage repair by treating HeLa cells expressing UBD sensors with the chromophore neocarzinostatin (NCS) for 30 min to allow initiation of DNA damage and its subsequent repair. The K63-specific GFP-RAP80 UIM was readily recruited to γ-H2Ax-containing foci, colocalizing with Ub (Figures 3A–3C). Nuclear localization of GFP-UBAN sensors was achieved by inserting the SV40 nuclear localization signal (NLS) (GFP-UBAN NLS). Both GFP-UBD sensors were expressed at comparable levels (Figure S2C), and insertion of an NLS did not interfere with UBAN poly-Ub binding (Figure S3). In contrast to the UIM sensor, nuclear-localized GFP-UBAN was not significantly enriched on damaged-induced γ-H2Ax- or Ub-positive foci (Figures 3A and 3B). This indicates that K63-specific sensors efficiently localize to sites where K63 chains are being generated and that linear-specific sensors, upon artificial introduction into the nucleus, retain their discriminative selectivity against alternative K63-linked chains.
Figure 3. Selective Monitoring of DNA Double-Strand Breaks by UBD Sensors.
(A) Confocal images of HeLa cells expressing GFP-RAP80 UIM that were treated with 200 ng/ml neocarzinostatin (NCS) for 30 min, followed by washout of NCS and recovery in normal medium for 2 hr. Cells were simultaneously stained with antibodies against ubiquitin (FK2) and γ-H2Ax, followed by incubation with Cy5- and Cy3-conjugated secondary antibodies. DNA was counterstained with 4′,6-diamidino-2-phenyl-indole (DAPI). Scale bars, 5 μm. Inset panels are enlarged from dashed boxes. Arrowheads emphasize foci containing Ub and γ-H2Ax staining.
(B) As in (A), but for GFP-UBAN.
(C) Quantification of colocalization. Fraction of nuclei displaying >10 GFP-positive, Ub- and γ-H2Ax-positive foci were quantified. n represents number of nuclei counted. Data are shown as mean.
Selective Sensing of Ubiquitin Signals in Mitophagy
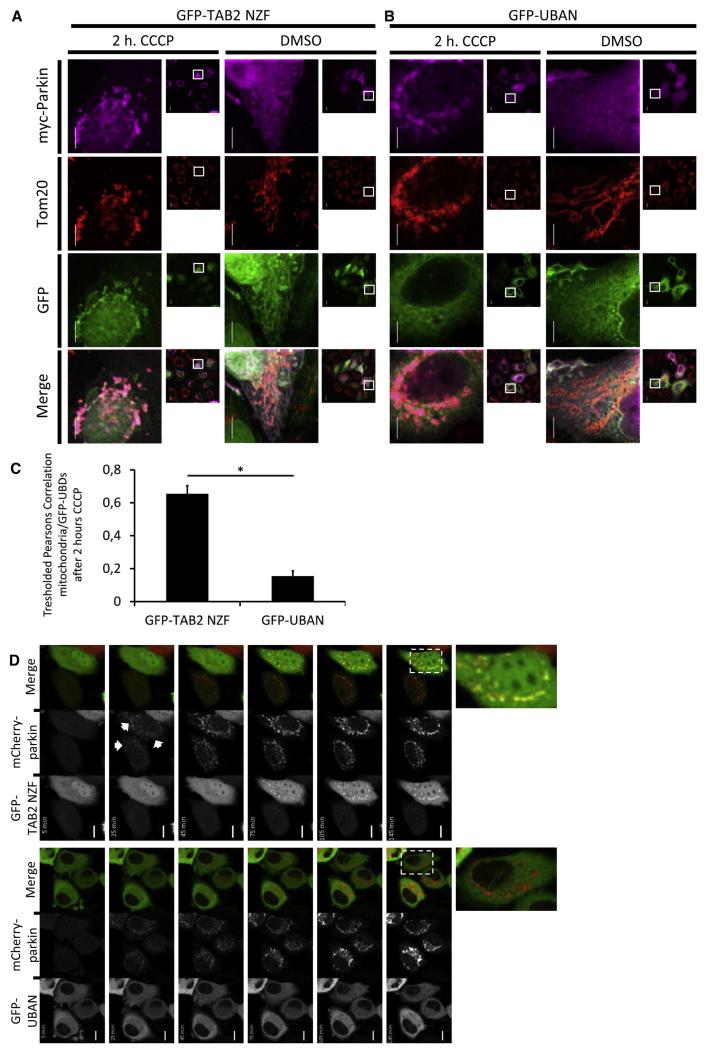
We next investigated sensor function in autophagy-mediated destruction of damaged mitochondria, in which the E3 ligase Parkin mediates the K63- and K27-linked chain-dependent mitochondrial clearance (Geisler et al., 2010; Vives-Bauza et al., 2010). The GFP-TAB2 NZF and GFP-UBAN sensors were coexpressed with myc-tagged Parkin, and mitochondrial depolarization was induced with carbonyl cyanide m-chlorophenylhydrazone (CCCP). CCCP treatment induced the characteristic fragmented and perinuclear cluster-like structures of mitochondria in a Parkin-dependent fashion (Figure 4A). GFP-TAB2 NZF was enriched on mitochondria in CCCP-treated cells expressing Parkin, colocalizing with the mitochondrial remnants, Parkin, and the mitochondrial marker Tom20 (Figures 4A and 4C). In contrast, GFP-UBAN was not detected on damaged mitochondria, suggesting that linear chains might not play prominent roles in mitophagic clearance (Figures 4B and 4C). These findings support the current model in which K63 Ub chains are prominently involved in removal of damaged mitochondria. It furthermore demonstrates that UBAN-based sensors display selectivity within the cytoplasm and discriminate against high local concentrations of lysine-linked Ub chains.
Figure 4. Selective Sensing of Ubiquitin Signals in Parkin-Mediated Mitophagy.
(A and B) Representative confocal images of HeLa cells coexpressing GFP-TAB2 NZF (A) or GFP-UBAN (B) with myc-tagged Parkin. Cells were treated with DMSO as vehicle control or with 25 μm CCCP for 2 hr and prepared for immunofluorescence. Stainings were performed simultaneously with antibodies against Tom20 and myc, followed by incubation with Cy3- and Cy5-conjugated secondary antibodies, respectively. Scale bars, 10 μm.
(C) Quantification of GFP-UBD colocalization with myc-Parkin-induced mitochondrial remnants after 2 hr of CCCP treatment. Tresholded Pearsons correlations were calculated between mitochondria and GFP-UBDs. Data are shown as mean ± standard error of the mean (SEM) of two independent experiments. Number of quantified cells is as follows: GFP-TAB2 NZF, n = 189 cells; GFP-UBAN, n = 89 cells. *p < 0.0005 by Student’s t test.
(D) Representative time-lapse imaging of UBD sensor recruitment to Parkin-associated CCCP-induced depolarizing mitochondria. HeLa cells coexpressing mCherry-parkin and GFP-TAB2 NZF (upper panels) or GFP-UBAN (bottom panels) were treated with 25 μm CCCP and time-lapse imaged for a total period of 2 hr. Images were taken every 10 min. Scale bar, 10 μm.
The GFP- TAB2 NZF and GFP-UBAN sensors were also used to monitor mitophagy in living cells. HeLa cells were cotransfected with GFP sensors and mCherry-tagged Parkin, followed by the addition of CCCP. Cells were subsequently imaged over a period of 2 hr. As demonstrated in Figure 4D and Movie S2, mCherry-tagged Parkin translocated to mitochondria 25 min after the addition of CCCP. GFP-TAB2 NZF displayed a delayed recruitment as compared to Parkin and could be detected after approximately 45 min following CCCP treatment (Figure 4D and Movie S2). As expected, GFP-UBAN was not recruited to depolarized mitochondria (Figure 4D and Movie S3). Recruitment of GFP-TAB2 NZF to mitochondria was dependent on Ub binding, since the TAB2 NZF E685A Ub-binding deficient sensor did not translocate to mitochondria (Figure S4A and Movie S4). Furthermore, recruitment of K63 sensors was dependent on the addition of CCCP (Figure S4B and Movie S5). These findings indicate a sequential order of recruitment to depolarized mitochondria, where Parkin relocates first, followed by Ub-dependent translocation of Ub sensors.
UBD Sensors Affect NF-κB Signaling
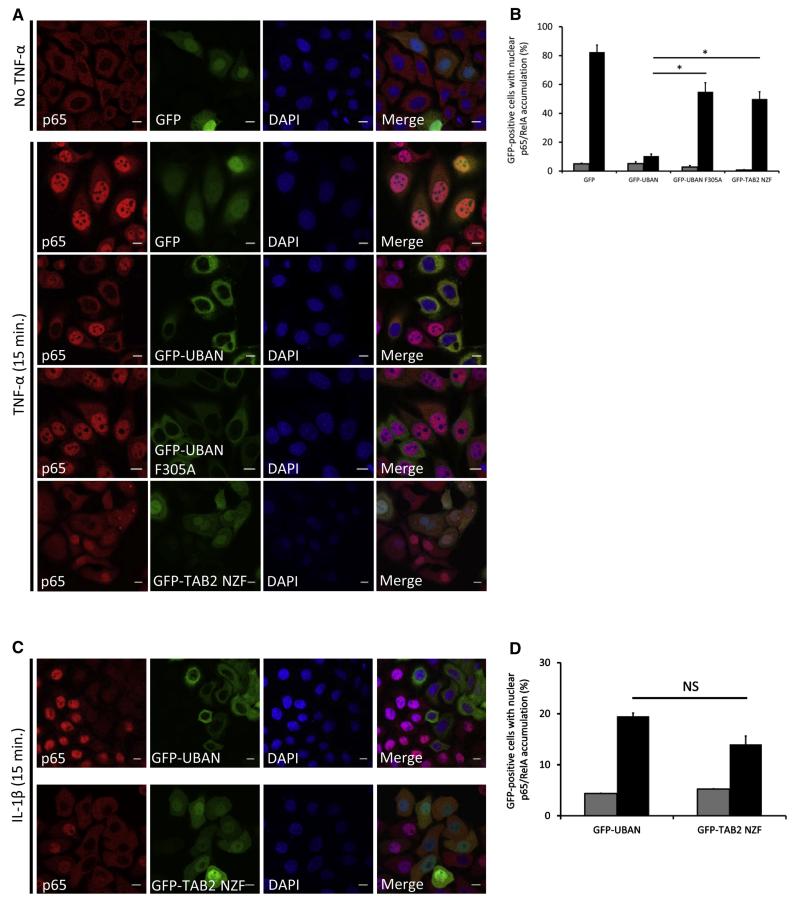
The specific localization of sensors at cytosolic Salmonella, DNA damage sites, and damaged mitochondria underscores their usefulness as genetic visualization reagents to better understand the spatial control of Ub signaling networks (Grabbe et al., 2011). These reagents could also have the potential to monitor the activity of Ub-regulated signaling pathways in vivo. Toward this end, we focused on TNFα-induced activation of NF-κB signaling, since distinct activation steps require K63, linear, and K48 chains, which are dynamically regulated by E3 ligases and deubiquitinating enzymes (DUBs) (Bianchi and Meier, 2009; Grabbe et al., 2011). Cells expressing GFP-UBDs were TNFα stimulated and stained for endogenous p65 (RelA), a NF-κB transcription factor that translocates to the nucleus where it activates target gene expression. In untreated cells, nuclear p65 accumulation was not detected (Figure 5A). Cells expressing GFP-UBAN displayed a prominent inhibition of nuclear accumulation of p65 after 15 min of TNFα-stimulation, whereas in cells expressing GFP alone or GFP-UBAN F305A, less inhibition of nuclear p65 localization was observed (Figures 5A and 5B). The sensors were expressed at equal levels, and therefore the observed effects cannot be explained solely by differences in abundances of the UBD-based sensors (Figure S2C). Direct interaction between GFP-UBDs and endogenous NEMO could not be detected (data not shown); therefore, we believe that the observed effects on NF-κB signaling are caused by sensor binding to endogenous linear Ub chains. These findings suggest that UBAN-based sensors, apart from acting as genetic visualization reagents, also exert functional inhibitory effects on endogenous signaling complexes. On the other hand, K63-binding sensors did not significantly block TNF-induced nuclear p65 accumulation compared to the UBAN sensor (Figures 5A and 5B), confirming previous observations that K63 Ub chains are not essential for efficient TNF-mediated NF-κB signaling (Xu et al., 2009; Kensche et al., 2012).
Figure 5. UBD Sensors Affect NF-κB Signaling Differentially.
(A) Confocal images of HeLa cells expressing indicated GFP or GFP-UBD constructs that were stimulated with 20 ng/ml TNF-α for 15 min. Staining was performed with p65/RelA antibody, followed by incubation with Cy3-labeled secondary antibody. DNA was counterstained with 4′, 6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm.
(B) Quantification of TNF-α-induced p65/RelA nuclear accumulation. Shown is fraction of GFP-positive cells with nuclear p65/RelA accumulation. Gray bars, unstimulated; black bars, TNF stimulated. Data are shown as mean ± SEM, n > 150 for each condition, *p < 0.0005 by Student’s t test.
(C) As in (A), but cells were stimulated with 10 ng/ml IL-1β for 15 min.
(D) As in (B), but cells expressing GFP-UBAN and GFP-TAB2 NZF were stimulated with IL-1β for 15 min. Data are shown as mean ± SEM, n > 150 cells for each condition. NS, nonsignificant.
Stimulation of the IL-1 receptor by IL-1β also involves differentially linked polymeric Ub chains (Iwai and Tokunaga, 2009; Tokunaga et al., 2011), but IL-1 receptor signaling relies more on K63-linked chains than TNF signaling does (Xu et al., 2009). In contrast to what is observed for TNF-mediated NF-κB signaling, upon IL-1β stimulation we could detect equal inhibition of nuclear p65 accumulation by K63-specific NZF sensors and GFP-UBAN (Figures 5C and 5D).
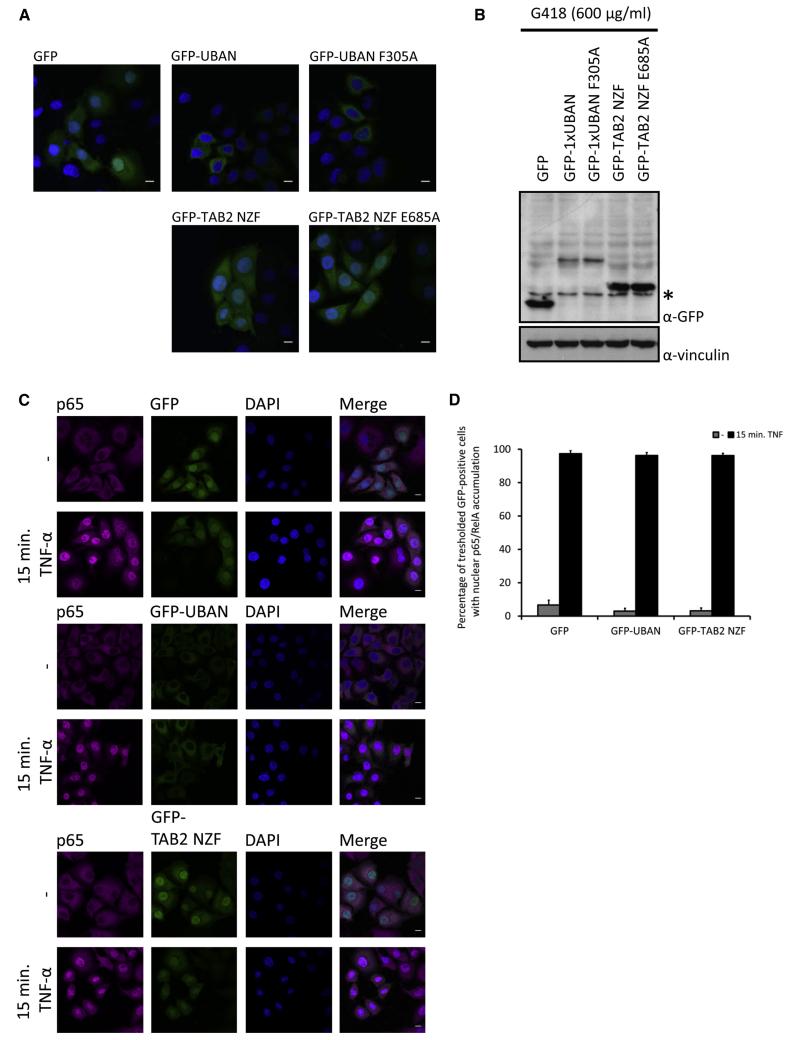
Transient overexpression of the sensors inhibits Ub-dependent processes. To expand the applicability of the sensors to long-term experiments or even whole-organism incorporation, we also studied the consequences of prolonged sensor exposure. HeLa cells stably expressing the UBD-based sensors at comparable levels were generated (Figures 6A and 6B). Notably, no obvious alterations in cell morphology were observed (Figure 6A), indicating that cells tolerate long-term expression of Ub-binding sensors and that toxicity associated with prolonged exposure is unlikely. Surprisingly, stable expression of GFP-UBAN or GFP-TAB2 NZF did not affect TNF-induced nuclear p65 translocation (Figures 6C and 6D), implying that cells stably expressing UBD sensors underwent a certain degree of adaptation to resist the inhibitory function of the sensors. The same cells were subsequently infected with SifA-deficient Salmonella, and stably expressing GFP-UBAN could still recognize cytosolic Salmonella (Figure S5).
Figure 6. Stable Expression of GFP-UBD Sensors Induces Resistance within the NF-κB Signaling Pathway.
(A) Representative confocal images of HeLa cells stably expressing indicated GFP-UBDs after a 28 day selection period with G418 (600 mg/ml). DNA was counterstained with DAPI. Scale bar, 10 μm.
(B) Expression levels of HeLa cells stably expressing the indicated GFP-UBDs. Total cell lysates were immunoblotted and probed with antibodies against GFP and vinculin (α-vinc) as loading control. Asterisk indicates a specific background band.
(C) Confocal images of HeLa cells stably expressing indicated GFP or GFP-UBD constructs that were stimulated with 20 ng/ml TNF-α for 15 min. Staining was performed with p65/RelA antibody, followed by incubation with Cy5-labeled secondary antibody. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm.
(D) Quantification of TNF-α-induced p65/RelA nuclear accumulation in HeLa cells stably expressing GFP, GFP-UBAN, and GFP-TAB2 NZF. Threshold for quantifying GFP-positive cells was set at four times the intensity of background cells. Shown is fraction of threshold GFP-positive cells with nuclear p65/RelA accumulation. Gray bars, unstimulated; black bars, TNF stimulated. Data are shown as mean ± SEM, n > 150 for each condition from two independent selection experiments.
These findings illustrate a differential role of short-term or chronic expression of UBAN sensors in monitoring localization or functional inhibition. Acute expression of sensors blocks endogenous signaling complexes, whereas stable expression (at moderate level) induces compensatory mechanisms that enable cells to adjust to the permanent presence of sensors. These observations also indicate that the sensors can be used to track Ub-dependent processes without altering Ub conjugation and deconjugation machineries.
DISCUSSION
Here we reveal two important features of UBD-based sensors (see the schematic model in Figure S6). The first is their usefulness to visualize local Ub chain production and accumulation upon Salmonella infection, DNA damage, and mitophagy. For certain chain types, antibodies have been developed and applied to monitor ubiquitin chains in vitro and in fixed cells (Matsumoto et al., 2010, 2011b; Newton et al., 2008). Compared to chain-type-specific antibodies, sensor-based visualization has several advantages. Sensors can be expressed genetically in isolated cells or in complete organisms, enabling flexibility to study ubiquitin-mediated processes in many settings. The binding properties of the sensors can be modulated by mutagenesis to decrease or increase affinity, such as described for the UBA domain of p62 (Matsumoto et al., 2011a). Furthermore, sensors provide better spatial resolution than currently available Ub chain antibodies and can be used to capture Ub chains and their substrates in combination with mass-spectrometric analysis (Ikeda et al., 2011), in a similar manner as that described for tandem-repeated ubiquitin-binding entities (TUBEs) (Hjerpe et al., 2009). Finally, sensors have also a potential to be applied in live-cell imaging combined with superresolution microscopy as well as with FRET and FRAP, to study Ub chain behavior in living cells.
Second, the fluorescence-labeled sensors may exhibit functional inhibitory activities on pathways that rely on Ub chains, like TNF- and IL-1β-mediated NF-κB signaling. Hitherto, deciphering the involvement of specific ubiquitin chains required genetic interference with the Ub conjugation machinery, i.e., deletion or mutation of E3 ligases (Emanuele et al., 2011) or the introduction of ubiquitin mutants (Xu et al., 2009). We also demonstrated that UBD-based sensors could functionally interfere with ubiquitin-mediated processes without the need to modify the ubiquitin-conjugation machinery genetically. Differences in functional inhibition of K63 and linear chain sensors might be explained by differential abundances of the target Ub chains in time and space (Grabbe et al., 2011). Activation of and signal propagation from the activated TNFR1 is thought to take place in a highly controlled manner (Haas et al., 2009; Grabbe et al., 2011). Within this dynamic window, many chain types have been found in the activated TNFR1 complex, such as linear, K63, and K11 chains, and many substrates become modified with one or multiple types of these chains (Bianchi and Meier, 2009; Gerlach et al., 2011; Ikeda et al., 2011; Dynek et al., 2010). The use of sensors that can specifically interfere with K11, K63, and linear pathways may provide new insights in quantitative and qualitative regulation in the NF-κB pathway.
Very recently, avidity-based triple fusions of the K63-specific UIMs of Vps27 and RAP80 have been shown to bind K63 chains selectively in vitro and in vivo (Sims et al., 2012). These reagents emphasize that tandem UBDs often exhibit increased Ub binding affinities and specificities compared to single UBDs (Hjerpe et al., 2009). Intriguingly, both the avidity-based sensors and single-UBD sensors recognized K63 chains in DNA damage repair and mitophagy. One aspect of sensors with ultra-high affinities is a longer residence time of sensor bound to a certain chain in the cell. This could potentially affect DUB access to the chains and hydrolysis. In contrast, single UBD-based sensor might display higher on/off rates and thereby allow a more dynamic access of additional factors to the chains. Additional experiments, including SILAC-based mass spectrometry in combination with AQUA approaches, are required to quantify alterations in mono-Ub and Ub chain pools in the presence of different sensors.
Cells that stably express GFP-UBD sensors were insensitive to the inhibitory effects of the sensors. Interestingly, in these cells we were able to detect sensor recruitment to the Ub coat of cytosolic Salmonella, indicating that the sensors lose their inhibitory effects upon long-term exposure but retain the ability to visualize Ub-enriched structures. This opens new possibilities to incorporate UBD-based sensors in complete organisms to track and monitor Ub signals in highly complex environments.
The described linear- and K63-specific sensors serve as prototypical systems to study the significance and physiology of Ub signals in many different biological settings. We anticipate that additional Ub sensors, targeting mono-Ub, K11, or K48 chains or other ubiquitin-like signals (e.g., SUMO or the autophagy modifier LC3), can be created and efficiently used in living cells.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Cells
Wild-type and F305A murine CoZi domain (referred to as UBAN), human TAB2 NZF, and human RAP80 UIM were subcloned into pGEX-4T-1 (GE Healthcare) and pEGFP-C1 (Clontech). The integrity of all plasmids was verified by DNA sequencing. HeLa and HEK293T (ATCC) cells were maintained under standard conditions and transfected using GeneJuice (Merck). Experimental details regarding plasmids, antibodies, cells, and additional reagents are specified in the Supplemental Experimental Procedures.
Recombinant Protein Expression and Purification
Expression of GST proteins was performed in Escherichia coli (BL21) and verified by resolving on SDS-PAGE and analyzed by Coomassie staining and immunoblotting (see the Supplemental Experimental Procedures).
Ubiquitin Pull-Downs
Linear and K63-linked ubiquitin chains of defined lengths and the K63-linked polyubiquitin mixture were produced as described previously (Reyes-Turcu et al., 2008). A mixture of linear polyubiquitin chains was generated in an in vitro ubiquitination reaction as reported earlier (Tokunaga et al., 2009). Additional technical background is described in the Supplemental Experimental Procedures.
Biophysical Analysis
Steady-state anisotropy was measured as described by (Squire et al., 2004). See the Supplemental Experimental Procedures for detailed experimental information for anisotropy and affinity measurements.
Immunofluorescence
Detailed technical information regarding Salmonella infections, DNA double-strand break repair, mitochondrial depolarization, cytokine stimulations, and immunofluorescence can be found in the Supplemental Experimental Procedures.
Live-Cell Imaging
Live-cell imaging was performed in LabTek dishes (NUNC), and time-lapse images were acquired on a Nikon Eclipse TE2000-E confocal microscope using a Nikon Plan Apochromat 60.03/1.40/0.21 spring-loaded oil-immersion objective or an Olympus wide-field fluorescence microscope (CellR IX81 Life Science Imaging Station) using an Olympus Plan Apochromat 60.0×/1.40/0.21 oil-immersion objective (Supplemental Experimental Procedures).
ACKNOWLEDGMENTS
We are grateful to Stefan Müller, Jaime Lopez-Mosqueda, and David McEwan for critical comments on the manuscript and Dirk Bumann (University of Basel, Switzerland) for providing Salmonella strains. This work was supported by the Cluster of Excellence “Macromolecular Complexes” of the Goethe University, Frankfurt am Main (EXC115), LOEWE Oncology Signaling Network and LOEWE Center for Cell and Gene therapy Frankfurt, and a European Research Council Advanced Grant (I.D.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures, five movies, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2012.06.017.
REFERENCES
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol. Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains—from structures to functions. Nat. Rev. Mol. Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D, Walker O. Exploring the linkage dependence of polyubiquitin conformations using molecular modeling. J. Mol. Biol. 2010;395:803–814. doi: 10.1016/j.jmb.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensche T, Tokunaga F, Ikeda F, Goto E, Iwai K, Dikic I. Analysis of NF-κB essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J. Biol. Chem. 2012 doi: 10.1074/jbc.M112.347195. Published online May 17, 2012. http://dx.doi.org/10.1074/jbc.M112.347195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H. Structural basis for recognition of diubiquitins by NEMO. Mol. Cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 2011a;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Matsumoto ML, Dong KC, Yu C, Phu L, Gao X, Hannoush RN, Hymowitz SG, Kirkpatrick DS, Dixit VM, Kelley RF. Engineering and structural characterization of a linear-polyubiquitin-specific antibody. J. Mol. Biol. 2011b;418:134–144. doi: 10.1016/j.jmb.2011.12.053. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Tokunaga F, Sakata S, Iwai K. Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 2006;351:340–347. doi: 10.1016/j.bbrc.2006.09.163. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr. Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Phu L, Izrael-Tomasevic A, Matsumoto ML, Bustos D, Dynek JN, Fedorova AV, Bakalarski CE, Arnott D, Deshayes K, Dixit VM, et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003756. http://dx.doi.org/10.1074/mcp.M110.003756, M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009a;28:2461–2468. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009b;28:3903–3909. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods. 2012;9:303–309. doi: 10.1038/nmeth.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire A, Verveer PJ, Rocks O, Bastiaens PI. Red-edge anisotropy microscopy enables dynamic imaging of homo-FRET between green fluorescent proteins in cells. J. Struct. Biol. 2004;147:62–69. doi: 10.1016/j.jsb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- Verhelst K, Verstrepen L, Carpentier I, Beyaert R. Linear ubiquitination in NF-kappaB signaling and inflammation: What we do understand and what we do not. Biochem. Pharmacol. 2011;82:1057–1065. doi: 10.1016/j.bcp.2011.07.066. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi: 10.1186/1741-7007-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol. Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv I, Matiuhin Y, Kirkpatrick DS, Erpapazoglou Z, Leon S, Pantazopoulou M, Kim W, Gygi SP, Haguenauer-Tsapis R, Reis N, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009753. http://dx.doi.org/10.1074/mcp.M111.009753, M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]