Coordination of adjacent domains mediates TACC3–ch-TOG–clathrin assembly and mitotic spindle binding (original) (raw)
Aurora A phosphorylation-induced interaction of TACC3 and clathrin coordinates adjacent domains in each protein to create a microtubule-binding interface, whereas a distinct site in TACC3 recruits ch-TOG to mitotic spindles.
Abstract
Acomplex of transforming acidic coiled-coil protein 3 (TACC3), colonic and hepatic tumor overexpressed gene (ch-TOG), and clathrin has been implicated in mitotic spindle assembly and in the stabilization of kinetochore fibers by cross-linking microtubules. It is unclear how this complex binds microtubules and how the proteins in the complex interact with one another. TACC3 and clathrin have each been proposed to be the spindle recruitment factor. We have mapped the interactions within the complex and show that TACC3 and clathrin were interdependent for spindle recruitment, having to interact in order for either to be recruited to the spindle. The N-terminal domain of clathrin and the TACC domain of TACC3 in tandem made a microtubule interaction surface, coordinated by TACC3–clathrin binding. A dileucine motif and Aurora A–phosphorylated serine 558 on TACC3 bound to the “ankle” of clathrin. The other interaction within the complex involved a stutter in the TACC3 coiled-coil and a proposed novel sixth TOG domain in ch-TOG, which was required for microtubule localization of ch-TOG but not TACC3–clathrin.
Introduction
The spindle apparatus is a dynamic array of microtubules (MTs) that is responsible for the alignment and segregation of duplicated chromosomes during mitosis. Numerous MT-binding proteins, including motor and nonmotor proteins, control spindle assembly and organization (Manning and Compton, 2008). Many of these proteins are under the control of mitotic kinases (Barr and Gergely, 2007). One such complex of nonmotor proteins consists of transforming acidic coiled-coil protein 3 (TACC3), colonic and hepatic tumor overexpressed gene (ch-TOG), and clathrin (Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010; Booth et al., 2011). TACC3–ch-TOG–clathrin is important for stabilizing MTs in the kinetochore fibers of the mitotic spindle by forming inter-MT bridges that cross-link adjacent MTs (Booth et al., 2011). Additionally, this complex may also stabilize the MTs via the MT polymerization activity of ch-TOG (Charrasse et al., 1998; Brouhard et al., 2008). TACC3 is a substrate of Aurora A and this activity is crucial for TACC3–clathrin interactions and spindle localization (Kinoshita et al., 2005; LeRoy et al., 2007; Lin et al., 2010; Fu et al., 2010; Hubner et al., 2010; Booth et al., 2011; Cheeseman et al., 2011). Understanding how the members of the complex bind one another and how the complex interacts with MTs could facilitate the design of targeted inhibitors to disrupt the function of the complex. Such inhibitors could have potential clinical implications because Aurora A, TACC3, and ch-TOG are dysregulated in several cancers (Charrasse et al., 1998; Barr and Gergely, 2007; Peset and Vernos, 2008; Singh et al., 2012) and TACC3 in particular is an attractive target for chemotherapy (Yao et al., 2012).
TACC3, ch-TOG, and clathrin are all found at the mitotic spindle, although the patterns of distribution differ slightly: ch-TOG is more pronounced at centrosomes, whereas TACC3 is highly enriched on spindle MTs, and clathrin is found on spindle MTs but also in the cytoplasm (Gergely et al., 2000a; Gergely et al., 2003; Royle et al., 2005; Foraker et al., 2012). Ch-TOG is important for spindle assembly and MT stabilization during mitosis, whereas clathrin and TACC3 are important for MT stability (Gergely et al., 2003; Cassimeris and Morabito, 2004; Royle et al., 2005). Ch-TOG has distinct mitotic functions at centrosomes and MTs, whereas in human cells TACC3 and clathrin appear to function primarily at spindle MTs as part of the TACC3–ch-TOG–clathrin complex (Holmfeldt et al., 2004; Barr and Gergely, 2008; Cassimeris et al., 2009; Lin et al., 2010; Booth et al., 2011). In some other species there are also critical functions for TACC proteins at centrosomes, which could perhaps be fulfilled by TACC1 and TACC2 in mammals (Gergely et al., 2000b; Peset et al., 2005).
Both TACC3 and clathrin are needed for MT localization of ch-TOG, likely mediated by a predicted direct interaction between ch-TOG and the conserved coiled-coil TACC domain at the C-terminal end of TACC3 (Lee et al., 2001; Conte et al., 2003). Intriguingly, TACC3 and clathrin were each reported to be essential for recruitment of the other to the spindle in different studies (Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010; Booth et al., 2011). TACC3 and clathrin interact directly once TACC3 is phosphorylated at serine 558 (S558) by Aurora A (Fu et al., 2010; Lin et al., 2010). This phosphorylated motif is thought to contact the “ankle” of the clathrin heavy chain (CHC; Lin et al., 2010). TACC3 phosphorylation is known to be essential for TACC3 recruitment to the spindle, and experiments involving Aurora A inhibition indicated that TACC3 phosphorylation also regulates clathrin recruitment (Kinoshita et al., 2005; LeRoy et al., 2007; Booth et al., 2011). This implies that phosphorylation influences localization via regulation of interaction with clathrin. Despite the need for TACC3 and clathrin for complex localization, neither protein interacts with MTs in vitro, suggesting that spindle recruitment of the TACC3–ch-TOG–clathrin complex may be more complicated than serial recruitment and anchoring via a single MT-binding subunit (Lin et al., 2010; Booth et al., 2011; Royle, 2012). Consistent with this, additional regions apart from the CHC ankle and TACC3 pS558 contribute to mitotic localization in cells (Gergely et al., 2000a; Royle et al., 2005).
In summary, it is unclear (a) how TACC3, ch-TOG, and clathrin interact with one another, (b) which members of the complex interact with MTs, and (c) how this is regulated by Aurora A. To answer these questions, we began by defining the regions of TACC3 and clathrin that are required for spindle recruitment. We identify a novel mode of MT binding in which domains in TACC3 and clathrin create a composite binding interface when coordinated in tandem. This coordination is regulated by Aurora A phosphorylation of TACC3, which triggers interaction of TACC3 with clathrin via regions of each protein that are distinct from their MT interaction surface. We also map the TACC3–ch-TOG interaction, propose an additional TOG domain, and show that ch-TOG is dispensable for MT localization of TACC3–clathrin.
Results
Two separate regions of TACC3 are required for spindle binding
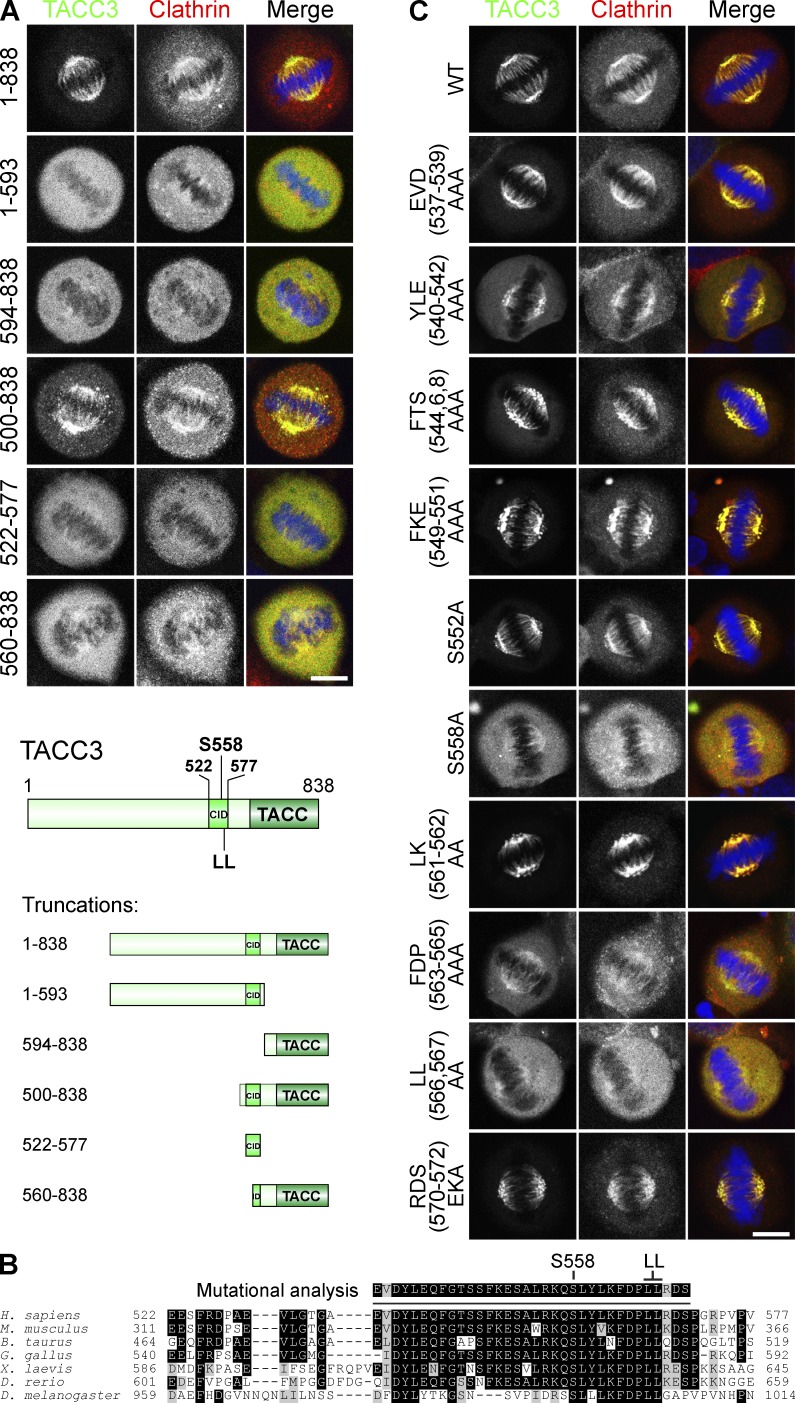
To investigate which regions of TACC3 are required for spindle binding, we examined the localization of GFP-tagged TACC3 truncation proteins in cells. These RNAi-resistant proteins were expressed in TACC3-depleted cells together with mCherry-tagged clathrin light chain a (mCherry-LCa) to visualize clathrin. Removal of the TACC domain (GFP-TACC3[1–593]) prevented spindle binding (Fig. 1 A), confirming the importance of the TACC domain for TACC3 localization (Peset et al., 2005). However, other regions of TACC3 were also required, as the TACC domain alone (GFP-TACC3[594–838]) was only weakly recruited. GFP-TACC3(500–838) was recruited as efficiently as full-length TACC3, indicating that this fragment contains all necessary residues for spindle binding and that residues 1–499 are not required (Fig. 1 A). This fragment contains the region aa 522–577, which was shown previously to bind CHC (Lin et al., 2010). Like the TACC domain, this clathrin interaction domain (CID) was necessary but not sufficient for spindle binding as GFP-TACC3(522–577) was not localized to the spindle. A truncation that disrupted the CID also prevented recruitment (GFP-TACC3[560–838]; Fig. 1 A). Spindle localization of clathrin matched that of TACC3 constructs, highlighting the interdependence of spindle recruitment of these two proteins. In summary, the first 499 residues are dispensable for localization of TACC3, but the TACC domain and CID are each necessary but not sufficient for spindle binding.
Figure 1.
Two regions in TACC3 are needed for spindle recruitment: the TACC domain and the CID comprising a dileucine motif and pS558. (A) Representative confocal micrographs of GFP-TACC3 constructs (green) coexpressed in TACC3-depleted HeLa cells with mCherry-LCa (red) and DNA (blue). (bottom) Schematic of truncation constructs. (B) Alignment of the CID from TACC3 homologues to show sequence conservation. (C) Representative confocal micrographs as in A, for TACC3 point mutants. Bars, 10 µm.
A dileucine motif in TACC3 is essential for spindle localization
To understand which residues within the CID are required for spindle localization, we made a series of GFP-TACC3 mutants and coexpressed them with mCherry-LCa. We targeted conserved residues (Fig. 1 B) and included S558A, which has reduced spindle localization, as a reference (Booth et al., 2011). Several TACC3 mutants had reduced recruitment relative to wild type (Fig. 1 C). The most striking was mutation of a dileucine motif (LL[566,567]AA), which strongly disrupted localization of TACC3, similar to S558A (Fig. 1 C). Mutation of residues immediately upstream of this dileucine motif (FDP[563–565]AAA) also perturbed TACC3 localization (Fig. 1 C). We also noted a subtle change in localization with mutation of YLE(540–542)AAA. S552 in addition to S558 is a known substrate of Aurora A (Fu et al., 2010), yet mutation S552A had little effect on localization. Again, the spindle localization of clathrin matched that of TACC3 in all cases. These results show that the CID of TACC3 contains a novel dileucine motif (L566,567) close to S558. Because the CID is known to interact with CHC in an Aurora A–dependent manner it seemed likely that these residues form part of the interface with CHC.
The TACC3 dileucine motif functions independently of phosphorylation
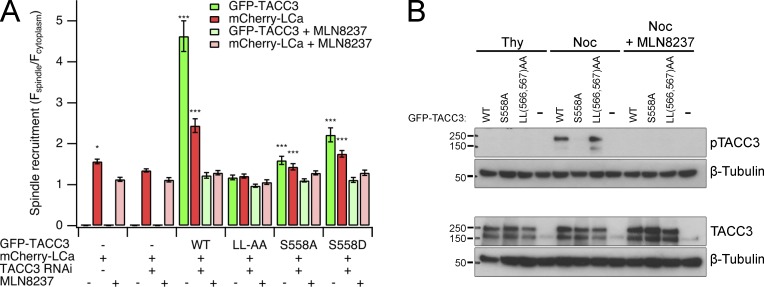
To determine the relative contributions to spindle localization of TACC3 by S558 and the dileucine motif within the CID, we again coexpressed TACC3 mutants with mCherry-LCa. MLN8237 (0.3 µM) was used to inhibit Aurora A activity as a reference for minimal recruitment (Booth et al., 2011). LL(566,567)AA was poorly recruited, with lower enrichment at spindles than S558A, underlining the importance of the dileucine motif (Fig. 2, A and B). The phosphomimetic S558D showed good spindle recruitment, albeit less than wild type. Inhibition of Aurora A completely disrupted spindle localization for all TACC3 constructs including S558 mutants. This argues that phosphorylation of another site on TACC3 by Aurora A occurs, at least when S558 is mutated, and has a minor role in localization. Again, spindle localization of clathrin always mirrored that of TACC3.
Figure 2.
The dileucine motif in CID mediates TACC3 recruitment independently of S558 phosphorylation. (A) Quantification of spindle recruitment of GFP-TACC3 mutants and mCherry-LCa, coexpressed in TACC3-depleted HEK293 cells. Values for mCherry-LCa alone in the presence or absence of endogenous TACC3 are included. Treatment with MLN8237 (0.3 µM) gave a minimal recruitment value for each construct. Depletion of TACC3 in HEK293 cells is ∼75% effective (Booth et al., 2011). Graph shows mean ± SEM spindle recruitment from at least 17 cells over three separate experiments. *, P < 0.05; ***, P < 0.001; one-way analysis of variance with Tukey’s post-hoc test compared with LL(566,567)AA + MLN8237. (B) Phosphorylation of GFP-TACC3 constructs expressed in TACC3-depleted HEK293 cells in S phase (Thy, thymidine) or mitosis (Noc, nocodazole) or in mitosis with Aurora A inhibition using MLN8237 (0.3 µM).
We tested whether mutation of the dileucine motif merely disrupts phosphorylation of the nearby S558 by immunoblotting cell lysates expressing GFP-TACC3[LL(566,567)AA] with a pS558 phospho-specific antibody (Fig. 2 B). This confirmed that S558 was phosphorylated, indicating that the dileucine motif contributes to recruitment independently of pS558 and that phosphorylation of S558 by itself is not sufficient for recruitment.
TACC3 phosphorylation and dileucine motif are required for interaction with clathrin
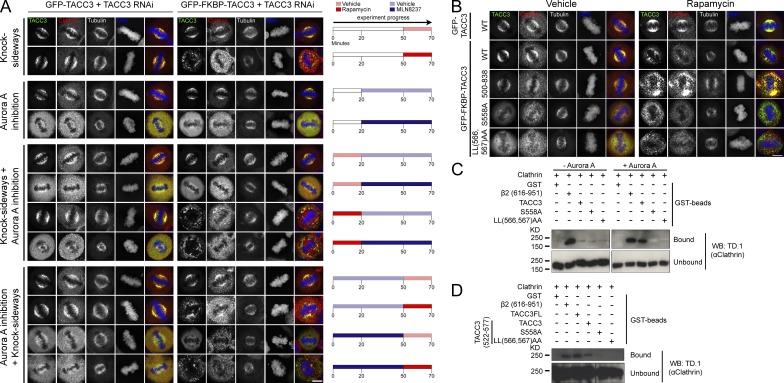
Are pS558 and the dileucine motif both part of a clathrin interaction site? To begin to address this question in cells, we used a method for inducible rerouting of expressed proteins to mitochondria (knocksideways [KS]) to examine interactions between clathrin and TACC3 mutants (Robinson et al., 2010; Cheeseman et al., 2013). In this approach, a GFP-TACC3 construct with an FKBP tag is coexpressed with a mitochondrially targeted protein containing an FRB tag (MitoTrap) in TACC3-depleted cells. Addition of rapamycin induces heterodimerization of the FKBP and FRB, meaning that FKBP-tagged proteins become stably rerouted and trapped at the mitochondria. In this way, we can test in cells whether or not TACC3 constructs and clathrin interact, as the corerouting of clathrin with TACC3 indicates a tight interaction (Cheeseman et al., 2013).
When MLN8237 treatment was performed before TACC3 KS, there was a reduction in CHC corecruitment compared with controls. Interestingly, treatment with MLN8237 after TACC3 KS also reduced clathrin enrichment at mitochondria (Fig. 3 A), suggesting that Aurora A activity is not completely restricted to the spindle poles. These results indicate that phosphorylation is required to maintain the TACC3–clathrin interaction in cells. Furthermore, corecruitment of endogenous clathrin to mitochondria upon KS was defective with both LL(566,567)AA and S558A TACC3 mutants compared with wild type (Fig. 3 B). This supports the idea that spindle recruitment of these mutants is defective because of reduced interaction with clathrin in cells.
Figure 3.
The dileucine motif and pS558 in the CID of TACC3 are important for interaction with CHC. (A and B) KS experiments. Representative confocal micrographs of TACC3-depleted HeLa cells coexpressing PAGFP-MitoTrap with either GFP-TACC3 (control) or GFP-FKBP-TACC3 (wild type or mutant). Cells were fixed and costained for CHC (red) and DNA (blue). (A) Inhibition of Aurora A using MLN8237 (0.7 µM) disrupts colocalization of CHC with wild-type TACC3 at mitochondria whether inhibited either before or after rapamycin addition. DMSO is vehicle control for MLN8237 and ethanol is vehicle control for rapamycin. (B) CHC is colocalized with TACC3 on mitochondria upon KS of GFP-FKBP-TACC3 but not S558A or LL(566,567)AA mutants. Bars, 10 µm. (C) GST-TACC3-His6 or β2 adaptin (616–951) control were phosphorylated with Aurora A/TPX2 (or BSA control) before incubation with purified clathrin triskelia and pull-down on glutathione beads. Coprecipitated clathrin was detected by TD.1 antibody imunoblotting. (D) GST-TACC3(522–577)-His6 was phosphorylated with Aurora A/TPX2 before incubation with purified clathrin triskelia and pull-down on glutathione beads. GST-TACC3-His6 (TACC3FL) and β2 adaptin (616–951) were included as controls. Coprecipitated clathrin was detected by TD.1 antibody.
To confirm these findings we studied direct interactions of purified proteins in vitro. GST-TACC3-His6 was phosphorylated with Aurora A, incubated with purified clathrin triskelia, and then affinity purified on glutathione beads. The appendage+hinge fragment of the β2 subunit of AP-2 was used as a positive control for clathrin binding (Edeling et al., 2006). Wild-type TACC3 coprecipitated clathrin, only after phosphorylation, but this binding was greatly reduced by either S558A or LL(566,567)AA mutations (Fig. 3 C). Thus, both pS558 and LL(566,567) are necessary but not sufficient for interaction with CHC. These findings support our conclusions from cell experiments. We obtained the same results using just the CID of TACC3 (GST-TACC3[522–577]-His6) to coprecipitate clathrin (Fig. 3 D). These experiments therefore revealed that the TACC domain is not needed for interaction with clathrin. Together the results point to a model where the CID of TACC3 is required exclusively for clathrin interaction and the TACC domain is required for MT binding.
Two separate regions of CHC are required for spindle binding
Having defined residues in TACC3 that are important for clathrin interaction and spindle recruitment, we turned our attention to clathrin. We previously found that the CHC N-terminal domain (NTD) and ankle are each essential for recruitment to the spindle (Royle and Lagnado, 2006; Hood and Royle, 2009). To identify the residues involved, we expressed GFP-tagged CHC mutants in CHC-depleted cells and examined their spindle recruitment (Fig. 4, A and B; and Fig. S1) and mitotic function (Fig. 4 C).
Figure 4.
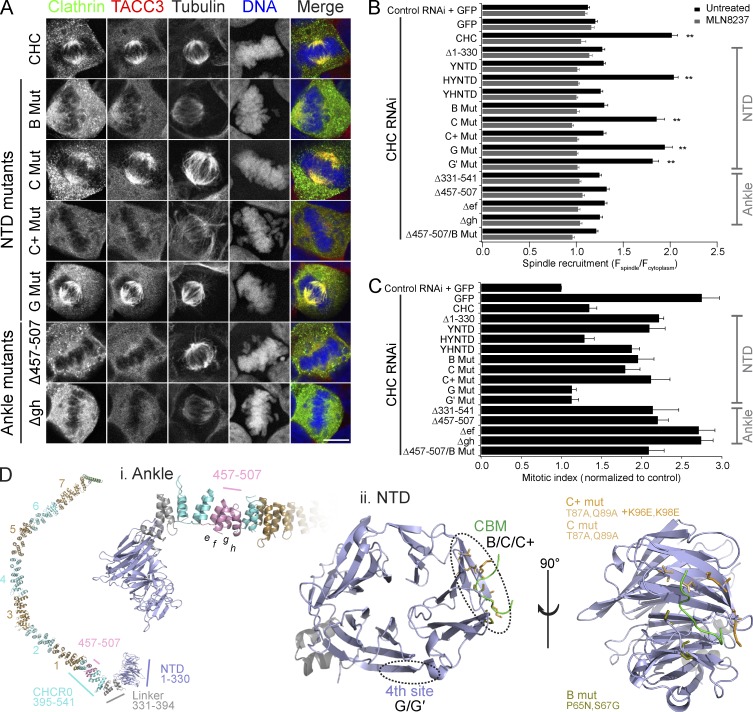
Two sites on clathrin are required for spindle localization and function. (A) Representative confocal micrographs of colocalization of GFP-CHC constructs with endogenous TACC3 in CHC-depleted HEK293 cells. A selection of mutants is shown; all mutants are shown in Fig. S1. Bar, 10 µm. (B) Quantification of GFP-CHC mutants at the mitotic spindle relative to the cytoplasm in CHC-depleted HEK293 cells. MLN8237 (0.3 µM) treatment gave a minimal recruitment value for each construct. Values are mean ± SEM calculated from Nuntreated = 55–90 and NMLN8237 = 30–52 cells from three experiments for each construct over a total of five experiments. **, P < 0.01 from one way analysis of variance using Dunnett’s post-hoc test compared with Δ1–330. (C) Rescue of increased mitotic index in CHC-depleted HEK293 cells by GFP-CHC mutants. Nexperiment = 3–7 and Ncell = 2185–9728. (D) Structure of a CHC leg. The locations of key mutations in the ankle region and in the NTD (1–330) are shown. NTD is shown with a CBM ligand (green) in the groove between blades 1 and 2.
A chimera in which the NTD of CHC was replaced with the NTD of the yeast CHC homologue, chc1p (YNTD), was not recruited to the mitotic spindle. This suggested that NTD residues that determine recruitment differ in chc1p. Further chimeras demonstrated that these are located within the first 100 aa of CHC (YHNTD and HYNTD; Fig. 4 B, Fig. S1, and Table S1).
The NTD contains four known sites for interactions, two of which are in the first 100 residues: the clathrin box motif (CBM)–binding site in a groove between blades 1 and 2 of the β-propeller and part of the “fourth” site between blades 6 and 7 (Fig. 4 D; Willox and Royle, 2012). We tested whether these sites mediate spindle recruitment using mutants in which they were disrupted. The fourth site was excluded because neither mutant G (E11K) nor mutant G′ (Q14D, Q16M, and N17S) affected localization (Fig. 4, A and B; and Fig. S1). The CBM-binding site mutant C (T87A and Q89A) did not significantly disrupt localization; however, mutation of additional residues within the same site in mutant C+ (T87A, Q89A, K96E, and K98E) was sufficient to prevent spindle enrichment (Fig. 4, A, B, and D; and Fig. S1). Mutant B (P65N and S67G), in which CHC residues are switched to chc1p counterparts, also disrupted recruitment (Fig. 4, B and D; and Fig. S1). Together these findings strongly suggest that an interaction at the CBM-binding site in CHC is required for spindle recruitment, thus explaining the role of the NTD.
The other critical region for spindle recruitment is in the ankle of CHC, between aa 457 and 507 (Fig. 4 D; Hood and Royle, 2009). To identify important residues here we expressed deletion constructs in clathrin-depleted cells. This region consists of the final four α helices (termed e_–_h) of CHC repeat 0 (CHCR0; Fotin et al., 2004). We deleted each pair of helices in turn (Δ_ef_ and Δ_gh_ mutants) and found that removal of either pair disrupts spindle enrichment as completely as Δ457–507 (Fig. 4, A and B; and Fig. S1). In keeping with a previous study (Lin et al., 2010), we found that deletion of a larger segment containing aa 457–507 (Δ331–541) also significantly disrupted clathrin localization (Fig. 4 B and Fig. S1). Disruption of the ankle site or the NTD each caused substantial loss of spindle localization. A combined mutant (Δ457–507 + mutation B) caused no additional loss of spindle localization (Fig. 4 B and Fig. S1). These experiments indicate that two separate regions on CHC are necessary for spindle localization and that disruption of either completely abrogates recruitment.
TACC3 spindle localization mirrored that of clathrin, again highlighting the interdependence of TACC3–clathrin recruitment (Fig. 4 A). As expected, all CHC mutations that caused mislocalization failed to rescue mitotic index (Fig. 4 C), confirming that coupled TACC3–clathrin spindle recruitment is essential for mitotic function.
The ankle but not the NTD of clathrin mediates interaction with TACC3
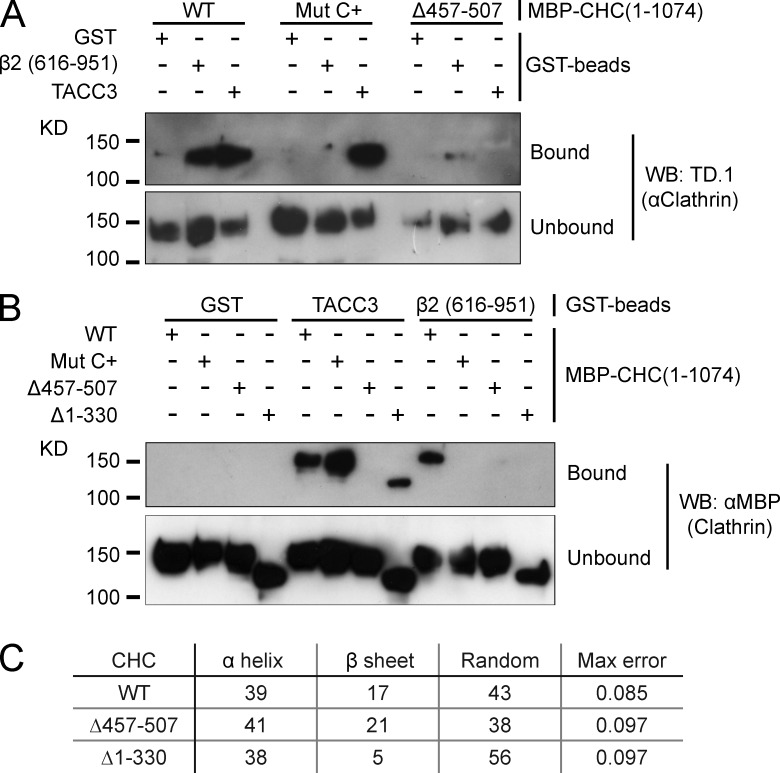
How is this coupling achieved? We tested whether the NTD and/or ankle of clathrin are required for binding TACC3. To do this, we again used the in vitro binding experiment, with a large recombinant CHC fragment (MBP-CHC[1–1074]-His6) rather than purified triskelia. First, we found that deletion of aa 457–507 virtually abolished coprecipitation of MBP-CHC(1–1074)-His6 with GST-TACC3-His6, confirming the importance of the ankle for interaction with TACC3 (Fig. 5 A). Second, the “C+” mutation that disrupted TACC3–clathrin localization in cells did not affect coprecipitation with GST-TACC3-His6, excluding the CBM-binding site from a role in TACC3 interaction (Fig. 5 A). In fact, deletion of the entire NTD had no effect on TACC3 interaction, demonstrating that the essential role of the CHC NTD for spindle localization is not via interaction with TACC3 (Fig. 5 B).
Figure 5.
The ankle site but not the NTD of clathrin binds TACC3. (A and B) GST-TACC3-His6 or β2 adaptin (616–951) control were phosphorylated with Aurora A/TPX2 before incubation with MBP-CHC(1–1074)-His6 (either wild type or mutants as indicated) and pull-down on glutathione beads. Coprecipitated clathrin fragments were detected by TD.1 antibody (A) or anti-MBP (B) immunoblotting. (C) Summary of protein composition determined by CD spectroscopy of MBP-CHC(1–1074)-His6 constructs. Protein secondary structure was analyzed from CD spectra using DichroWeb server (Whitmore and Wallace, 2008). Results are discarded if maximum error is >0.227. Average traces from a single experiment are shown in Fig. S2.
The lack of involvement of the NTD in TACC3–clathrin interactions was surprising to us as the TACC3 dileucine motif conforms to a potential CBM (LLRDS[566–570]). The lack of TACC3 interaction with the Δ457–507 clathrin fragment is in agreement with previous findings (Lin et al., 2010). However, we wanted to verify that this fragment was folded normally because this deletion had also interfered with the clathrin–β2 interaction (Fig. 5, A and B), which was unexpected. Circular dichroism (CD) spectroscopy showed that Δ457–507 did not significantly interfere with protein structure (Fig. 5 C and Fig. S2).
TACC3 binds via a positive patch and hydrophobic pocket on the ankle of CHC
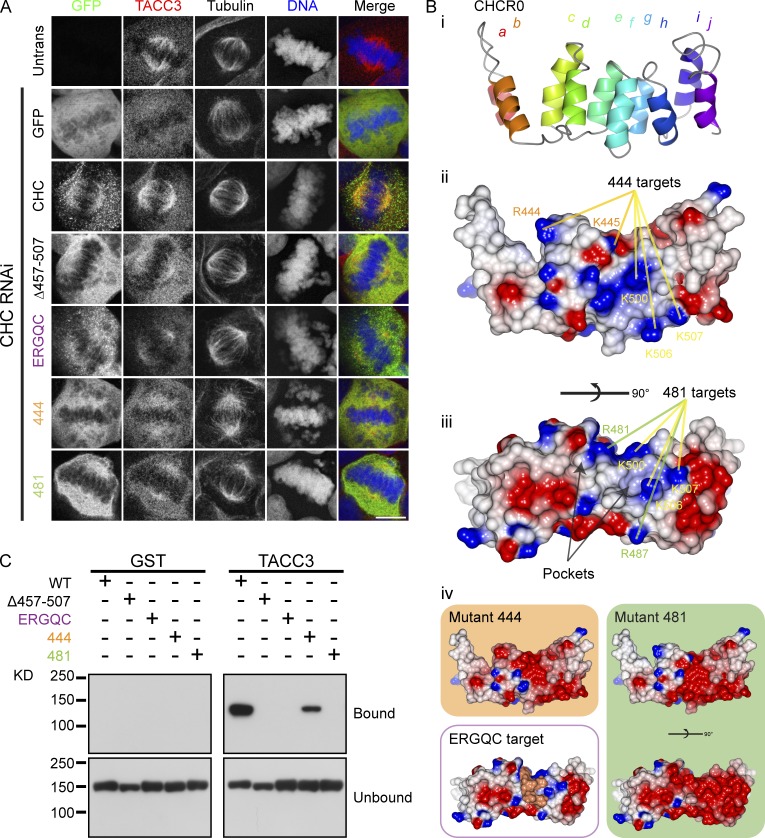
We next investigated the CID–ankle interaction in more detail. To understand how pS558 and the dileucine motif may bind to the ankle, another series of mutant GFP-CHC constructs were expressed in clathrin-depleted cells and their localization was examined together with that of endogenous TACC3 (Fig. 6 A). A model of CHCR0 was used to direct the mutation of positive charges on the ankle surface to which pS558 may bind (Fig. 6 B). Mutation of five arginine and lysine residues to glutamic acid (Table S1, mutants 444 and 481) was sufficient to disrupt spindle localization of clathrin and TACC3 (Fig. 6 A).
Figure 6.
TACC3 binds clathrin via a positive patch and hydrophobic pocket on the ankle of CHC. (A) Representative confocal micrographs showing the colocalization of GFP-CHC with endogenous TACC3 in CHC-depleted HEK293 cells. Bar, 10 µm. (B) Structural model of CHCR0. (i) Helices a_–_j are shown. (ii) Same view of CHCR0 with surfaces colored for electrostatic potential (positive [+10 kT e−1; blue] to negative [−10 kT e−1; red]) using CCP4mg (McNicholas et al., 2011). (iii) Rotated along the ankle axis by 90° to show two pockets bordered by the loop between helices f and g. Location of positively charged residues that are mutated in 444 and 481 are shown in ii and iii. Common residues are indicated in yellow, 444-specific residues are in orange and 481-specific residues are in green. (iv) Predicted change in surface potential in the model induced by 444 or 481 mutations and a view to show the targets of the LRANV sequence mutated in the ERGQC mutant. (C) GST or GST-TACC3-His6 was phosphorylated with Aurora A/TPX2 before incubation with MBP-CHC(1–1074)-His6 (either wild type or mutants as indicated) and pull-down on glutathione beads. Coprecipitated clathrin fragments were detected by anti-MBP antibody. Details of mutations are given in Table S1.
The model also revealed two pockets on the upper surface of CHCR0 facing the NTD that were potential sites for binding the TACC3 dileucine motif (Fig. 6 B). Both of these pockets would have been disrupted in the Δ_ef_ and Δ_gh_ mutants described above (Fig. 4). We therefore tested whether the loop between CHCR0 helix g and h was required for localization (Fig. 6 B). When this loop was mutated to the corresponding sequence in CHCR3, the most similar CHCR to CHCR0 (Table S1, mutant ERGQC), we found that clathrin and TACC3 were no longer localized to the spindle (Fig. 6 A). To test whether these mutations in clathrin interfered with TACC3 binding, we used the in vitro binding assay described above. These experiments showed that disruption of the positive charges or the hydrophobic pockets prevented TACC3–clathrin binding (Fig. 6 C). The effect of mutation 444 was weaker than that of mutation 481 despite both mutants being mislocalized in cells.
Our results so far demonstrate that two parts of CHC (NTD and ankle) and two parts of TACC3 (CID and TACC domain) are needed for TACC3–clathrin to localize to the mitotic spindle. The CHC ankle and TACC3 CID regions do this by mediating the TACC3–clathrin interaction, but the TACC domain and NTD must contribute to spindle recruitment in a different way.
NTD-TACC tandem domain coordination permits association of TACC3–clathrin with MTs
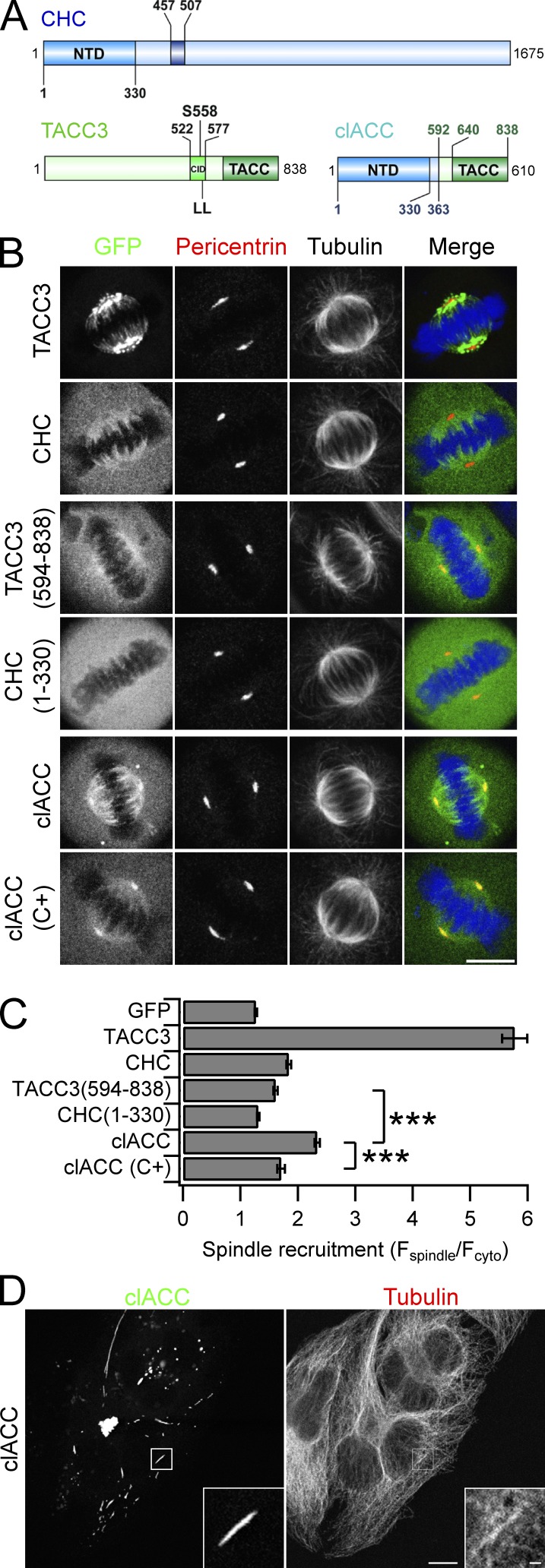
One possibility is that the NTD and TACC domain together form a composite MT interaction surface, with these two domains being coordinated in tandem by the interaction between CID and ankle. We reasoned that if this were the case, we could reconstitute activity by fusing together the NTD and TACC domain as a single protein, thus bypassing the CID–ankle interaction. We generated a GFP-tagged construct in which CHC(1–363) was fused to aa 592–838 of TACC3 (Fig. 7 A). This portmanteau protein was termed clACC. We found that GFP-clACC was enriched on the mitotic spindle and centrosomes compared with TACC domain alone or to CHC(1–330) (Fig. 7, B and C). Introduction of C+ mutation into GFP-clACC caused its mislocalization (Fig. 7, B and C). Therefore, combination of the NTD and TACC domains in a single fusion recapitulated the localization of TACC3–clathrin in cells.
Figure 7.
Coordination of NTD and TACC domains in tandem allows binding to MTs. (A) Schematic diagram of clACC to show tandem NTD-TACC domains. (B) Representative confocal micrographs of GFP-clACC, GFP-CHC, or GFP-TACC3 expressed in HeLa cells. Cells were fixed and stained for pericentrin (red), tubulin, and DNA (blue). (C) Bar graph of spindle recruitment for each construct. Values are mean ± SEM calculated from Ncell = 52–81 over three experiments. ***, P < 0.001 from a Kruskal-Wallis test with Dunn’s multiple comparisons test. (D) Representative confocal micrographs of GFP-clACC in interphase HeLa cells stained for tubulin. Bars, 10 µm.
The clACC fusion also bypassed Aurora A regulation of the CID–ankle interaction. Inhibition of Aurora A activity using MLN8237 (0.3 µM) caused the loss of endogenous TACC3 from spindles; however, in the same cells, clACC remained bound to the spindle (Fig. S3 A). This experiment also showed that spindle binding by clACC did not require endogenous TACC3 and was likely direct.
To test if the coordination of NTD-TACC domains in tandem created a MT-binding surface, we examined the localization of GFP-clACC in interphase cells. We found that clACC was enriched at a subset of MT bundles in some interphase cells, suggesting that the NTD-TACC domains in tandem can interact directly with MTs, and in some cases, bundle them (Fig. 7 D). However, clACC clearly did not decorate all MTs in interphase cells indicating some specificity of targeting. Depolymerization of MTs using nocodazole (10 µM, 1 h) resulted in the loss of the clACC-MT bundles (Fig. S3 B). However, imaging bundles undergoing depolymerization at shorter time points revealed some degree of protection of the bundles possibly conferred by clACCs (Fig. S3 C).
Reconstitution of MT binding and bundling in vitro using purified components
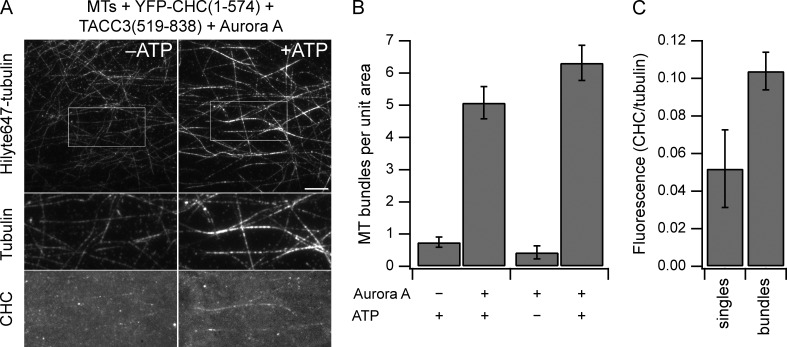
We next wanted to assess whether TACC3 and clathrin together bind MTs. Our first attempts at clACC cosedimentation with MTs were unsuccessful because of the instability of recombinant clACC protein. Instead, we used total internal reflection fluorescence microscopy to visualize binding of proteins to MTs. YFP-CHC(1–574) and mCherry-TACC3(519–838) were incubated together with Aurora A and ATP to form a subcomplex. Adding this mixture to preformed MTs caused single MTs to bundle together (Fig. 8 A). This bundling was not seen when either Aurora A or ATP were excluded from the mixture (Fig. 8 B). The bundles were weakly decorated with YFP-CHC(1–574), indicating that these proteins could bind directly to the MTs (Fig. 8 A). Quantification of YFP-CHC(1–574) fluorescence at bundles versus single MTs showed a twofold preference for binding MT bundles and ruled out the possibility that the binding observed was a function of the MT density (Fig. 8 C). These observations support the idea that (a) clathrin and TACC3 cannot individually bind MTs, (b) they must be phosphorylated by Aurora A to bind, and (c) together they form a composite surface for direct interaction with MTs. Moreover, they suggest that the TACC3–CHC fragment pair favors binding to MT bundles and promotes MT bundling, which is in keeping with its proposed role in inter-MT bridging (Booth et al., 2011). Note that MT binding is much weaker than that observed for established MAPs, such as MAP4 (Hawkins et al., 2013). This may be because of our use of TACC3 and CHC fragments and it is possible that this assay can be further optimized.
Figure 8.
Direct MT binding and bundling by TACC3–clathrin subcomplex. (A) MT bundles (labeled by Hilyte647-tubulin) formed after flow-in of 1 µM YFP-CHC(1–574), 1 µM mCherry-TACC3(519–838), and 25 nM Aurora A in the presence of 1 mM ATP (right), but not in the absence of ATP (left). Bar, 10 µm. Boxed region is expanded (2×) below to show localization of YFP-CHC(1–574). Images are maximum projections of 100 images acquired at 2 fps, followed by background subtraction. (B) Quantification of MT bundling efficiency as the number of MT bundles of >10-µm length per field of view. n = 13–20. Bars show mean ± SEM. (C) Quantification of MT binding by the TACC3–clathrin subcomplex. The ratio of YFP-CHC(1–574) intensity versus tubulin intensity is shown for MT bundles and individual MTs. Bundles contained 3.5 MTs on average. n = 50 from 10 fields. Bars show mean ± SEM.
MT localization of TACC3–ch-TOG–clathrin is not via ch-TOG
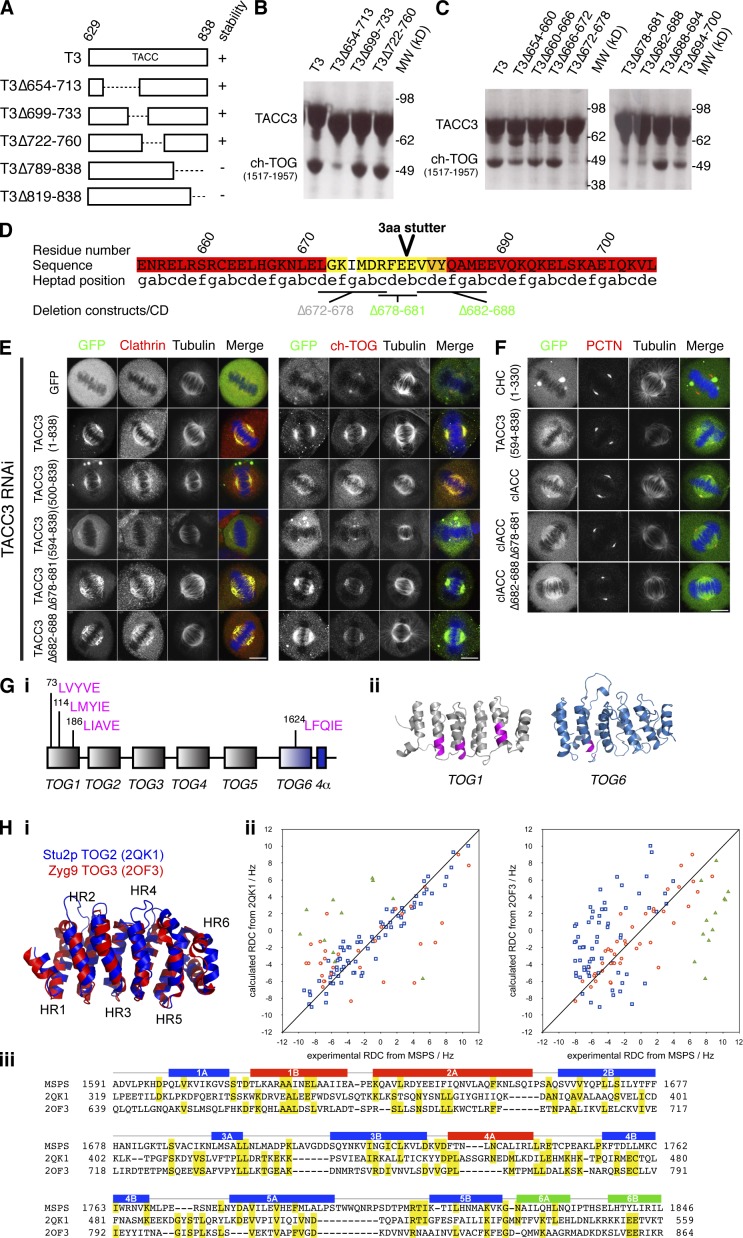
What is the role of ch-TOG in the TACC3–ch-TOG–clathrin complex? Our results so far indicated that ch-TOG is not involved in MT binding at the spindle and ch-TOG depletion was shown previously to have only a modest effect on TACC3 spindle localization (Gergely et al., 2003; Booth et al., 2011). However, because ch-TOG may have affinity for the MT lattice and its TOG domains may bind tubulin dimers (Widlund et al., 2011), it was important to investigate how ch-TOG is integrated into the TACC3–ch-TOG–clathrin complex. We generated a series of recombinant TACC3 TACC domain (residues 629–838) deletion mutants and tested for binding to a C-terminal ch-TOG fragment (1517–1957) that is predicted to interact with TACC3. The first set of deletions identified a region between 654 and 713 that was required for binding (Fig. 9, A and B). Within this region, three deletions individually disrupted the interaction with ch-TOG C-terminal domain (Fig. 9 C). These three deletions mapped to a predicted 3-aa stutter in the coiled-coil region of the TACC domain. We assessed whether the deletions had disrupted folding of the protein by CD spectroscopy and found two mutants (Δ678–681 and Δ682–688) had normal structure, whereas one (Δ672–678) was disrupted (Fig. 9 D and Fig. S4). Using this information, we generated the same deletions in full-length GFP-TACC3 and expressed them in TACC3-depleted cells to examine the localization of the mutants together with clathrin and ch-TOG (Fig. 9 E). Neither deletion affected the spindle localization of TACC3 or clathrin. We found that the amount of ch-TOG on the spindle MTs was reduced compared with wild type, but was not as low as depletion of TACC3 (Fig. 9 E). Note that centrosomal recruitment of ch-TOG was unaffected. We also generated the same deletions in GFP-clACC and found that localization to spindle MTs was unaffected (Fig. 9 F). Interestingly, wild-type clACC often localized to centrosomes similarly to ch-TOG and this was disrupted by either deletion. This observation argues that both deletions disrupt TACC–ch-TOG interactions in cells. Moreover, this centrosomal enrichment of clACC, which is not seen with full-length or endogenous TACC3, must be caused by deregulated binding of ch-TOG. These results argue that the interaction between TACC3 and ch-TOG is not required for spindle localization of TACC3 and clathrin.
Figure 9.
The interaction between TACC3 and ch-TOG is not required for MT localization of TACC3–ch-TOG–clathrin. (A) Schematic diagram of TACC3 TACC domain truncation proteins and their stability. (B and C) ch-TOG(1517–1957) and His-NusA–tagged TACC3 deletion proteins pulled-down on Ni resin. Stable deletions from A are tested in B. Deletions 654–700 are tested in C. (D) Annotated sequence of TACC3 in the vicinity of the deletions that abrogate ch-TOG binding. The position of a predicted 3-aa stutter is marked (Lupas et al., 1991). The sequence is colored by the probability of coiled coil (pcc), from red (>0.75) through orange (0.50–0.75) and yellow (0.25–0.50) to white (<0.25). Marked deletions are colored by their structural integrity (Fig. S4): gray, misfolded; green, folded. (E) Representative confocal micrographs of TACC3-depleted HeLa cells expressing GFP or GFP-TACC3 constructs. Cells were fixed and stained for tubulin, DNA (blue), and clathrin (red; left) or for GFP (green), tubulin, DNA (blue), and ch-TOG (red; right). Note, methanol fixation decreased GFP fluorescence and required staining. Bars, 10 µm. (F) Representative confocal micrographs of HeLa cells expressing GFP-tagged TACC domain or clACC (wild type or deletion mutants). Cells were fixed and stained for pericentrin (PCTN; red), tubulin, and DNA (blue). Bar, 10 µm. (G, i) Putative clathrin-binding motifs in ch-TOG are located in TOG1 and the potential TOG6 domain. (ii) These motifs (magenta) are mapped onto models of TOG1 and TOG6. (H, i) Superposition of the two TOG domain models used to evaluate the Msps TOG6 RDC data. (ii) Plots of experimental versus calculated RDCs for each helical residue in Msps TOG6 region versus the two TOG domain crystal structures. Red circles show values for helices 1B, 2A, and 4A; blue boxes show values for helices 1A, 2B, 3A, 3B, 4B, 5A, and 5B; and green triangles show values for helices 6A and 6B. The RDC experiment was repeated twice using two different sample dilutions (3 and 5 mg/ml), and the results were in good agreement. The data shown is from the sample at 5 mg/ml. (iii) Primary sequence alignment of Msps TOG6, Stu2p TOG2, and Zyg9 TOG3, based on the superposed coordinates of the models and crystal structures. Identities are highlighted in yellow. The positions of the α helices (A and B) that comprise the six HEAT repeats of Msps TOG6 are shown above the sequences and are colored as defined in H (ii).
The possibility remained that clathrin interacts with ch-TOG to mediate association with spindle MTs. If this were the case, the interaction would involve a CBM in ch-TOG binding to the NTD of CHC between blades 1 and 2 (Fig. 4 D). Four CBM sequences are present in ch-TOG, but the first three are folded in the TOG1 domain and would not be available for binding CHC as linear motifs (Fig. 9 G [i]). The other, LFQIE, was found in the C-terminal region of ch-TOG. However, a ch-TOG mutant of this motif (AAQAK) and clathrin were both found on the mitotic spindle, arguing against a possible interaction (Fig. S5 A).
We next investigated the structure of the TACC3-binding region of ch-TOG. Using nuclear magnetic resonance (NMR) spectroscopy, we probed the structure of a C-terminal fragment of the ch-TOG Drosophila melanogaster homologue minispindles (Msps). Msps was chosen in preference to ch-TOG for technical reasons, because the recombinant protein was smaller in size and more stable, allowing assignment of 85% of the backbone resonances. Measurement of the dynamics of this region revealed three subregions, which had properties consistent with two independently mobile folded domains connected by a flexible linker. The larger and more conserved of the two-folded domains in Msps C-terminal fragment has 12 α helices, consistent with the arrangement of a TOG domain, as is the equivalent region in ch-TOG (Fig. S5, B and C). This domain encompasses the LFQIE motif described above, such that this CBM is also folded in an α helix and unavailable for binding. The four motifs within ch-TOG that resemble CBMs were all found in structurally related positions, at the C-terminal end of the B helices in HEAT repeats (Fig. 9 G [ii]), indicating that these two very different functional motifs have similar sequence preferences.
To provide experimental validation of the TOG6 domain in Msps, we measured residual dipolar couplings (RDCs; Tjandra and Bax, 1997) for Msps TOG6. RDCs arise from aligning a protein in a liquid crystal and are dependent on the orientation of NH bond vectors relative to the alignment axis. The resulting values allow a robust evaluation of protein folds based on the relative orientation of secondary structure elements (Fowler et al., 2000). We compared the experimental RDCs of Msps TOG6 domain to values calculated for homology models of Msps TOG6 (Zweckstetter, 2008) based on the crystal structures of two different TOG domains that, although highly similar, vary in the precise orientations of α helices (Fig. 9 H): TOG2 from Stu2p (PDB accession no. 2QK1) and TOG3 from Zyg9 (PDB accession no. 2OF3). We focused on the α-helical residues that would be expected to show similar orientations between different TOG domains, whereas the loops would be expected to be highly variable. Overall, the orientations of the backbone residues in Msps were a good fit to the Stu2p TOG2 model, and fitted Zyg9 TOG3 less well (Fig. 9 H [ii]). We classified the α helices of Msps TOG6 domain (Fig. 9 H [iii]): seven helices that best fit Stu2p TOG2 (blue), three helices that best fit Zyg9 TOG3 (red), and two helices of HR6 that were a poor fit to either model (green). The orientation of HR6 relative to the other repeats varies between TOG domains, as illustrated by Stu2p TOG2 and Zyg9 TOG3 (Fig. 9 H [i]), and it is not surprising that HR6 of Msps does not align with the equivalent regions of other TOG models. Thus, the number and orientations of the α helices of Msps (1591–1850) are consistent with the presence of a sixth TOG domain in ch-TOG homologues.
To make an initial comparison of the Msps TOG6 domain with TOG domains of known structure, their primary sequences were aligned (Fig. 9 H [iii]). Although the overall pairwise identities of the three sequences are low (<20%), there are regions of sequence conservation within and between several of the HEAT repeats. The prominent divergent features are HR6 of all three proteins, which is poorly conserved in structure and sequence, and the extended loop region between the A and B helices of HR5 of Msps TOG6.
Discussion
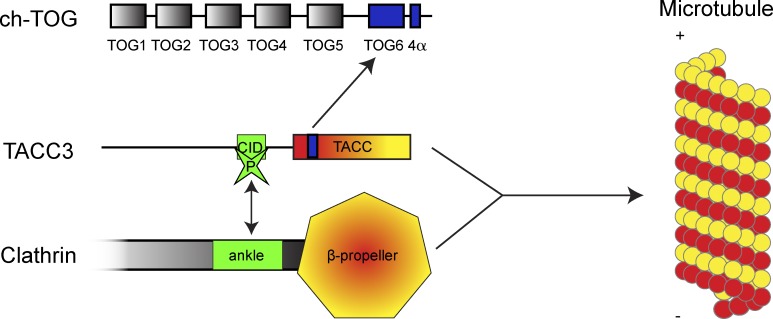
We have identified a novel mode of MT binding in which domains in two separate proteins create a composite binding interface when coordinated in tandem. This coordination is regulated by Aurora A phosphorylation. Our results explain the previously observed interdependence of recruitment of TACC3 and clathrin to the mitotic spindle, where neither TACC3 nor CHC associate efficiently with the spindle without the other (Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010; Booth et al., 2011; Royle, 2012). Finally, we mapped the TACC3–ch-TOG interaction, proposed a new sixth TOG domain, and showed that ch-TOG is dispensable for MT localization of TACC3–clathrin. The interactions and a new scheme for TACC3–ch-TOG–clathrin recruitment to spindle MTs are shown in Fig. 10.
Figure 10.
Schematic diagram to summarize our findings. TACC3 and clathrin interact via the CID and ankle regions, respectively (green). This is regulated by Aurora A phosphorylation of CID at S558. This interaction coordinates the NTD (β-propeller) and TACC domain (coiled-coil) in tandem to make a composite MT interaction surface (yellow-red). TACC3 binds ch-TOG via a break in the coiled-coil of TACC3 and α region of ch-TOG containing TOG6 and 4α domains.
Clathrin and TACC3 interact directly when TACC3 S558 is phosphorylated by Aurora A (Fu et al., 2010; Lin et al., 2010). Previous work narrowed down this interaction to residues 522–577 of TACC3 and residues 331–542 of CHC, with phosphorylation of S558 by Aurora A being important (Lin et al., 2010). It was unclear whether other residues within TACC3(522–577) contributed to binding, but the data presented here demonstrate that several residues between aa 540 and 567 are required for this interaction and resultant spindle recruitment in cells. Our model of CHCR0 and experimental tests of this model indicate that the dileucine motif on TACC3 binds at a hydrophobic pocket bordered by the loop between helices f and g of CHCR0, with pS558 of TACC3 being coordinated by positive charge on the “side” surface of CHCR0. The TACC3 residues involved are conserved from mammals to flies (Fig. 1 B), consistent with a conserved mitotic function for the TACC3–ch-TOG–clathrin complex (Fu et al., 2010). The CHC ankle-binding site for TACC3 is distinct from that for the β2 subunit of the AP-2 complex, meaning that mitosis-specific clathrin inhibitors could be developed without necessarily disrupting endocytosis.
We showed that coordination of clathrin NTD and TACC domains in tandem is sufficient for MT association, via a synthetic clACC fusion protein. The organization of a β-propeller/WD40 domain in tandem with a coiled-coil domain is found in other proteins, e.g., coronin, Nedd1, and POCs, that bind MTs and/or centrosomes (Goode et al., 1999; Woodland and Fry, 2008; Zhang et al., 2009). This pairing of domains could therefore represent a general MT interaction surface. An alternative interpretation is that TACC3–clathrin interaction results in two individually weak MT-binding surfaces that can bind to MTs via increased avidity, but are not necessarily coordinated in space. It is difficult to formally exclude this possibility, but we favor the interpretation of tandem domain coordination. Clathrin triskelia have three NTDs (putative weak MT interaction domains) per molecule and yet cannot interact with MTs (Lin et al., 2010; Booth et al., 2011). Coordination of one NTD with one TACC domain within clACC permits MT binding and this binding is lost upon C+ mutation of the NTD. The most parsimonious conclusion here is that a mix and match of two different domains is required and this argues against a simple mechanism based on increased avidity. A structural explanation for precisely how the NTD and TACC domain bind MTs is an important question for the future.
At centrosomes, ch-TOG is critical for MT assembly and maintenance of MT attachments (Holmfeldt et al., 2004; Barr and Gergely, 2008; De Luca et al., 2008; Cassimeris et al., 2009). This activity is seemingly independent of TACC3–clathrin in human cells. The role of ch-TOG in the TACC3–ch-TOG–clathrin complex remains enigmatic. It is curious that ch-TOG requires TACC3–clathrin for interaction with spindle MTs because TOG domains have a conserved interaction with tubulin. In addition, a MT lattice-binding motif, identified in XMAP215, is also partially conserved in ch-TOG (Spittle et al., 2000; O’Brien et al., 2005; Widlund et al., 2011). Homologues of ch-TOG and TACC3 are known to bind in lower species but so far, in humans, only a direct interaction between ch-TOG and TACC1 had been demonstrated (Conte et al., 2003; Peset and Vernos, 2008). We mapped the direct interaction between human ch-TOG and TACC3 to a specific break in the coiled-coil of TACC3 and the C-terminal region of ch-TOG. This interaction is required for ch-TOG spindle recruitment, but is not required for assembly of the TACC3–clathrin complex on spindle MTs. Our analysis identified new structural features of the XMAP215 family of MT polymerases: the conserved C-terminal region of ch-TOG has a potential novel TOG6 domain. Future work will characterize the structure and MT-binding properties of this domain.
Materials and methods
Antibodies and inhibitors
Primary antibodies were: (1) mouse monoclonals TACC3, α-tubulin DM1A (Abcam), CHC TD.1, X22 (produced in-house), Aurora A 4/IAK (BD), and MBP 8G1 (2396; Cell Signaling Technology); (2) rabbit polyclonals TACC3 H300 (Santa Cruz Biotechnology, Inc.), β-tubulin, pericentrin (Abcam), and pS558 TACC3 (raised in-house). Secondary antibodies were HRP conjugates (GE Healthcare) or conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 633 (Molecular Probes). MLN8237 was obtained from Selleck Chemicals. Rapamycin was obtained from Sigma-Aldrich. Restriction endonucleases were purchased from New England Biolabs, Inc. All other reagents were obtained from Sigma-Aldrich unless specified otherwise.
Molecular biology
For replacement of endogenous protein with expressed constructs, the shRNA is encoded on the same plasmid (pBrain) as the protein encoding sequence, which is rendered resistant to RNAi (Royle et al., 2005). Most plasmids were available from previous work, including clathrin-replacement plasmids (pBrain-GFP-CHC-shCHC mutants), TACC3-replacement plasmids (pBrain-GFP-TACC3-shTACC3), KS plasmids (pBrain-GFP-FKBP-TACC3-shTACC3 and pMito-PAGFP-FRB), plasmids for bacterial expression of CHC fragments (MBP-CHC(1–1074)-His6 and MBP-CHC(1–1074)(Δ457–507)-His6), and mCherry-LCa (Royle et al., 2005; Royle and Lagnado, 2006; Hood and Royle, 2009; Booth et al., 2011; Willox and Royle, 2012; Cheeseman et al., 2013). New pBrain-GFP-CHC-shCHC constructs containing ΔEF, ΔGH, mutant B (P56N and S67G), mutant 444 (R444E, K445E, K500E, K506E, and K507E), mutant 481 (R481E, K487E, K500E, K506E, and K507E), and ERGQC (L480E, A482G, N483Q, and V484C) were made using site-directed mutagenesis. CHC(Δ331–541) was amplified by PCR and exchanged for the equivalent full-length region in pBrain-GFP-CHC-shCHC. pBrain-GFP-CHC(Δ457-507 + B Mut)-shCHC was made by cutting and pasting an equivalent fragment from pBrain-GFP-CHC(mutant B)-shCHC into pBrain-GFP-CHC(Δ457-507)-shCHC. pBrain-GFP-TACC3-shTACC3 truncations were generated by PCR from an IMAGE clone (6148176) and insertion into pBrain-GFP-shTACC3, whereas point mutants and Δ678–681 and Δ682–688 were generated by site-directed mutagenesis. pBrain-GFP-FKBP-TACC3-shTACC3 mutants were made by cutting and pasting equivalent TACC3 fragments from pBrain-GFP-TACC3-shTACC3 versions into pBrain-GFP-FKBP-TACC3-shTACC3. To generate GFP-clACC, CHC(1–363) was PCR amplified from a Kasuza clone (KIA00034) and inserted into pEGFP-C1, and then TACC3(592–838) was PCR amplified from the IMAGE clone and inserted immediately downstream of the CHC fragment, with an EcoRI site linking the two fragments in a single reading frame. C+, Δ678–681, and Δ682–688 mutants of GFP-clACC were generated by cutting and pasting equivalent fragments from the pBrain-GFP-CHC-shCHC and pBrain-GFP-TACC3-shTACC3 mutants into GFP-clACC or by site-directed mutagenesis. β2 adaptin (616–951) was PCR amplified from an IMAGE clone (3532472) and inserted into pGEX6p1 for expression with a GST tag. GST-TACC3-His6 was generated by PCR amplification to include a 3′ 6×His tag and inserted into pGEX6p1. S558A and LL(566,567)AA versions of this construct were made by cutting and pasting equivalent fragments from pBrain-GFP-TACC3-shTACC3 mutants into GST-TACC3-His6. GST-TACC3(522–577) and GST-TACC3(522–577)S558A were made by PCR from pBrain-GFP-TACC3-shTACC3 plasmids and insertion into pGEX6p1. GST-TACC3(522–577)LL(566,567)AA was generated by site-directed mutagenesis. C+, 444, 481, and ERGQC mutant versions of MBP-CHC(1–1074)-His6 were generated by site-directed mutagenesis or cutting and pasting an equivalent fragment of pBrain-GFP-CHC(C+ Mut)-shCHC into MBP-CHC(1–1074)-His6. MBP-CHC(Δ1–330) was generated by PCR amplification and insertion into pMal-Pre-His. TACC3(629–838) was PCR amplified from cDNA provided by F. Gergely (Cancer Research UK Cambridge Institute, Cambridge, UK) after the silent mutation of two internal NcoI sites by site-directed mutagenesis. TACC3(629–838) was subcloned into pETM6T1 for expression with a His-NusA tag. TACC3 internal deletion constructs were made by PCR using pETM6T1 TACC3(629–838) as a template. TACC3(629–788 and 629–818) were subcloned from pETM6T1 TACC3(629–838) into pET30TEV for expression with a His6 tag. Ch-TOG(1517–1957) was amplified from Kasuza clone KIAA0097 and subcloned into pETM6T1. Msps(1591–1941) was amplified from cDNA clone LP04448 (Source Biosciences) and inserted into pETM6T1. TACC3(519–838) was subcloned into a modified version of pMAX (Lonza) for high level expression in human cells as an Strep-mCherry–tagged protein. YFP and CHC(1–574) were cloned into pET30TEV to assemble a His6-YFP-CHC(1–574) fusion for expression in Escherichia coli. Msps(1591–1850) was subcloned into pETM6T1 using NcoI–EcoRI and the Msps(1591–1941) construct as a template. All constructs were verified by restriction digest and automated DNA sequencing (GATC; Eurofins MWG Operon).
Biochemistry
For Western blotting, HEK293 cells were incubated for 48 h posttransfection before treatment with 50 ng/ml nocodazole or 2 mM thymidine for 16 h, and then 0.3 µM MLN8237 for the final 20 min where indicated. Cells were washed twice with ice cold PBS and lysed on ice, for 30 min, with lysis buffer A (20 mM Tris.Cl, pH 7.5, 150 mM NaCl, 1% Igepal, 1% Triton X-100, 15 µg/ml DNase I, 5 mM Na3VO4, 30 mM NaF, 100 nM okadaic acid, and Roche protease inhibitor cocktail). Lysates were spun at 14,000 rpm at 4°C for 15 min and the supernatant was collected. Protein concentration of lysates was performed using BCA assay kit (Sigma-Aldrich) following the manufacturer’s instructions and 20 µg of protein was analyzed by Western blot.
MBP-CHC(1–1074)-His6 proteins and GST-TACC3-His6 proteins were purified using Ni/NTA agarose (QIAGEN) according to the manufacturer’s instructions. GST–β2 adaptin was purified using GS4B (GE Healthcare) according to the manufacturer’s instructions. For purification via MBP tag, amylose resin was used according to the manufacturer’s instructions. BL21pLysS E. coli were transfected with plasmid then grown in 2× TY plus 2% glucose containing antibiotics until OD600 of 0.6–0.8. Expression was induced using 0.8 mM IPTG overnight at 20°C. Protein was incubated with amylose resin, washed, and eluted using maltose at concentrations from 10 to 50 mM.
Clathrin was purified from clathrin-coated vesicle (CCV) preparations from rat livers (Campbell et al., 1984). In brief, three to six rat livers were washed in cold HKM buffer (25 mM Hepes, pH 7.4, 125 mM potassium acetate, and 5 mM magnesium acetate with 1 mM DTT) and cut into pieces, supplemented with protease cocktail set III (total vol 40 ml; EMD Millipore), and homogenized in a dounce homogenizer, all steps on ice or 4°C. Insoluble material was removed by two spins for 20 min at 3,800 _g_av. Supernatant was ultracentrifuged at 149,000 _g_av for 40 min. Pellet was resuspended in 10 ml of HKM and homogenized. An equal volume of HKM containing Ficoll (12.5% wt/vol) and sucrose (12.5% wt/vol) was added and mixed. After a 20-min, 46,000-_g_av spin, CCVs are in the supernatant; these were diluted with 3 vol of HKM and pelleted at 96,000 _g_av for 60 min. Pellet was resuspended in 15 ml HKM, homogenized, and incubated on ice for 20 min. Insoluble material was pelleted by spinning at 17,000 _g_max for 10 min. 5 ml of supernatant was layered over a 5-ml cushion of 8% (wt/vol) sucrose/HKM and spun at ∼80,000 _g_max in a swing-out rotor (SW41; Beckman Coulter) for 2 h. The CCV pellet was resuspended in 0.5 ml of uncoating buffer (1 M Tris-HCl, pH 7.4, with 1 mM DTT) and rotated for 1 h at 4°C; vesicles were pelleted at 96,000 _g_av for 25 min and supernatant was snap frozen and stored at −80°C.
Other His6- and His-NusA tagged proteins were expressed in CodonPlus RIL E. coli. Cells were grown in LB media with the addition of antibiotics until OD600 of ∼0.6 was reached. Expression was induced by 0.6 mM IPTG overnight at 21°C. Tagged protein was purified by immobilized metal ion affinity chromatography using a HiTrap Chelating Sepharose HP column (GE Healthcare) as per the manufacturer’s instructions. The expression tag was removed by overnight TEV cleavage. Immobilized metal ion affinity chromatography was repeated to remove the TEV protease and expression tag. In the case of proteins expressed in pETM6T1, Q-Sepharose chromatography (GE Healthcare) was performed according to the manufacturer’s instructions to improve protein purity.
HEK293F cells were grown in suspension culture in Freestyle 293 expression medium (Gibco) in a 5% CO2 atmosphere and transfected with the pMAXI Strep2-mCherry-TACC3(519–838) plasmid at a density of 106 cells/ml using 2 µg/ml polyethylenimine. Cells were harvested after 2 d and lysed in TBS. The expression protein was captured on a 1-ml StrepTrap column (GE Healthcare), which was then washed with TBS and eluted with 3 mM Desthiobiotin.
Stable isotope-labeled Msps(1591–1941) protein was produced by CodonPlus RIL E.coli cells transformed with plasmid. Cells were grown in deuterated M9 media supplemented with 15N ammonium chloride, 13C d-glucose (Goss Scientific Instruments Ltd), kanamycin, and chloramphenicol. Cultures were grown at 37°C until an OD600 of 1.25 was reached. Recombinant protein expression was induced by the addition of 0.6 mM IPTG and overnight incubation at 21°C. Stable isotope-labeled Msps(1591–1850) was generated as for Msps(1591–1941) using E.coli OD2 N media (Silantes) with induction at an OD600 of ∼0.6. Tagged Msps protein was purified by Q-Sepharose chromatography (GE Healthcare) as per the manufacturer’s instructions. The His-NusA tag was removed by overnight TEV cleavage. The resulting protein was passed through a further Q-Sepharose column to remove the TEV protease and His-NusA tag. Msps-containing fractions were concentrated before size exclusion chromatography on a HiPrep Sephacryl-S300 HR column (GE Healthcare) into 20 mM potassium phosphate, pH 7.0, 50 mM NaCl, 1 mM DTT, and 0.02% sodium azide. Purified Msps was concentrated to ∼400 µM for NMR analysis.
For in vitro binding assays, 50 µg of GST-tagged protein was incubated with 2 µg/ml Aurora A kinase (EMD Millipore) or BSA, 2 µg/ml GST-TPX2(1–43), and 10 mM MgATP for 2 h at 30°C in reaction buffer (50 mM Tris.Cl, pH 7.5, 150 mM NaCl, and 0.1 mM EGTA). For CHC mutant-binding experiments, TACC3 was phosphorylated before incubation with MBP-CHC mutants. Phosphorylated GST protein was incubated with 30 µl glutathione sepharose 4B in a total volume of 200 µl NET-2 buffer (50 mM Tris.Cl, pH 7.5, 150 mM NaCl, and 0.5% NP-40 substitute [Lin et al., 2010]) containing 0.1 mg/ml of clathrin or clathrin mutants. Proteins were incubated overnight with rotation at 4°C, and then spun at 10,000 g for 2 min. Supernatant was retained and beads were washed four times with 1 ml NET-2. 30 µl of 2× Laemmli buffer was added to the beads; they were denatured at 100°C for 5 min and half was analyzed by Western blotting along with 5 µl of the supernatant.
In vitro binding assays between purified His-NusA TACC3 proteins and ch-TOG 1517–1957 were performed at 4°C. 100 µg His-NusA TACC3 was incubated with 20 µl of Nickel Sepharose resin (GE Healthcare) for 2 h with rotation in reaction buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 40 mM imidazole, and 0.1% Tween 20). Beads were washed three times with 1 ml of reaction buffer after which 100 µg ch-TOG was added and incubated with the beads for a further 2 h. The beads were washed three times with 1 ml of reaction buffer and analyzed by SDS-PAGE after the addition of 20 µl SDS-loading buffer.
CD spectroscopy was performed on a spectropolarimeter (J-715; Jasco) with temperature control. Clathrin protein samples were diluted to ∼0.1 mg/ml in 20 mM Tris.Cl and 20 mM NaCl, pH 7.4. Far ultraviolet scans were taken from 250 to 190 nm at 30, 50, 66, and 90°C. Protein composition was determined using DichroWeb (Whitmore and Wallace, 2004, 2008). TACC3 protein samples were at ∼150 µM in 20 mM potassium phosphate, pH 7.0, 50 mM NaCl, and 1 mM DTT, and data were analyzed using a custom-written Mathematica macro designed to compare reference spectra for α helix, β sheet, and random coil to the experimental data (see cdfit.ma in online supplemental material).
Cell biology
Cells were maintained in DMEM plus 10% fetal bovine serum and 100 U/ml penicillin and streptomycin at 37°C in 5% CO2. HEK293 cells were transfected using calcium phosphate. In brief, 1 h before transfection the medium is replaced with 1 ml of fresh medium. For one well of a six-well plate, 5 µl of 2.5 M CaCl2 and 1.25 µg DNA were mixed with 0.1× TE buffer (1 mM Tris.Cl and 0.1 mM EDTA, pH 7.6) to make up to 50 µl. 50 µl of 2× Hepes buffer (140 mM NaCl, 1.5 mM Na2HPO4, and 50 mM Hepes, pH 7.05) was added and left for 1 min at 37°C to allow precipitate to form. Precipitate was added to the cells and the media changed 6 h later. HeLa cells were transfected by lipofection using Genejuice (Merck), according to the manufacturer’s instructions. For CHC and TACC3 RNAi, HEK293 cells were transfected 3 d before analysis. For TACC3 RNAi HeLa, cells were transfected 2 d before analysis. For Aurora A kinase inhibition, cells were incubated with 0.3 µM MLN8237 for 30 (Fig. 2) or 40 min (Fig. S3 A) or with 0.7 µM MLN8237 for 50 min (Fig. 3).
Cells were fixed with PTEMF (50 mM Pipes, pH 7.2, 10 mM EGTA, 1mM MgCl2, 0.2% Triton X-100, and 4% paraformaldehyde) for 15 min at RT, and then permeabilized (PBS and 0.5% Triton X-100) for 10 min. Cells were blocked (PBS, 3% BSA, and 5% goat serum) for 1 h, and then incubated with primary antibodies for 2 h. Cells were then rinsed with PBS, before incubation with secondary antibodies for 30 min. Cells were rinsed with PBS and mounted with mowiol containing DAPI. For ch-TOG staining, cells were fixed with methanol at –20°C for 10 min and washed three times with PBS, and blocking was performed with 3% BSA only to allow costaining with sheep anti–GFP antibody (GFP fluorescence was reduced by methanol fixation).
For KS experiments (Cheeseman et al., 2013), pMito-PAGFP-FRB (MitoTrap) and pBrain-GFP-FKBP-TACC3-shTACC3 (or mutants) were transfected into HeLa cells. The MitoTrap was invisible during imaging but the PAGFP was activated by the multiphoton laser, which was the last line scanned in all cases. This allowed verification by eye that the cells captured did express this construct. 2 d after transfection, cells were treated with either 200 nM Rapamycin or an equivalent volume of ethanol (vehicle only control) for 20 min at 37°C. For combination of KS with Aurora A inhibition, 0.7 µM MLN8237 or an equal volume of DMSO (vehicle control) was added for 50 min at 37°C, either before or after KS. Therefore, cells that were treated with MLN8237 before KS were exposed to MLN8237 for a total of 70 min, whereas those treated after KS were exposed to Rapamycin for a total of 70 min.
For MT depolymerization experiments, HeLa cells were transfected with GFP-clACC constructs and after 48 h were treated with 10 µM nocodazole (1 h) before fixation and staining.
Microscopy and quantification
Confocal microscopy was performed on an SP2 (Leica) with a 63× (1.4 NA) oil immersion objective. GFP and Alexa Fluor 488 were excited using an Ar/Kr 488-nm laser, mCherry and Alexa Fluor 568 were excited with the 543 line of a He/Ne laser, and Alexa Fluor 633 was excited using a He/Ne 633-nm laser. DAPI was excited using a multiphoton laser. Images were averages of four scans. Epifluorescence images were captured with a microscope (TE2000U; Nikon) with 60× (1.4 NA) oil immersion objective with standard filter sets for DAPI, GFP, Alexa Fluor 568, and Alexa Fluor 633. For quantification, identical settings were used. For GFP-clACC and mCherry-tubulin live cell imaging, cells in CO2-independent media were imaged on glass-bottom dishes in a humidified chamber (Okolab) at 37°C. Images were captured with a DS-Qi1Mc camera and NiS Elements AR software.
Spindle enrichment analysis was essentially performed as described elsewhere (Booth et al., 2011). In brief, clathrin or TACC3 images were captured using identical settings, and then either a 9 × 9-pixel (1.82 µm2; Fig. 2) or a 5 × 5-pixel (0.56 µm2; Fig. 4) region of interest was measured either over the spindle (away from poles), in the cytoplasm, or in a noncell region of background, using ImageJ. Background was subtracted from the spindle and cytoplasm measurements, and, where indicated, spindle fluorescence was then divided by cytoplasm fluorescence to give a spindle enrichment ratio. For analysis of mitotic index, GFP-expressing cells were assessed as being in either interphase or mitosis based on chromosome morphology, visualized using DAPI staining. The results of at least three independent experiments for each construct are shown. These were each normalized to the control from that experiment before calculating the mean of these normalized values. Statistical testing is described in the figure legends. Graphing was done using IgorPro (Wavemetrics). Figures were assembled using Adobe Photoshop and Illustrator.
For MT-binding experiments, tubulin was prepared from pig brains according to published protocols (Gell et al., 2011). In brief, brains are homogenized and then clarified by centrifugation and filtration. Tubulin is purified by cycles of polymerization (in the presence of GTP), centrifugation, and depolymerization. Labeled tubulin was obtained from Cytoskeleton, nucleotides were obtained from Jena Biosciences, and all other chemicals were obtained from Sigma-Aldrich unless otherwise indicated. YFP-CHC(1–574) and mCherry-TACC3(519–838) were dialyzed into MRB80 (80 mM Pipes, pH 6.8, with KOH, 1 mM EGTA, and 4 mM MgCl2) supplemented with 50 mM KCl and adjusted to a concentration of 5 µM. MT seeds were assembled from tubulin, biotin-tubulin, and Hilyte647-tubulin at a molar ratio of 25:1:1 in the presence of 1 mM GMP-CPP in MRB80 for 1 h at 37°C, diluted 20-fold with MRB80 with 2 µM Taxol, and stored at RT. A 100-µm deep flow chamber was made from a slide and a hydrochloric acid–treated coverslip using double-sided tape (Scotch; 3M) and passivated with PLL-PEG-50% biotin (Susos AG). Seeds were attached to this surface using streptavidin and blocked with 1 mg/ml k-casein. A reaction mix containing 14 µM tubulin, 50 mM KCl, 1 mM GTP, 0.6 mg/ml k-casein, 0.2% methyl cellulose, 4 mM DTT, 0.2 mg/ml catalase, 0.4 mg/ml glucose oxidase, and 50 mM glucose in MRB80, supplemented with 0.3 µM Hilyte647-tubulin, was clarified for 8 min at 190,000 g in an airuge (Beckman Coulter) and the supernatant was added to the flow chamber and temporarily closed with sticky tape. MT assembly was allowed to occur for 30–60 min at 32°C under the microscope. A cleared protein mix containing 1 µM YFP-CHC(1–574), 1 µM mCherry-TACC3(519–838), 25 nM Aurora A, 1 mM ATP, 50 mM KCl, 4 mM DTT, 0.2 mg/ml catalase, 0.4 mg/ml glucose oxidase, 50 mM glucose, and 2 µM Taxol in MRB80 was then flowed into the chamber. YFP-CHC(1–574) and MTs were observed on a total internal reflection fluorescence system (Olympus) using a 100× NA 1.49 objective, 1.6× additional magnification, 488- and 640-nm laser lines, respectively, and an ImageEM-1k back-illuminated EM-CCD camera (Hamamatsu Photonics) under the control of xcellence software.
NMR spectroscopy and structural modeling
The structural model of CHCR0 was built using MODELLER 9.9 and an aligned CHCR0 sequence to search for homology to known CHC structures. The best model was merged with residues 395–483 from chain B of 1BPO and further refined using MolProbity (University of California San Francisco). Mutations were generated in Pymol and represented using CCP4mg (McNicholas et al., 2011). The structural models of TOG1 and TOG6 were produced using the PHYRE2 server (for template 2OF3) and SwissModel (for template 2QK1) and figures were generated in Pymol.
For NMR spectroscopy, 85% of backbone resonances were assigned using 15N- and 2H/13C/15N -labeled samples of Msps (aa 1591–1941). The assignment allowed the collection of secondary structure specific NOESY cross-peaks and secondary chemical shifts (Cα, Cβ, C’, and Hα) in addition to 15N R1 and R2 relaxation rates as well as 1H-15N heteronuclear nuclear Overhauser effects. All of these were combined to identify the two-folded domains and the secondary structure elements in these domains. The putative TOG domain fold in Msps 1591–1850 (TOG6) was further explored by collection of RDCs (Tjandra and Bax, 1997) by aligning the protein in a solution of 5 mg/ml Pf1 phage. From a total of 220 assigned residues (out of 260 in the Msps 1591–1850 construct), RDCs could be obtained for 200 residues. The experimental values were compared with values calculated with PALES (Zweckstetter, 2008) for homology models for Msps 1591–1850 based on crystal structures 2QK1 and 2OF3.
Online supplemental material
Fig. S1 shows the localization of GFP-CHC mutants in mitotic HEK293 cells depleted of endogenous CHC. Fig. S2 shows normal folding of MBP-CHC(1–1074)-His6 mutants. Fig. S3 shows that GFP-clACC does not require TACC3 or Aurora A kinase activity for mitotic spindle localization and that interphase MT bundles decorated with clACC have short-term protection from depolymerization. Fig. S4 shows normal folding of TACC3 629–838 Δ678–681 and TACC3 629–838 Δ682–688. Fig. S5 shows the identification of TOG6 domain in ch-TOG homologues. The text file contains the custom-written Mathematica macro for fitting CD data. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211127/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201211127.dv.
Supplementary Material
Supplemental Material
Acknowledgments
We thank Anja Winter, Maria Blixt, and Nancy Powell for technical assistance. F.E. Hood thanks Ian Prior for support. Spectroscopic data were collected in the King’s College London Centre for Biomolecular Spectroscopy and the National Henry Wellcome Building-NMR Biomolecular NMR facility in Birmingham. We wish to thank Andrew Atkinson and Sara Whittaker for help and support.
This work was funded by a Career Establishment Award from Cancer Research UK (C25425/A8722) to S.J. Royle, by grants to R. Bayliss from the Medical Research Council (G0800021) and Cancer Research UK (C24461/A12772), and a Marie Curie Cancer Care program grant to A. Straube. S.J. Royle is a Senior Cancer Research Fellow of Cancer Research UK and R. Bayliss is a Royal Society Research Fellow.
Footnotes
Abbreviations used in this paper:
CBM
clathrin box motif
CCV
clathrin-coated vesicle
CD
circular dichroism
CHC
clathrin heavy chain
CHCR0
CHC repeat 0
ch-TOG
colonic and hepatic tumor overexpressed gene
CID
clathrin interaction domain
KS
knocksideways
MT
microtubule
NMR
nuclear magnetic resonance
NTD
N-terminal domain
RDC
residual dipolar coupling
TACC3
transforming acidic coiled-coil protein 3
References
- Barr A.R., Gergely F. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Barr A.R., Gergely F. 2008. MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol. Cell. Biol. 28:7199–7211 10.1128/MCB.01040-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth D.G., Hood F.E., Prior I.A., Royle S.J. 2011. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30:906–919 10.1038/emboj.2011.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C., Squicciarini J., Shia M., Pilch P.F., Fine R.E. 1984. Identification of a protein kinase as an intrinsic component of rat liver coated vesicles. Biochemistry. 23:4420–4426 10.1021/bi00314a028 [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590 10.1091/mbc.E03-07-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Becker B., Carney B. 2009. TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motil. Cytoskeleton. 66:535–545 10.1002/cm.20359 [DOI] [PubMed] [Google Scholar]
- Charrasse S., Schroeder M., Gauthier-Rouviere C., Ango F., Cassimeris L., Gard D.L., Larroque C. 1998. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 111:1371–1383 [DOI] [PubMed] [Google Scholar]
- Cheeseman L.P., Booth D.G., Hood F.E., Prior I.A., Royle S.J. 2011. Aurora A kinase activity is required for localization of TACC3/ch-TOG/clathrin inter-microtubule bridges. Commun. Integr. Biol. 4:409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman L.P., Harry E.F., McAinsh A.D., Prior I.A., Royle S.J. 2013. Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. J. Cell Sci. 126:2102–2113 10.1242/jcs.124834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte N., Delaval B., Ginestier C., Ferrand A., Isnardon D., Larroque C., Prigent C., Séraphin B., Jacquemier J., Birnbaum D. 2003. TACC1-chTOG-Aurora A protein complex in breast cancer. Oncogene. 22:8102–8116 10.1038/sj.onc.1206972 [DOI] [PubMed] [Google Scholar]
- De Luca M., Brunetto L., Asteriti I.A., Giubettini M., Lavia P., Guarguaglini G. 2008. Aurora-A and ch-TOG act in a common pathway in control of spindle pole integrity. Oncogene. 27:6539–6549 10.1038/onc.2008.252 [DOI] [PubMed] [Google Scholar]
- Edeling M.A., Mishra S.K., Keyel P.A., Steinhauser A.L., Collins B.M., Roth R., Heuser J.E., Owen D.J., Traub L.M. 2006. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell. 10:329–342 10.1016/j.devcel.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Foraker A.B., Camus S.M., Evans T.M., Majeed S.R., Chen C.Y., Taner S.B., Corrêa I.R., Jr, Doxsey S.J., Brodsky F.M. 2012. Clathrin promotes centrosome integrity in early mitosis through stabilization of centrosomal ch-TOG. J. Cell Biol. 198:591–605 10.1083/jcb.201205116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S.C., Kirchhausen T., Walz T. 2004. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 432:573–579 10.1038/nature03079 [DOI] [PubMed] [Google Scholar]
- Fowler C.A., Tian F., Al-Hashimi H.M., Prestegard J.H. 2000. Rapid determination of protein folds using residual dipolar couplings. J. Mol. Biol. 304:447–460 10.1006/jmbi.2000.4199 [DOI] [PubMed] [Google Scholar]
- Fu W., Tao W., Zheng P., Fu J., Bian M., Jiang Q., Clarke P.R., Zhang C. 2010. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 123:3645–3651 10.1242/jcs.075911 [DOI] [PubMed] [Google Scholar]
- Gell C., Friel C.T., Borgonovo B., Drechsel D.N., Hyman A.A., Howard J. 2011. Purification of tubulin from porcine brain. Methods Mol. Biol. 777:15–28 10.1007/978-1-61779-252-6_2 [DOI] [PubMed] [Google Scholar]
- Gergely F., Karlsson C., Still I., Cowell J., Kilmartin J., Raff J.W. 2000a. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA. 97:14352–14357 10.1073/pnas.97.26.14352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Kidd D., Jeffers K., Wakefield J.G., Raff J.W. 2000b. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 19:241–252 10.1093/emboj/19.2.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Draviam V.M., Raff J.W. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341 10.1101/gad.245603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Wong J.J., Butty A.C., Peter M., McCormack A.L., Yates J.R., Drubin D.G., Barnes G. 1999. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144:83–98 10.1083/jcb.144.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T.L., Sept D., Mogessie B., Straube A., Ross J.L. 2013. Mechanical properties of doubly stabilized microtubule filaments. Biophys. J. 104:1517–1528 10.1016/j.bpj.2013.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P., Stenmark S., Gullberg M. 2004. Differential functional interplay of TOGp/XMAP215 and the KinI kinesin MCAK during interphase and mitosis. EMBO J. 23:627–637 10.1038/sj.emboj.7600076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood F.E., Royle S.J. 2009. Functional equivalence of the clathrin heavy chains CHC17 and CHC22 in endocytosis and mitosis. J. Cell Sci. 122:2185–2190 10.1242/jcs.046177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N.C., Bird A.W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189:739–754 10.1083/jcb.200911091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K., Noetzel T.L., Pelletier L., Mechtler K., Drechsel D.N., Schwager A., Lee M., Raff J.W., Hyman A.A. 2005. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170:1047–1055 10.1083/jcb.200503023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.J., Gergely F., Jeffers K., Peak-Chew S.Y., Raff J.W. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649 10.1038/35083033 [DOI] [PubMed] [Google Scholar]
- LeRoy P.J., Hunter J.J., Hoar K.M., Burke K.E., Shinde V., Ruan J., Bowman D., Galvin K., Ecsedy J.A. 2007. Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 67:5362–5370 10.1158/0008-5472.CAN-07-0122 [DOI] [PubMed] [Google Scholar]
- Lin C.H., Hu C.K., Shih H.M. 2010. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J. Cell Biol. 189:1097–1105 10.1083/jcb.200911120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Manning A.L., Compton D.A. 2008. Structural and regulatory roles of nonmotor spindle proteins. Curr. Opin. Cell Biol. 20:101–106 10.1016/j.ceb.2007.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas S., Potterton E., Wilson K.S., Noble M.E. 2011. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67:386–394 10.1107/S0907444911007281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien L.L., Albee A.J., Liu L., Tao W., Dobrzyn P., Lizarraga S.B., Wiese C. 2005. The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol. Biol. Cell. 16:2836–2847 10.1091/mbc.E04-10-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I., Vernos I. 2008. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 18:379–388 10.1016/j.tcb.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Peset I., Seiler J., Sardon T., Bejarano L.A., Rybina S., Vernos I. 2005. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol. 170:1057–1066 10.1083/jcb.200504037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.S., Sahlender D.A., Foster S.D. 2010. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell. 18:324–331 10.1016/j.devcel.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle S.J. 2012. The role of clathrin in mitotic spindle organisation. J. Cell Sci. 125:19–28 10.1242/jcs.094607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle S.J., Lagnado L. 2006. Trimerisation is important for the function of clathrin at the mitotic spindle. J. Cell Sci. 119:4071–4078 10.1242/jcs.03192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle S.J., Bright N.A., Lagnado L. 2005. Clathrin is required for the function of the mitotic spindle. Nature. 434:1152–1157 10.1038/nature03502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A., Liu E.M., Reichel J., Porrati P., Pellegatta S., et al. 2012. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 337:1231–1235 10.1126/science.1220834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittle C., Charrasse S., Larroque C., Cassimeris L. 2000. The interaction of TOGp with microtubules and tubulin. J. Biol. Chem. 275:20748–20753 10.1074/jbc.M002597200 [DOI] [PubMed] [Google Scholar]
- Tjandra N., Bax A. 1997. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 278:1111–1114 10.1126/science.278.5340.1111 [DOI] [PubMed] [Google Scholar]
- Whitmore L., Wallace B.A. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L., Wallace B.A. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 89:392–400 10.1002/bip.20853 [DOI] [PubMed] [Google Scholar]
- Widlund P.O., Stear J.H., Pozniakovsky A., Zanic M., Reber S., Brouhard G.J., Hyman A.A., Howard J. 2011. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. USA. 108:2741–2746 10.1073/pnas.1016498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willox A.K., Royle S.J. 2012. Functional analysis of interaction sites on the N-terminal domain of clathrin heavy chain. Traffic. 13:70–81 10.1111/j.1600-0854.2011.01289.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H.R., Fry A.M. 2008. Pix proteins and the evolution of centrioles. PLoS ONE. 3:e3778 10.1371/journal.pone.0003778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Natsume Y., Saiki Y., Shioya H., Takeuchi K., Yamori T., Toki H., Aoki I., Saga T., Noda T. 2012. Disruption of Tacc3 function leads to in vivo tumor regression. Oncogene. 31:135–148 10.1038/onc.2011.235 [DOI] [PubMed] [Google Scholar]
- Zhang X., Chen Q., Feng J., Hou J., Yang F., Liu J., Jiang Q., Zhang C. 2009. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 122:2240–2251 10.1242/jcs.042747 [DOI] [PubMed] [Google Scholar]
- Zweckstetter M. 2008. NMR: prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 3:679–690 10.1038/nprot.2008.36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material