Expression of the secondary granule proteins major basic protein 1 (MBP-1) and eosinophil peroxidase (EPX) is required for eosinophilopoiesis in mice (original) (raw)
Key Points
- The loss of the two most abundant eosinophil granule proteins disrupts the production of blood eosinophils from marrow progenitors.
- Knockout animals deficient for both MBP-1 and EPX represent a novel strain of mice with a specific and congenital loss of eosinophils.
Abstract
Eosinophil activities are often linked with allergic diseases such as asthma and the pathologies accompanying helminth infection. These activities have been hypothesized to be mediated, in part, by the release of cationic proteins stored in the secondary granules of these granulocytes. The majority of the proteins stored in these secondary granules (by mass) are major basic protein 1 (MBP-1) and eosinophil peroxidase (EPX). Unpredictably, a knockout approach targeting the genes encoding these proteins demonstrated that, unlike in mice containing a single deficiency of only MBP-1 or EPX, the absence of both granule proteins resulted in the near complete loss of peripheral blood eosinophils with no apparent impact on any other hematopoietic lineage. Moreover, the absence of MBP-1 and EPX promoted a concomitant loss of eosinophil lineage-committed progenitors in the marrow, identifying a specific blockade in eosinophilopoiesis as the causative event. Significantly, this blockade of eosinophilopoiesis is also observed in ex vivo cultures of marrow progenitors and is not rescued in vivo by adoptive bone marrow engraftment, suggesting a cell-autonomous defect in marrow progenitors. These observations implicate a role for granule protein gene expression as a regulator of eosinophilopoiesis and provide another strain of mice congenitally deficient of eosinophils.
Introduction
The identification of eosinophils and subsequent discussions of eosinophil-associated activities are invariably linked with the prominent groups of proteins stored in the numerous secondary granules contained within the cytoplasm of these leukocytes.1,2 Three abundant groups of proteins exist among the numerous proteins stored in secondary granules (reviewed by Lee et al3), including eosinophil major basic proteins (human and mouse: MBP-1 and MBP-24-7), eosinophil-associated ribonucleases (human: eosinophil-derived neurotoxin8 and eosinophil cationic protein,9 mouse: eosinophil-associated ribonucleases [Ear-1, -2, -6/7, -5/1110,11]), and eosinophil peroxidase (human and mouse: EPX12,13).
The majority of the proteins stored in the secondary granules of eosinophils (by mass) is easily accounted for by MBP-1 and EPX, comprising ∼50% and ∼25%, respectively.14,15 However, despite their abundance and their relative effects on the morphology of the secondary granules,16,17 studies defining specific roles for these proteins in hematopoiesis and/or as mediators of eosinophil effector function in vivo remain inconclusive. Specifically, mice deficient for either MBP-116 or EPX17 showed no hematopoietic perturbations at baseline, and effects on the pathologies linked to eosinophil-associated diseases remain controversial.18-21
The formation of eosinophils occurs during a 3- to 4-day process of proliferation and differentiation within the hematopoietic compartment from initially pluripotent stem cells that yield eosinophil lineage-committed progenitor (EoP) cells.22 An early period of granulopoiesis in EoPs is a hallmark event characterized by robust expression of the genes encoding a multitude of granule proteins (reviewed by Wintrobe et al23). The granules containing MBP-1 and EPX themselves appear to form as small unit vesicles at the Golgi complex, which aggregate to form larger structures the sizes of which are multiples of this unit granule.24,25 Thus, expression of MBP-1 and EPX, as well as the formation of eosinophil secondary granules, is hypothesized to be a consequence of events in leukocytes whose lineage commitment toward the eosinophil cell subtype is already established.
Our demonstration that the singular loss of either MBP-1 or EPX in gene knockout mice (MBP-1−/− or EPX−/−, respectively) had no impact on the pulmonary pathologies linked with allergen provocation16,17 led us to hypothesize that the contributions of these granule proteins to disease pathology may occur via their combined activities. Thus, our intent was to cross the MBP-1−/− and EPX−/− single knockout mice to address the potential role of overlapping activities of these abundant granule proteins. Unexpectedly, MBP-1−/−/EPX−/− double knockout mice were viable and apparently healthy, yet they displayed significantly fewer circulating peripheral blood eosinophils relative to wild-type controls. Moreover, unlike wild-type animals with resident populations of eosinophils in various tissue compartments, MBP-1−/−/EPX−/− mice also displayed significantly fewer eosinophils in each of these compartments (eg, intestines, thymus, uterus26). Further studies examining the marrow hematopoietic compartment of MBP-1−/−/EPX−/− mice revealed that this loss of peripheral eosinophils resulted from a targeted disruption of eosinophilopoiesis. In summary, our data suggest a previously underappreciated link between granule formation and the maturation/terminal differentiation of eosinophils from marrow progenitors. Moreover, these data provide evidence of yet another eosinophil-less strain of mice that are otherwise healthy despite the nearly complete loss of eosinophil lineage-committed cells.
Methods
Mice
Studies were performed with strains of mice that included C57BL/6J wild-type controls (The Jackson Laboratory; Bar Harbor, ME), MBP-1−/− mice16,17 backcrossed onto C57BL/6J (>12 generations), EPX−/− mice16,17 backcrossed onto C57BL/6J (>12 generations), eosinophil-less PHIL mice27 backcrossed onto C57BL/6J (>20 generations), and NJ.1638 interleukin-5 (IL-5) transgenic mice28 backcrossed onto C57BL/6J (>20 generations). MBP-1−/−/EPX−/− double knockout mice were generated either by intercross of double heterozygotes (MBP-1−/+/EPX−/+ x MBP-1−/+/EPX−/+) or directly through crosses of double knockout mice (MBP-1−/−/EPX−/− x MBP-1−/−/EPX−/−). All mice were maintained in ventilated microisolator cages housed in a specific pathogen-free animal facility. Protocols and studies involving animals were performed in accordance with National Institutes of Health and Mayo Clinic Institutional Animal Care and Use Committee.
Hematologic assays: cytospins/smear preparations and cell counts/differentials
Films (blood), brush smears (marrow and spleen), or cytospins (marrow progenitor cell cultures) were stained with Diff-Quik Stain Set (Siemens Healthcare Diagnostics, Newark, DE). Summaries and full descriptions of the hematologic methods noted above are outlined in detail in McGarry et al.29
Flow cytometry analysis
Single-cell suspensions were stained for 25 minutes on ice with cell type–specific antibodies after blockade of Fc receptors by using 1 μg/μL of Fc blocker (CD16/32; BD Biosciences). The cell surface definitions of various leukocyte populations described here include basophils (as described previously by Obata et al30): B220-PE− (RA3-6B2; BD Biosciences), CD4-PE− (GK1.5; BD Biosciences), CD8-PE− (53-6.7; eBioscience, San Diego, CA), GR1-PE-Cy7− (RB6-8C5; eBioscience), c-kit-Brilliant Violet 421− (2B8; BD Biosciences), CD49b-APC**+** (DX5; eBioscience), and FcεRIα-FITC**+** (MAR-1; eBioscience); differentiated eosinophils: GR1-PE-Cy7**+** (RB6-8C5), IL-5Rα-FITC**+** (T21; BD Biosciences), CCR3-Alexa 647**+** (83101; BD Biosciences), and Siglec-F-PE**+** (E50-2440; BD Biosciences). EoP, granulocyte/monocyte progenitor, and common myeloid progenitor cells were identified as described previously by Iwasaki et al31 and de Bruin et al.32 In summary, the lineage cocktail used to exclude other cells and to identify these progenitors included antibodies (eBioscience) specific for CD3 (145-2C11), CD4 (GK1.5), CD8α (53-6.7), B220 (RA3-6B2), CD19 (1D3), Gr1 (RB6-8C5), and Ter119 (Ly-7d). Additional antibodies used included Sca1-PE-Cy7 (D7; eBioscience), CD34-FITC (RAM34; eBioscience), CD16/32-eFluor 450 (93; eBioscience), c-kit-APC (2B8; eBioscience), and IL-5Rα-PE (T21; BD Biosciences). For assessments of chimerism, CD45.1-PE-Cy7 (A20; eBioscience) and CD45.2-PE (104; eBioscience) were used. Flow cytometry was performed on a cytofluorimeter (CyAn; Dako, Carpinteria, CA). Data acquisition and analysis were performed by using Summit version 4.3 software (Dako).
Assessments of apoptosis
EoP cells from wild-type, MBP-1−/−/EPX−/−, and PHIL mice were identified by flow cytometry of total marrow leukocytes as described above: lineage negative (CD3−, CD4−, CD8α−, B220−, CD19−, Gr1−, Ter119−), Sca1−, and positive for CD34+, cKit+, and IL-5Rα+. In addition, YO-PRO-1 iodide (Y3603; Molecular Probes, Eugene, OR) and propidium iodide (PI/RNase staining buffer; BD Biosciences) were added to the antibody-stained cell suspensions to identify apoptotic cells and exclude dead cells, respectively. Subsequent flow cytometric assessments were performed with an LSR Fortessa cytofluorimeter (BD Biosciences), and the data were analyzed by using FACSDiva Version 6.2 software (BD Biosciences).
Transmission electron microscopy
Peripheral blood leukocytes were sorted for cells staining positive for IL-5Rα-FITC, Siglec-F-PE, Gr1-PE-Cy7, and CCR3-Alexa 647 (83103; BD Biosciences) on a cytofluorimeter (FACSAria; BD Biosciences) using FACSDiva Version 6.1.1 software (BD Biosciences). As described in earlier studies by Ochkur et al,33 after flow cytometric sorting, eosinophils were fixed in Trump’s fixative (1% glutaraldehyde, 4% formaldehyde, 0.1 M phosphate buffer; pH 7.2) prior to preparation for electron microscopy and photographic evaluations.
Immunohistochemical detection of tissue eosinophils
The presence of bone marrow–derived eosinophils was determined by immunohistochemistry by using a rat monoclonal antibody (mAb) (MT3-25.1.1) recognizing an epitope common to the eosinophil-associated ribonucleases sequestered in the secondary granules of eosinophil lineage-committed leukocytes.34
Partial bone marrow engraftment
Partial (∼70%) bone marrow chimeras were generated by irradiating (3 Gy whole-body irradiation) 3-month-old wild-type C57BL/6J control mice prior to adoptive transfer of marrow derived from double knockout MBP-1−/−/EPX−/− animals.33 Briefly, 1 × 107 bone marrow cells from MBP-1−/−/EPX−/− (CD45.2_+) donors were transferred by tail vein injection into irradiated wild-type C57BL/6J (CD45.1+) recipients. Cell counts and differential assessments were performed on mice following a 60-day recovery period. Donor cell engraftment of ∼70% was achieved as determined by fluorescence activated cell sorting assessments of various leukocyte subpopulations by the presence of cells presenting the donor-derived CD45.2+_ marker.
Ex vivo generation of eosinophils by targeted proliferation/differentiation of unselected bone marrow progenitors
The targeted differentiation and proliferation of eosinophil lineage-committed cells ex vivo was achieved by using a marrow culture system35 with modifications that have been previously described.36 Briefly, whole marrow was flushed from tibias and femurs into supplemented RPMI,35 red blood cells were lysed, and the suspension was resuspended at 106 cells/mL in RPMI further supplemented with 100 ng/mL stem cell factor and FLT3 ligand. Then, 12.5 × 106 marrow leukocytes were transferred to a 25-cm2 culture flask and cultured until day 4, at which point the cells were washed and resuspended in RPMI supplemented with 10 ng/mL IL-5 and returned to culture. On days 8, 10, and 12 of culture, the cells were washed, and half the media was replaced with fresh media supplemented with 20 ng/mL IL-5.
Statistical analysis
Data were analyzed and graphed by using the GraphPad Prism statistics program (GraphPad Prism Software, San Diego, CA). Results are presented as means ± standard error of the mean. Statistical analysis was performed by using Student t tests, with differences between means considered significant at P < .05.
Results
The concomitant loss of MBP-1 and EPX in double knockout mice results in a significant loss of peripheral eosinophils, creating an effectively eosinophil-deficient strain of mice
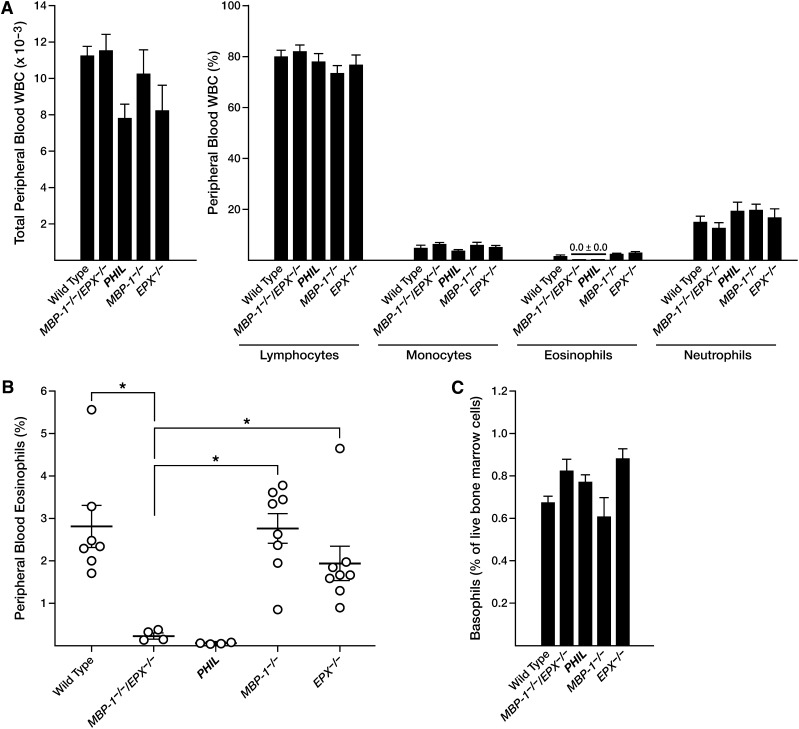
Single-gene knockout mice with targeted deletions within the MBP-116,17 and EPX16,17 loci (Figure 1) were crossed to yield double knockout (MBP-1−/−/EPX−/−) animals for investigating potential functional redundancies of these cationic and abundant eosinophil secondary granule proteins. However, white blood cell (WBC) differential counts (Figure 2A) immediately established that MBP-1−/−/EPX−/− mice were deficient for peripheral blood eosinophils to an extent comparable to previously established eosinophil-deficient mouse strains such as ΔdblGATA37 and PHIL.27 This observation was confirmed by flow cytometric assessments of peripheral blood (Figure 2B) that used a mouse eosinophil-specific cell surface profile (CCR3+, IL-5Rα+). Similar reductions in eosinophils are also observed in lympho-hematopoietic compartments such as the spleen (supplemental Figure 1). In contrast, the number of basophils in the bone marrow of MBP-1−/−/EPX−/− mice (defined by flow cytometric methods; supplemental Figure 2) was unchanged relative to the other experimental cohorts examined, including wild-type as well as MBP-1−/− and EPX−/− single knockout mice (Figure 2C). Assessments of peripheral blood hematocrits from the different experimental cohorts (mean ± standard error of the mean) also showed that the loss of MBP-1 and EPX in double knockout mice had no impact on erythrocyte levels (wild-type, 45.3% ± 0.3%; MBP-1−/−/EPX−/−, 42.0% ± 3.0%; PHIL, 46.0% ± 0.4%; MBP-1−/−, 46.5% ± 0.9%; and EPX−/−, 46.0% ± 1.0%.
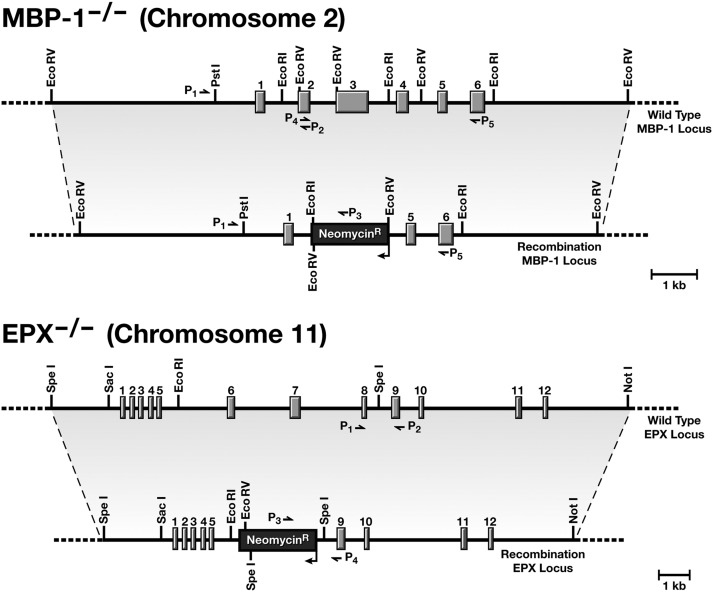
Figure 1.
The targeted disruptions of the mouse eosinophil MBP-1 and EPX genes. Restriction maps of the targeting constructs and the MBP-1 and EPX loci before and after the homologous recombination events occurring in the embryonic stem cells that were used to generate the corresponding knockout strains of MBP-1−/− and EPX−/− mice, respectively. As noted, the genes encoding these abundant secondary granule proteins are located on different mouse chromosomes, facilitating the production of double knockout mice through selective breeding strategies of the single knockout strains of mice; offspring genotypes are subject to Mendelian inheritance patterns based on the genotype of the breeding dam and sire. Various polymerase chain reaction (PCR) primer combinations used to identify and confirm the genotype of individual mice are shown for each loci (P1-P5, MBP-1 and P1-P4, EPX).
Figure 2.
The loss of both MBP-1 and EPX gene expression in MBP-1−/−/EPX−/− mice leads to a peripheral blood eosinophil deficiency that is both definitive and specific. (A) Cell counts and hematologic cell differentials showed that MBP-1−/−/EPX−/− double knockout mice have an eosinophil deficiency (without effects on the composition of the other prominent leukocytes; mean ± standard error of the mean; n = 5 to 8 animals per group) that was equivalent to the deficiency observed in an engineered transgenic mouse model congenitally devoid of eosinophils (ie, PHIL mice). (B) Flow cytometric assessments of circulating WBCs derived from individual mice confirm that eosinophils are virtually absent in the blood of MBP-1−/−/EPX−/− mice, similar to PHIL mice, and are significantly lower than eosinophil numbers observed in either wild-type controls or MBP-1−/− or EPX−/− single knockout animals. (C) The loss of eosinophils in MBP-1−/−/EPX−/− mice had no effect on basophils. Flow cytometric analysis (supplemental Figure 2) of bone marrow–derived leukocytes demonstrated again that the loss of eosinophils in MBP-1−/−/EPX−/− mice, similar to PHIL mice, had no effect on basophil populations. *P < .05.
The few eosinophils detected in MBP-1−/−/EPX−/− mice have eosinophilic features but remain deficient in MBP-1 and EPX
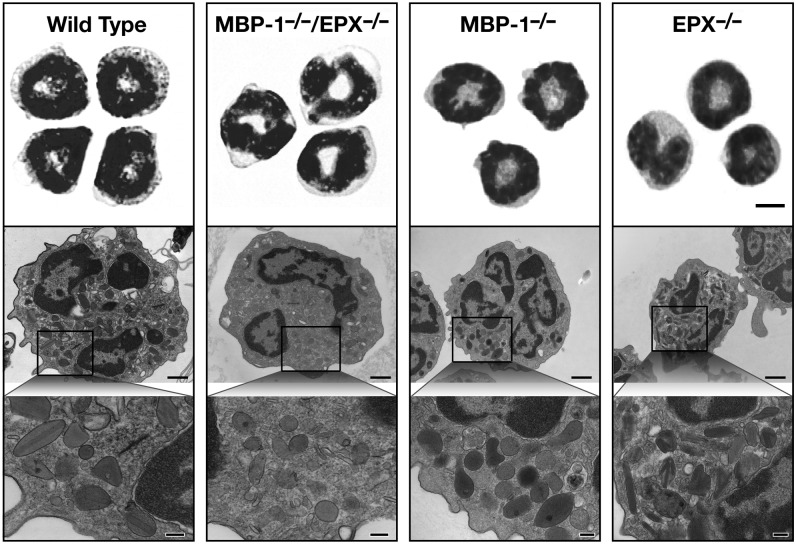
The 0% to 0.2% eosinophils that remain in the blood of MBP-1−/−/EPX−/− mice were notably low side scatter. Nonetheless, these cells displayed an inventory of cell surface markers characteristic of mouse peripheral blood eosinophils, including IL-5Rα, CCR3, Gr1, and Siglec-F (supplemental Figure 3). Romanowsky staining of sorted eosinophils recovered from MBP-1−/−/EPX−/− mice displayed nuclear morphologies characteristic of eosinophils. However, these leukocytes have lost any semblance of the prominent eosin-staining granularity characteristic of eosinophils (Figure 3, top panels). As previously reported, transmission electron microscopy of blood eosinophils from MBP-1−/− and EPX−/− single knockout mice (Figure 3, lower panels) display characteristic morphologies that include the loss of the electron-dense core of the secondary granules (MBP-1−/−)16,17 and the reduction of the electron-translucent matrices of these granules (EPX−/−).16,17 In contrast, the rare circulating MBP-1−/−/EPX−/− eosinophils demonstrated that while secondary granule-like vesicles remain, the secondary granules of these cells have lost both the electron-dense cores and much of the visual densities of the electron-translucent matrices (Figure 3, lower panels).
Figure 3.
Peripheral blood leukocytes that have staining characteristics and subcellular morphologies consistent with eosinophils deficient in MBP-1 and EPX are present in MBP-1−/−/EPX−/− mice. Flow cytometric assessments of WBCs (supplemental Figure 3) from mice of each genotype (ie, wild-type, MBP-1−/−/EPX−/−, MBP-1−/−, and EPX−/−) were used to sort cells, isolating eosinophils that were subsequently cytospun onto slides and stained with a Romanowsky dye set (top panels). Scale bar = 5 µm. The electron microscopic morphology of the eosinophils that remain in MBP-1−/−/EPX−/− mice (lower panels) showed that these cells retain the membrane-bound vesicles (ie, granules) found in wild-type, MBP-1−/−, or EPX−/− mice. As expected, these granules appear devoid of their electron-dense cores (MBP-1) and much of the electron-translucent matrices (characteristic of EPX) of these granules. Scale bars in panels highlighted by a single eosinophil = 1 µm. Scale bars in the insert panels representative of the cytoplasm = 200 nm.
MBP-1−/−/EPX−/− mice displayed severely reduced levels of marrow-derived eosinophilopoiesis
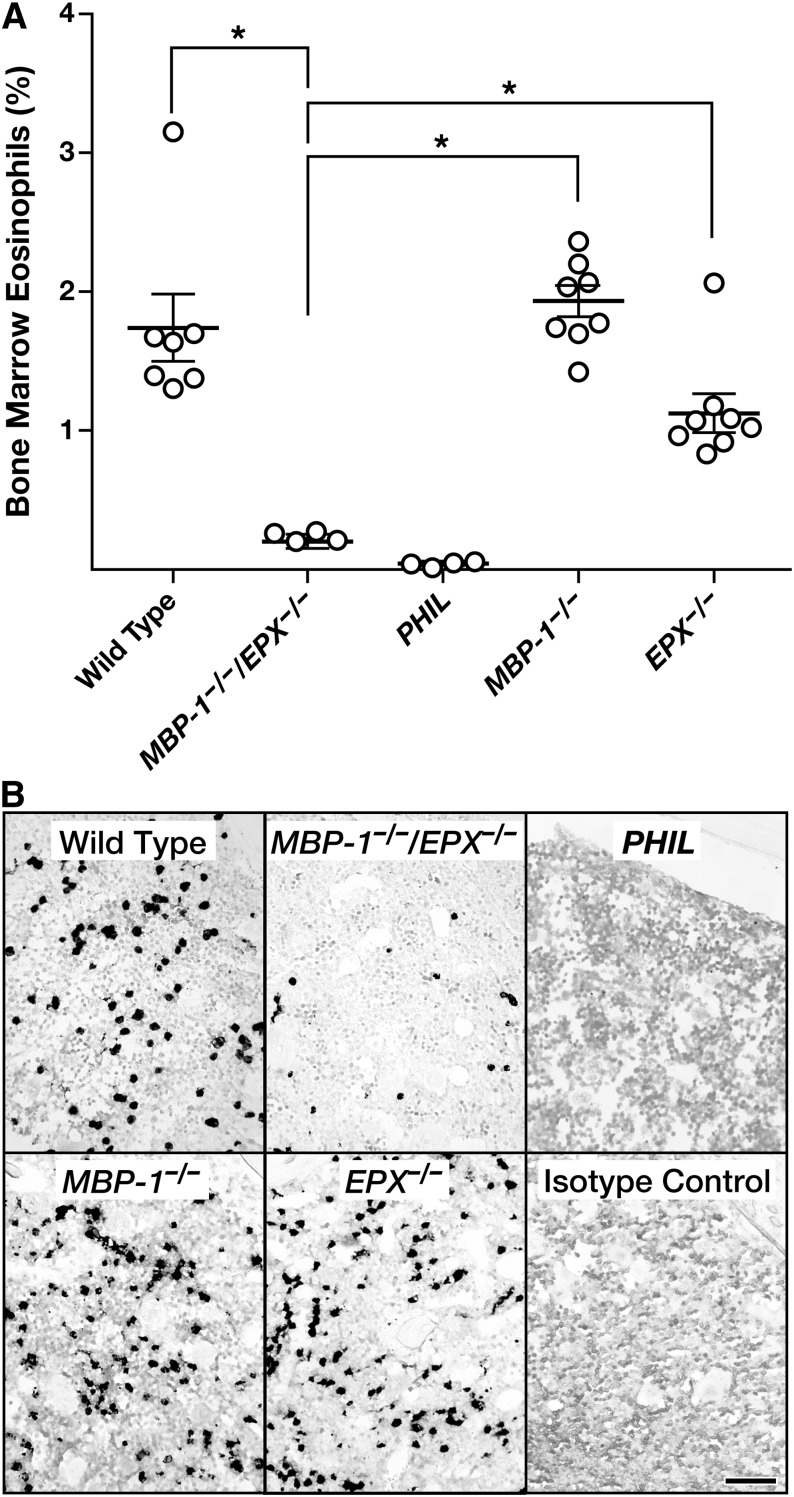
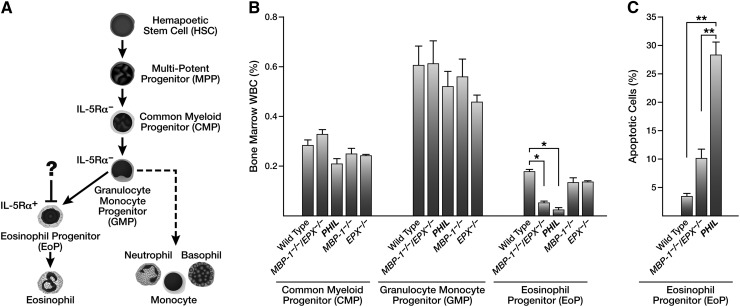
Flow cytometry performed on the bone marrow of MBP-1−/−/EPX−/− mice showed that the number of mature eosinophils and EoPs were reduced relative to both wild-type and single-granule protein gene knockout mice (Figure 4A). Interestingly, one can detect eosinophil lineage-committed cells in the bone marrow of MBP-1−/−/EPX−/− mice, unlike in PHIL mice in which the number of marrow eosinophils and progenitors was virtually zero. This reduction of (but not the complete absence of) marrow eosinophils was also confirmed by immunohistochemical staining of formalin-fixed and paraffin-embedded sections of femurs using an eosinophil-specific rat mAb (Ear mAb) recognizing a shared epitope of the eosinophil-associated ribonuclease cluster (Figure 4B). Collectively, these marrow assessments suggested a specific and unique defect in eosinophilopoiesis that was likely limited to EoPs and not earlier myeloid lineage stem cells (Figure 5A). Subsequent flow cytometric assessments of various lineage-committed marrow progenitors26,27 (supplemental Figure 4), confirmed that the definitive loss of progenitor cells was limited to EoPs because the numbers of common myeloid progenitors and granulocyte monocyte progenitors in the marrow of MBP-1−/−/EPX−/− mice remained unchanged relative to either wild-type or MBP-1−/− and EPX−/− single knockout animals (Figure 5B). These data also confirmed the fractional character of EoP depletion in MBP-1−/−/EPX−/− mice relative to the more complete loss of progenitors observed in PHIL mice. Significantly, assessments of apoptosis (Figure 5C) demonstrated that the fractional character of EoP depletion in MBP-1−/−/EPX−/− mice relative to the loss of EoPs in PHIL mice is reflected in fractional increases of apoptosis observed in these populations relative to wild-type animals (threefold and eightfold, respectively). This implicates that EoP survival is a key kinetic parameter responsible for the loss of eosinophils in MBP-1−/−/EPX−/− mice.
Figure 4.
The loss of both MBP-1 and EPX gene expression in MBP-1−/−/EPX−/− mice significantly reduces the steady-state number of eosinophils and their progenitors in the bone marrow. (A) Flow cytometric assessments of bone marrow–derived leukocytes from individual mice showed that the number of eosinophils (ie, CCR3+ and IL-5Rα+ cells) is significantly lower in MBP-1−/−/EPX−/− mice relative to either wild-type controls or MBP-1−/− or EPX−/− single knockout animals. *P < .05. (B) Immunohistochemical staining (dark-staining cells) of femur sections using a rat mAb (MT3 25.1.1) recognizing Ear-1, -2, -6/7, and -5/11 confirmed our flow cytometric data showing that the number of Ear+ eosinophils in MBP-1−/−/EPX−/− mice was significantly lower than the number of eosinophils observed in the marrow of either wild-type, MBP-1−/−, or EPX−/− mice. However, unlike the marrow of mice devoid of eosinophils and their progenitors (ie, PHIL mice), the marrow of MBP-1−/−/EPX−/− mice maintained a finite steady-state population of these eosinophil lineage-committed cells. Isotype control: rat normal serum immunoglobulin G (IgG). Scale bar = 100 µm.
Figure 5.
The concomitant loss of MBP-1 and EPX led to a targeted reduction in EoP cells without similar effects on other less committed myeloid progenitor populations. (A) Hematopoietic differentiation pathways leading to eosinophils in mice (adapted from Lee et al3). The unique and specific loss of eosinophils displayed by MBP-1−/−/EPX−/− mice suggested the blockade in eosinophilopoiesis (?) occurred at the EoP stage as opposed to earlier myeloid progenitors (ie, common myeloid progenitors [CMP] or granulocyte monocyte progenitors [GMP]). (B) Flow cytometric assessments of marrow leukocytes confirmed that the defect in eosinophilopoiesis observed in MBP-1−/−/EPX−/− mice is limited to the EoP population with no observed effects on either the CMP or GMP populations. (C) Flow cytometric assessments showed that the EoP populations of both PHIL (n = 3) and MBP-1−/−/EPX−/− (n = 7) mice each displayed significant increases in apoptosis (eightfold and threefold increases, respectively) relative to wild-type (n = 7). *P < .05; **P < .002.
IL-5–dependent expansion of eosinophils failed to occur in the marrow of transgenic mice (in vivo) as well as in ex vivo cultures of MBP-1−/−/EPX−/− bone marrow
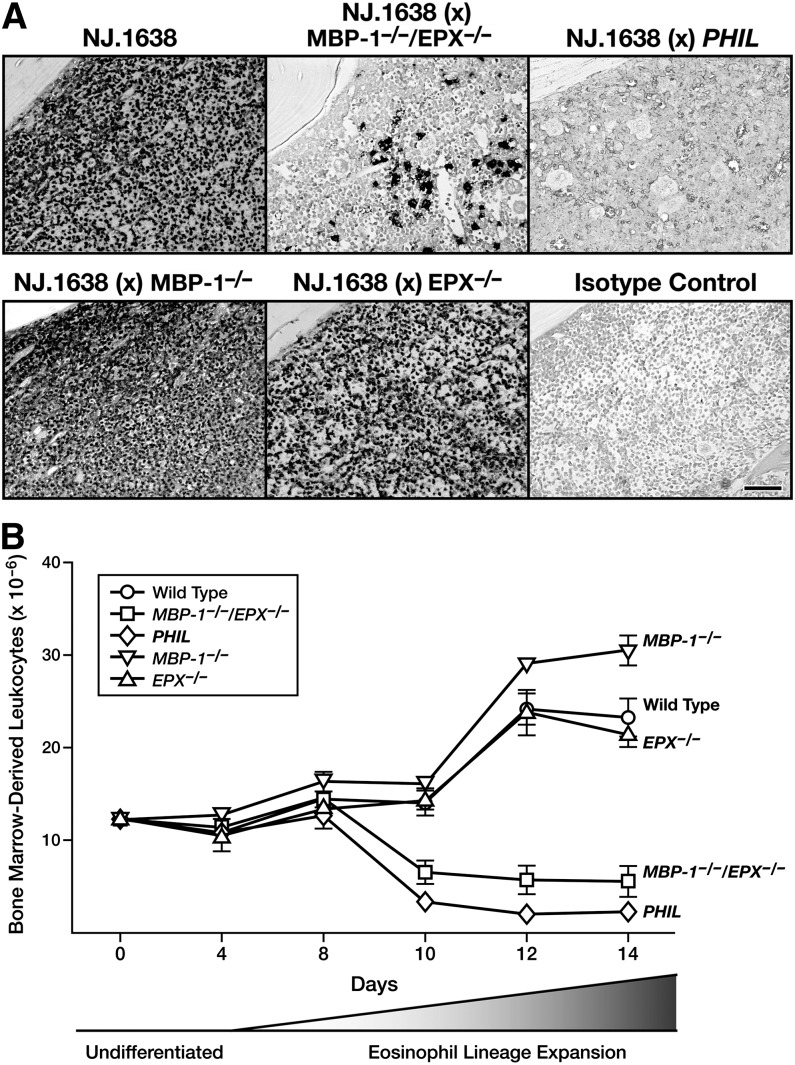
Our observation that only the IL-5Rα+ EoP population of the marrow was affected in MBP-1−/−/EPX−/− mice suggested that even the hematopoietic pressure of ectopic constitutive overexpression of the eosinophil proliferative cytokine IL-528 would fail to promote the production and accumulation of eosinophils. This hypothesis was tested by examining eosinophils derived from femoral marrow in each experimental cohort using immunohistochemistry and an mAb directed against a shared epitope of the eosinophil-associated ribonucleases cluster (ie, Ear mAb). The photomicrographs in Figure 6A show that an overwhelming expansion of marrow eosinophil lineage-committed cells occurred in the parental IL-5 transgenic line of mice (NJ.163828) as well as in compound transgenic/single knockout animals (ie, NJ.1638/MBP-1−/− and NJ.1638/EPX−/−). In contrast, the loss of both MBP-1 and EPX in transgenic double knockout mice (ie, NJ.1638/MBP-1−/−/EPX−/−) resulted in a loss of two to three orders of magnitude of eosinophil lineage-committed cells in the bone marrow (Figure 6A). However, similar to marrow in MBP-1−/−/EPX−/− animals, the marrow of NJ.1638/MBP-1−/−/EPX−/− mice was not devoid of eosinophils.
Figure 6.
IL-5–dependent expansion of EoPs is blocked in the marrow of MBP-1−/−/EPX−/− mice (ie, in vivo) as well as in cell cultures of marrow (ie, ex vivo) derived from these double granule protein gene knockout animals. (A) Constitutive high-level ectopic expression of IL-5 from mature T cells is unable to rescue the significant loss of marrow eosinophil lineage-committed cells observed in MBP-1−/−/EPX−/− mice. Immunohistochemical staining (dark-staining cells) of femur sections using a rat anti-mouse Ear mAb (MT3 25.1.1) confirmed our flow cytometric data showing a unique loss of eosinophil lineage-committed cells in MBP-1−/−/EPX−/− mice. This immunohistochemical staining also revealed that, unlike the marrow of IL-5 transgenic mice devoid of eosinophils and their progenitors (ie, NJ.1638/PHIL mice), the marrow of NJ.1638/MBP-1−/−/EPX−/− mice maintained a finite steady-state population of these eosinophil lineage-committed cells. Isotype control: rat normal serum IgG. Scale bar = 100 µm. (B) Eosinophil differentiation was significantly limited in ex vivo bone marrow cultures using hematopoietic stem cells/progenitors from MBP-1−/−/EPX−/− mice. Biphasic bone marrow cultures initially expanding undifferentiated stem cells/progenitors (undifferentiated) prior to IL-5–mediated expansion of EoPs (eosinophil lineage expansion) showed that similar to observations in vivo, EoP cells in PHIL and MBP-1−/−/EPX−/− mice were unable to undergo IL-5–dependent expansion. Thus, while bone marrow cultures of wild-type and single granule protein knockout mice (ie, MBP-1−/− and EPX−/−) each displayed a significant expansion in total cell number with a resulting compositional shift to >98% eosinophils, the marrow of PHIL and MBP-1−/−/EPX−/− mice failed to undergo this expansion.
IL-5–dependent eosinophilopoiesis in MBP-1−/−/EPX−/− mice was also examined in ex vivo bone marrow progenitor cultures (Figure 6B). Given the lack of observed effects on non-EoPs in the marrow of double knockout mice, it was not surprising that total bone marrow progenitor cultures from these animals displayed no kinetic or steady-state population differences relative to the wild-type progenitors prior to the application of recombinant IL-5 and the proliferation of EoPs. However, substantive differences were observed when eosinophilopoietic pressure (ie, exposure of the cells to IL-5–containing media) was applied to the cultures. Wild-type as well as single MBP-1−/− and EPX−/− knockout mice each displayed significant expansion in total eosinophil numbers with the differential composition of the cultures being >95% eosinophils with fractional percentages of mononuclear cells, neutrophils, and stromal cells. In contrast, the loss of MBP-1 and EPX in double knockout mice led to a specific blockade of eosinophil expansion (ie, following the shift to IL-5–containing media) in cultured marrow from MBP-1−/−/EPX−/− mice. However, significant effects on proliferation/differentiation are unlikely explanations for the lack of cell expansion in MBP-1−/−/EPX−/− marrow cultures because the differential composition of the cells resulting after the expansion phase remained similar to that in the control groups (ie, >90% eosinophils with fractional percentages of mononuclear cells, neutrophils, and stromal cells).
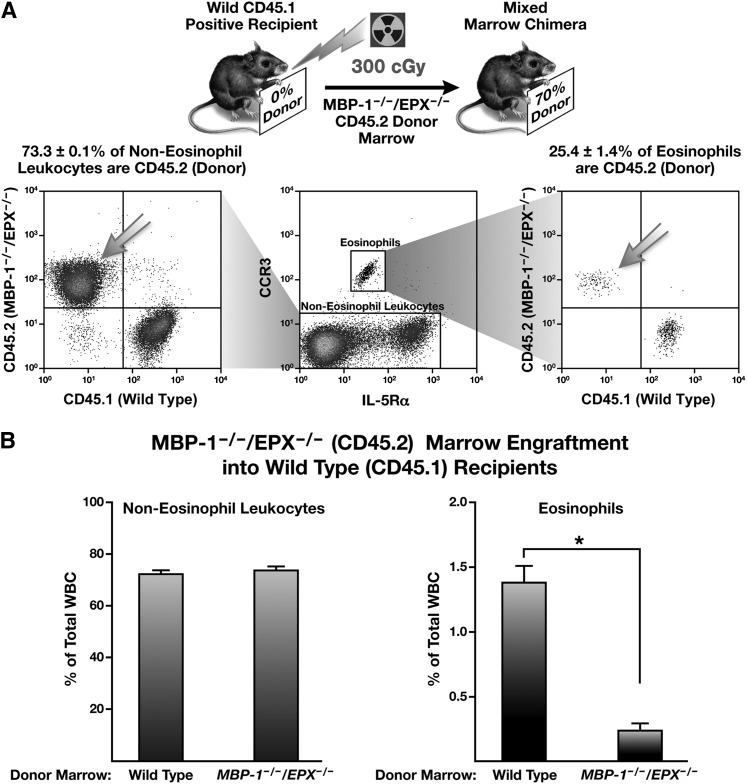
Partial bone marrow engraftment demonstrated that the defect in MBP-1−/−/EPX−/− marrow is cell autonomous
Partial bone marrow chimeras were created to determine whether the loss of eosinophilopoiesis in MBP-1−/−/EPX−/− mice resulted from either a cell-autonomous defect in MBP-1−/−/EPX−/− EoP cells or whether proliferation also required interaction with trans-acting soluble factor(s) secreted by wild-type marrow progenitor and/or stromal cells. Mixed-marrow chimeric mice were achieved by γ irradiation (3 Gy) of CD45.1+ recipient wild-type mice followed by adoptive intravenous transfer of CD45.2+ donor marrow from MBP-1−/−/EPX−/− mice (Figure 7A). These data showed that partial engraftment of MBP-1−/−/EPX−/− marrow into wild-type recipients resulted in mixed chimeric animals that were 70% donor marrow as determined from the non-eosinophil leukocyte populations. However, the donor MBP-1−/−/EPX−/− marrow cells had only a limited ability to generate peripheral blood eosinophils. Quantitative flow cytometric evaluations comparing the ability of wild-type vs MBP-1−/−/EPX−/− marrow to colonize the progenitor populations of recipient wild-type mice are presented in Figure 7B. These evaluations showed that MBP-1−/−/EPX−/− progenitors colonize the marrow of chimeric wild-type recipients and contributed to non-eosinophil leukocyte populations at levels identical to the levels observed following the transfer of wild-type marrow. In contrast, MBP-1−/−/EPX−/− donor marrow in these mixed chimeric studies was unable to generate peripheral blood eosinophils to the same level, displaying a >80% decrease relative to the contribution achievable with wild-type donor marrow (Figure 7B).
Figure 7.
Partial bone marrow engraftment of wild-type recipient mice with marrow from MBP-1−/−/EPX−/− marrow donors showed that the loss of eosinophilopoiesis induced by the concomitant loss of both MBP-1 and EPX was a cell-autonomous effect. (A) Representative flow cytometric assessments of peripheral WBC populations from the resulting mixed-marrow chimeric mice are shown; they demonstrated that while non-eosinophil leukocytes displayed a 70% donor/30% recipient distribution of CD45.2/CD45.1, respectively, contributions to overall steady-state eosinophil populations in these mixed-marrow chimeric animals by donor MBP-1−/−/EPX−/− (CD45.2) eosinophils was limited. (B) Flow cytometric assessments of peripheral blood from mixed-marrow chimeras generated by γ irradiation of CD45.1+ recipient wild-type mice and subsequent adoptive transfer of either CD45.2+ wild-type or MBP-1−/−/EPX−/− donor marrow. These data showed that the non-eosinophil blood leukocytes of donor origin (% of total WBC) remained unchanged when using wild-type vs MBP-1−/−/EPX−/− donor marrow. However, the eosinophil populations of these mixed-marrow chimeras displayed a differential distribution of donor origin (>80% lower) when comparing recipient mice that had received wild-type vs MBP-1−/−/EPX−/− marrow. *P < .05.
Discussion
The expression and storage of unique proteins in the secondary granules of eosinophils represents a defining characteristic of these leukocytes both in terms of cell lineage identification and proposed effector functions (reviewed by Lee et al2). In particular, the release of these proteins and other mediators stored within the secondary granules of eosinophils (ie, degranulation) has been a highlighted activity that is almost universally linked with these granulocytes.38 The underlying assumption has been that the expression of the genes encoding the abundant eosinophil secondary granule proteins was a consequence of eosinophil lineage commitment of previously pluripotent hematopoietic precursors (reviewed by Mori et al22). Our previous demonstration that the only baseline consequence of losing either MBP-1 or EPX in single knockout mice is the generation of peripheral eosinophils devoid of the respective secondary granule protein16,17 is consistent with this assumption. In light of this perceived mechanism of lineage commitment followed by differentiation/proliferation and cell-specific gene expression, we could not explain our observation that the combined loss of both MBP-1 and EPX gene expression resulted in the collapse of eosinophilopoiesis. This result suggested that granule protein gene expression and/or possibly the process of granule formation was not simply a passive consequence of cell lineage commitment but instead may have a larger hematopoietic role. The data demonstrated that the loss of both MBP-1 and EPX expression led directly to an increase in apoptosis among EoPs. This observation suggests that similar to the mechanism proposed for the loss of eosinophils in PHIL mice,27 effects on the survival of EoPs in MBP-1−/−/EPX−/− mice are a principle kinetic regulator limiting the steady-state number of marrow EoPs and, in turn, peripheral eosinophils.
The demonstration that this blockade of eosinophilopoiesis was a cell-autonomous event that occurred in vivo (ie, the continued absence of MBP-1−/−/EPX−/− eosinophils in wild-type double knockout bone marrow chimeras) as well as in ex vivo cultures of MBP-1−/−/EPX−/− marrow progenitors, ruled out two significant potential mechanisms. Namely, EoP survival is not the result of the release of MBP-1 and EPX by resident eosinophils in the marrow,39 modifying or creating a hematopoietic niche as part of a positive autocrine feedback loop supporting progenitor survival. In addition, the demonstration that wild-type hematopoietic stem cells could not rescue eosinophilopoiesis from MBP-1−/−/EPX−/− progenitors eliminated the possibility that the combined loss of these granule proteins prevented (and/or abolished) the release of trans-acting soluble factors that support EoP survival.
Two global mechanisms that potentially link EoP survival and the coexpression of MBP-1 and EPX are noteworthy. However, it is also of note that the available data only tangentially support many of these mechanisms. Nonetheless, the available data do provide important insights and a framework suggesting the likelihood and probable relevance of given mechanisms.
Granule protein gene expression and/or granule formation is a checkpoint for survival of EoPs
This hypothesis (and three variations in descending order of probability) potentially provides the most likely explanation for the loss of EoP cells in MBP-1−/−/EPX−/− mice, in part, because of the available data and similar observations in the literature.
Variation 1: Granule formation is a prerequisite for EoP cell survival.
This hypothesis suggests that the combined expression of MBP-1 and EPX and the subsequent granule formation that accompanies this expression is a cell signaling node and/or a checkpoint for the survival of EoPs. The fact that all of the surviving EoPs and mature eosinophils in MBP-1−/−/EPX−/− mice have granules is consistent with a direct link between granule formation and eosinophilopoiesis. In addition, the observed granules in double knockout mice appear to be structurally different relative to eosinophil granules from wild-type mice, which suggests that the formation of granules has been partially affected by the loss of MBP-1 and EPX. Collectively, these observations may explain the partial, as opposed to the complete, ablation of EoPs in double knockout mice. Unfortunately, despite the correlative data in support of this hypothesis, a direct link of granule formation and EoP cell survival remains to be established.
Variation 2: Sequestration of a toxicant.
In this hypothesis, the loss of MBP-1 and EPX expression in EoPs leads to either the aberrant secretion of toxic granule components or the aberrant trafficking of toxic granule components into the cytoplasm. Given the toxic character of many of the eosinophil granule proteins (reviewed in Lee and Lee38), it is difficult to ignore or dismiss this mechanism. As noted earlier, MBP-1−/−/EPX−/− eosinophils do display abnormalities in granule structure. However, the data also showed that the defect induced by the loss of MBP-1 and EPX expression is a cell-autonomous event and thus the aberrant release of a granule toxicant (eg, Ears) is an unlikely cause for the loss of EoPs. In contrast, even if only a small fraction of granule components such as Ears are released into the cytoplasm, this may be sufficient to underlie the observed cell-autonomous defect in MBP-1−/−/EPX−/− mice. Clearly, further studies investigating the specific trafficking of these proteins into granules vs the cytoplasm will be required to resolve this possibility.
Variation 3: Unfolded protein response.
Neutrophils are an abundant granulocyte population that follow a path of differentiation with some similarity to that of eosinophils. Cases of primary neutropenia occur in humans at a rate of approximately 1 in 106 subjects,40 and several mechanisms have been proposed to account for severe congenital neutropenia (SCN). Approximately half the observed SCN cases are linked to mutations in ELA2, the gene encoding the neutrophil granule protein elastase.40 It has been hypothesized that these mutations lead to an unfolded protein response in the endoplasmic reticulum resulting in blockade of differentiation and proliferation of progenitor cells (reviewed in Klein et al41). In addition, it is noteworthy that knock-in mice with single ELA2 mutations found in patients do not result in a baseline neutropenia in these animals.42,43 This implies that effects on multiple granule proteins may be required for this effect in neutrophils. However, to date we have not been able to show that the engineered MBP-1 and EPX loci16,17 actually generate truncated or aberrant polypeptides (our unpublished observations) and the suggestion that specific events need to coordinately happen in two genes limits the probability of this explanation.
The loss of concomitant MBP-1 and EPX expression in progenitors disrupts instructive gene regulatory mechanisms contributing to the continued appearance and/or survival of EoPs
The recent demonstration that micro RNAs (miRNAs) have potential effects on eosinophilopoiesis44 and that the noncoding RNA EGO regulates the translation of several secondary granule proteins during eosinophil development45 suggests the possibility that noncoding RNAs and/or one or more miRNAs critical for eosinophil development are encoded within the MBP-1 and EPX loci. The direct and straightforward character of these potential mechanisms for the deficiency of eosinophils in MBP-1−/−/EPX−/− mice promote them as attractive explanations. Unfortunately, detailed examinations of the sequences at each loci have failed to reveal the presence of noncoding RNAs (our unpublished observations) nor have searches of available miRNA databases (eg, www.miRBase.org) identified any occult miRNAs within the deleted regions of the MBP-1 or EPX knockout loci. Moreover, the likelihood of these explanations is severely dampened by the observation that the genetic manipulation of two loci (MBP-1 and EPX) is required. This suggests that while it is possible that the genetic manipulations of the MBP-1 and EPX genes during the generation of these knockout mice induced effects on the expression of small regulatory RNAs encoded at these loci, this possibility is an unlikely explanation for the loss of EoP cells in MBP-1−/−/EPX−/− mice.
The specific mechanism(s) responsible for the eosinophil deficiency in MBP-1−/−/EPX−/− mice remains elusive; nonetheless, these animals have immediate value to the research community as a specific and targeted eosinophil-less mouse model. Relative to other strains of eosinophil-less mice (ie, PHIL27 and ΔdblGATA37), the specific genetic aberrations associated with the MBP-1−/−/EPX−/− genotype appear to elicit a targeted deletion of the eosinophil lineage with fewer logistical caveats that would limit the usefulness of this mouse model. For example, PHIL mice employ a cytocidal mechanism (ie, eosinophil-specific expression of a diphtheria toxin subunit); thus, it has been suggested that the presence of this diphtheria toxin–mediated killing may contribute to baseline inflammation in this mouse relative to wild-type mice.46 The eosinophil-deficient character of ΔdblGATA mice results from a targeted mutation of a high-affinity GATA transcription factor binding site within the GATA-1 promoter itself. However, activities of GATA-1 are not confined to eosinophils37 (reviewed by Ferreira et al47), and this mutation has been shown to have additional unintended consequences as demonstrated by perturbations observed in red cell numbers.37 Our assessments of MBP-1−/−/EPX−/− mice showed that neither of these issues appear to be relevant; total leukocyte counts (aside from eosinophils) are normal in these mice, and no other cell lineage (aside from eosinophils) appears to be affected. Thus, while future studies are necessary to understand the link between granule protein gene expression and the survival of committed progenitors, the resulting eosinophil-deficient mouse model will be helpful in gaining a better understanding of eosinophil activities as part of studies of human disease in the mouse.
Supplementary Material
Supplemental Figures
Acknowledgments
The authors thank members of Lee Laboratories who reviewed various drafts of this manuscript and who helped in the data collection and the organization/infrastructure needed to complete the studies presented. We also acknowledge the efforts of the small animal facility and flow cytometry core staff, the invaluable assistance of the Mayo Clinic Arizona medical graphic artist, Marv Ruona, and the excellent administrative support provided to Lee Laboratories by Linda Mardel and Shirley (“Charlie”) Kern.
This work was supported by the Mayo Foundation, by National Institutes of Health grants HL058723 (N.A.L.), HL065228, and RR0109709 (J.J.L.), and grant AI000941 (H.F.R.) from the National Institute of Allergy and Infectious Diseases, Division of Intramural Research. Support for A.D.D. was provided by a Mayo Clinic Sidney Luckman Family Pre-doctoral Fellowship.
These funding sources had no involvement in study design, data collection (including analysis and interpretation), the writing of the manuscript, or the decision to submit for publication.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.D.D., J.J.L., and N.A.L. conceived, developed, and wrote this manuscript; A.D.D., E.A.J., S.I.O., and M.P.M. were the primary investigators who collected and/or analyzed data; M.P.M., K.G.S., D.T.C.N., C.P., J.K., J.N., and K.P.S. provided technical support to perform studies and assisted with the analysis of data; D.C. managed the animal colony providing support for the generation and genotyping of subject mice used in these studies; and K.D.D. and H.F.R. performed ex vivo kinetic studies of bone marrow growth and differentiation in addition to providing critical reviews of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nancy A. Lee, Mayo Clinic College of Medicine, Department of Biochemistry and Molecular Biology, Mayo Clinic Collaborative Research Building, 2-206, Mayo Clinic Arizona, 13400 E. Shea Blvd, Scottsdale, AZ 85259; e-mail: nlee@mayo.edu.
References
- 1.Erlich P. Ueber die Specifischen granulationen des Blutes. [in German] Arch Anat Physiol. 1879;3:571–579. [Google Scholar]
- 2.Lee JJ. In: Eosinophils in Health and Disease. Rosenberg HF, editor. Waltham, MA: Elsevier; 2012. [Google Scholar]
- 3.Lee JJ, Jacobsen EA, Ochkur SI, et al. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J Allergy Clin Immunol. 2012;130(3):572–584. doi: 10.1016/j.jaci.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker RL, Gundel RH, Gleich GJ, Checkel JL, Loegering DA, Pease LR, Hamann KJ. Acidic polyamino acids inhibit human eosinophil granule major basic protein toxicity. Evidence of a functional role for ProMBP. J Clin Invest. 1991;88(3):798–805. doi: 10.1172/JCI115379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson KA, Horton MA, Madden BJ, Gleich GJ, Lee NA, Lee JJ. The identification and cloning of a murine major basic protein gene expressed in eosinophils. J Immunol. 1995;155(6):3002–3012. [PubMed] [Google Scholar]
- 6.Macias MP, Welch KC, Denzler KL, Larson KA, Lee NA, Lee JJ. Identification of a new murine eosinophil major basic protein (mMBP) gene: cloning and characterization of mMBP-2. J Leukoc Biol. 2000;67(4):567–576. doi: 10.1002/jlb.67.4.567. [DOI] [PubMed] [Google Scholar]
- 7.Plager DA, Loegering DA, Weiler DA, et al. A novel and highly divergent homolog of human eosinophil granule major basic protein. J Biol Chem. 1999;274(20):14464–14473. doi: 10.1074/jbc.274.20.14464. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg HF, Tenen DG, Ackerman SJ. Molecular cloning of the human eosinophil-derived neurotoxin: a member of the ribonuclease gene family. Proc Natl Acad Sci USA. 1989;86(12):4460–4464. doi: 10.1073/pnas.86.12.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg HF, Ackerman SJ, Tenen DG. Human eosinophil cationic protein. Molecular cloning of a cytotoxin and helminthotoxin with ribonuclease activity. J Exp Med. 1989;170(1):163–176. doi: 10.1084/jem.170.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormier SA, Larson KA, Yuan S, et al. Mouse eosinophil-associated ribonucleases: a unique subfamily expressed during hematopoiesis. Mamm Genome. 2001;12(5):352–361. doi: 10.1007/s003350020007. [DOI] [PubMed] [Google Scholar]
- 11.Larson KA, Olson EV, Madden BJ, Gleich GJ, Lee NA, Lee JJ. Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc Natl Acad Sci USA. 1996;93(22):12370–12375. doi: 10.1073/pnas.93.22.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton MA, Larson KA, Lee JJ, Lee NA. Cloning of the murine eosinophil peroxidase gene (mEPO): characterization of a conserved subgroup of mammalian hematopoietic peroxidases. J Leukoc Biol. 1996;60(2):285–294. doi: 10.1002/jlb.60.2.285. [DOI] [PubMed] [Google Scholar]
- 13.Ten RM, Pease LR, McKean DJ, Bell MP, Gleich GJ. Molecular cloning of the human eosinophil peroxidase. Evidence for the existence of a peroxidase multigene family. J Exp Med. 1989;169(5):1757–1769. doi: 10.1084/jem.169.5.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ackerman SJ, Loegering DA, Venge P, Olsson I, Harley JB, Fauci AS, Gleich GJ. Distinctive cationic proteins of the human eosinophil granule: major basic protein, eosinophil cationic protein, and eosinophil-derived neurotoxin. J Immunol. 1983;131(6):2977–2982. [PubMed] [Google Scholar]
- 15.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 16.Denzler KL, Farmer SC, Crosby JR, et al. Eosinophil major basic protein-1 does not contribute to allergen-induced airway pathologies in mouse models of asthma. J Immunol. 2000;165(10):5509–5517. doi: 10.4049/jimmunol.165.10.5509. [DOI] [PubMed] [Google Scholar]
- 17.Denzler KL, Borchers MT, Crosby JR, et al. Extensive eosinophil degranulation and peroxidase-mediated oxidation of airway proteins do not occur in a mouse ovalbumin-challenge model of pulmonary inflammation. J Immunol. 2001;167(3):1672–1682. doi: 10.4049/jimmunol.167.3.1672. [DOI] [PubMed] [Google Scholar]
- 18.O’Connell AE, Hess JA, Santiago GA, Nolan TJ, Lok JB, Lee JJ, Abraham D. Major basic protein from eosinophils and myeloperoxidase from neutrophils are required for protective immunity to Strongyloides stercoralis in mice. Infect Immun. 2011;79(7):2770–2778. doi: 10.1128/IAI.00931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wehling-Henricks M, Sokolow S, Lee JJ, Myung KH, Villalta SA, Tidball JG. Major basic protein-1 promotes fibrosis of dystrophic muscle and attenuates the cellular immune response in muscular dystrophy. Hum Mol Genet. 2008;17(15):2280–2292. doi: 10.1093/hmg/ddn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Specht S, Saeftel M, Arndt M, et al. Lack of eosinophil peroxidase or major basic protein impairs defense against murine filarial infection. Infect Immun. 2006;74(9):5236–5243. doi: 10.1128/IAI.00329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forbes E, Murase T, Yang M, et al. Immunopathogenesis of experimental ulcerative colitis is mediated by eosinophil peroxidase. J Immunol. 2004;172(9):5664–5675. doi: 10.4049/jimmunol.172.9.5664. [DOI] [PubMed] [Google Scholar]
- 22.Mori Y, Iwasaki H, Akashi K. Eosinophil Lineage-Committed Progenitors. In: Lee JJ, Rosenberg HF, editors. Eosinophils in Health and Disease. Waltham, MA: Academic Press, Elsevier; 2012. pp. 89–97. [Google Scholar]
- 23.Wintrobe MM, Greer JP. Wintrobe's clinical hematology. 12th ed. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2009. [Google Scholar]
- 24.Hartmann J, Scepek S, Lindau M. Regulation of granule size in human and horse eosinophils by number of fusion events among unit granules. J Physiol. 1995;483(Pt 1):201–209. doi: 10.1113/jphysiol.1995.sp020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammel I, Lagunoff D, Galli SJ. Regulation of secretory granule size by the precise generation and fusion of unit granules. J Cell Mol Med. 2010;14(7):1904–1916. doi: 10.1111/j.1582-4934.2010.01071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsen EA, Helmers RA, Lee JJ, Lee NA. The expanding role(s) of eosinophils in health and disease. Blood. 2012;120(19):3882–3890. doi: 10.1182/blood-2012-06-330845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JJ, Dimina D, Macias MP, et al. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004;305(5691):1773–1776. doi: 10.1126/science.1099472. [DOI] [PubMed] [Google Scholar]
- 28.Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, Lee JJ. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol. 1997;158(3):1332–1344. [PubMed] [Google Scholar]
- 29.McGarry MP, Protheroe CA, Lee JJ. Mouse Hematology. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Laboratory Press; 2009. [Google Scholar]
- 30.Obata K, Mukai K, Tsujimura Y, et al. Basophils are essential initiators of a novel type of chronic allergic inflammation. Blood. 2007;110(3):913–920. doi: 10.1182/blood-2007-01-068718. [DOI] [PubMed] [Google Scholar]
- 31.Iwasaki H, Mizuno S, Mayfield R, et al. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005;201(12):1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bruin AM, Buitenhuis M, van der Sluijs KF, van Gisbergen KP, Boon L, Nolte MA. Eosinophil differentiation in the bone marrow is inhibited by T cell-derived IFN-gamma. Blood. 2010;116(14):2559–2569. doi: 10.1182/blood-2009-12-261339. [DOI] [PubMed] [Google Scholar]
- 33.Ochkur SI, Jacobsen EA, Protheroe CA, et al. Coexpression of IL-5 and eotaxin-2 in mice creates an eosinophil-dependent model of respiratory inflammation with characteristics of severe asthma. J Immunol. 2007;178(12):7879–7889. doi: 10.4049/jimmunol.178.12.7879. [DOI] [PubMed] [Google Scholar]
- 34.Cormier SA, Yuan S, Crosby JR, et al. T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am J Respir Cell Mol Biol. 2002;27(6):678–687. doi: 10.1165/rcmb.4882. [DOI] [PubMed] [Google Scholar]
- 35.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol. 2008;181(6):4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle AD, Jacobsen EA, Ochkur SI, et al. Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J Leukoc Biol. 2013;94:17–24. doi: 10.1189/jlb.0213089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195(11):1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JJ, Lee NA. Eosinophil degranulation: an evolutionary vestige or a universally destructive effector function? Clin Exp Allergy. 2005;35(8):986–994. doi: 10.1111/j.1365-2222.2005.02302.x. [DOI] [PubMed] [Google Scholar]
- 39.Butterfield JH, Ackerman SJ, Scott RE, Pierre RV, Gleich GJ. Evidence for secretion of human eosinophil granule major basic protein and Charcot-Leyden crystal protein during eosinophil maturation. Exp Hematol. 1984;12(3):163–170. [PubMed] [Google Scholar]
- 40.Donadieu J, Fenneteau O, Beaupain B, Mahlaoui N, Chantelot CB. Congenital neutropenia: diagnosis, molecular bases and patient management. Orphanet J Rare Dis. 2011;6:26. doi: 10.1186/1750-1172-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein C. Genetic defects in severe congenital neutropenia: emerging insights into life and death of human neutrophil granulocytes. Annu Rev Immunol. 2011;29:399–413. doi: 10.1146/annurev-immunol-030409-101259. [DOI] [PubMed] [Google Scholar]
- 42.Grenda DS, Johnson SE, Mayer JR, McLemore ML, Benson KF, Horwitz M, Link DC. Mice expressing a neutrophil elastase mutation derived from patients with severe congenital neutropenia have normal granulopoiesis. Blood. 2002;100(9):3221–3228. doi: 10.1182/blood-2002-05-1372. [DOI] [PubMed] [Google Scholar]
- 43.Nanua S, Murakami M, Xia J, Grenda DS, Woloszynek J, Strand M, Link DC. Activation of the unfolded protein response is associated with impaired granulopoiesis in transgenic mice expressing mutant Elane. Blood. 2011;117(13):3539–3547. doi: 10.1182/blood-2010-10-311704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu TX, Lim EJ, Besse JA, et al. MiR-223 deficiency increases eosinophil progenitor proliferation. J Immunol. 2013;190(4):1576–1582. doi: 10.4049/jimmunol.1202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner LA, Christensen CJ, Dunn DM, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109(12):5191–5198. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wills-Karp M, Karp CL. Biomedicine. Eosinophils in asthma: remodeling a tangled tale. Science. 2004;305(5691):1726–1729. doi: 10.1126/science.1104134. [DOI] [PubMed] [Google Scholar]
- 47.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures