Stochastic Processes Influence Stationary-Phase Decisions in Bacillus subtilis (original) (raw)
Abstract
It has recently been proposed that phenotypic variation in clonal populations of bacterial species results from intracellular “noise,” i.e., random fluctuations in levels of cellular molecules, which would be predicted to be insensitive to selective pressure. To test this notion, we propagated five populations of Bacillus subtilis for 5,000 generations with selection for one phenotype: the decision to sporulate. In support of the noise hypothesis, we report that none of the populations responded to selection by improving their efficiency of sporulation, indicating that intracellular noise is independent of heritable genotype.
It has recently been proposed that phenotypic variation in clonal populations, observed in a number of prokaryotic species (5, 16, 17, 24), results from a stochastic element in gene expression and biochemical reactions within the cell (5, 11, 13, 17, 20). Such random fluctuations, dubbed intracellular noise, could be responsible for triggering the decision of subpopulations of cells to embark on differing developmental pathways, resulting ultimately in a phenotypically heterogeneous population of cells with little or no genetic diversity.
The response of genetically clonal populations to adverse environments by spontaneously fragmenting into heterogeneous subpopulations would, in principle, benefit these populations by increasing their phenotypic variability, thus placing them in a better position for survival. In evolutionary terms such a strategy may represent a significant adaptation to environments that are themselves in constant flux, as the probability of individual cells from the population surviving and proliferating no matter what the future environment may entail is increased. In this adaptive model, constant environmental conditions might be expected to reduce intracellular noise because population fragmentation would no longer be adaptive, resulting in one specific developmental response dominating the population.
Clonal populations of the bacterium Bacillus subtilis are good models for studying the stochastic element in gene expression and how it relates to adaptation and survival. Upon exhaustion of local nutrients and entrance into stationary phase, single cells within a clonal B. subtilis population have the potential to embark on a number of different developmental pathways, each of which may promote survival depending on the environment. These include synthesis of extracellular polymer-degrading enzymes (15), competence for DNA uptake (4), motility and chemotaxis (7, 14), biofilm and fruiting body formation (2), adaptive mutagenesis (25), and cellular differentiation into dormant and resistant spores (18, 19). However, there is mounting evidence that in B. subtilis only a certain fraction of cells within a population actually embark on a particular pathway. Only about 5 to 10% of cells induced for competence actually become competent (9, 23), and only a small fraction of stationary-phase cells exhibit adaptive mutagenesis (25). Furthermore, in B. subtilis laboratory populations induced for the sporulation response, only a subpopulation of cells expressed genes needed for entrance into sporulation and completed the process of sporulation (3). Because the entrance into many of these developmental pathways is dependent on threshold levels of certain key regulator molecules (3, 4, 8, 18), random fluctuations in the concentration of these molecules among individual cells may result in phenotypic fragmentation of populations. These experiments suggest that the variation in decisions made by individual B. subtilis cells upon entrance into stationary phase may be the result of stochastic processes instead of underlying genetic variation.
If stochastic variation in gene expression within a clonal population of cells is not encoded within the genome, one would predict that strong directional selection for one particular developmental response would be unsuccessful, because selection would not be able to act on random molecular fluctuations. Using this logic to test the intracellular-noise hypothesis, we propagated five populations of B. subtilis in the laboratory for greater than 5,000 generations and at approximately seven-generation intervals imposed a total of over 800 selection events for the formation of heat-resistant spores. Our results suggest that the proximate decision to embark on the sporulation pathway is not encoded in the genome, as none of the five populations showed any response to selection after over 5,000 generations of directional selection for enhanced efficiency of spore formation.
Five replicate populations of B. subtilis strain WN628 (trpC2 amyE::cat) were subjected to strong selection for spore formation at approximately seven-generation intervals by the following protocol. Spores were inoculated into 10 ml of Schaeffer's sporulation medium (SSM) (22) and cultivated aerobically at 37°C. Spores germinated, and cells grew exponentially for approximately seven generations (∼4 to 5 h) and then entered the stationary phase, during which the decision whether or not to initiate sporulation is made. Approximately 24 h after inoculation, an aliquot of each population was subjected to a heat shock (80°C, 10 min) which is lethal to vegetative cells but which spores readily survive. The heat-shocked culture was then diluted 1:100 into fresh SSM, and the process was repeated. In each of the five populations only those cells that had embarked on and completed the sporulation pathway would survive the heat selection and be propagated in the next growth cycle.
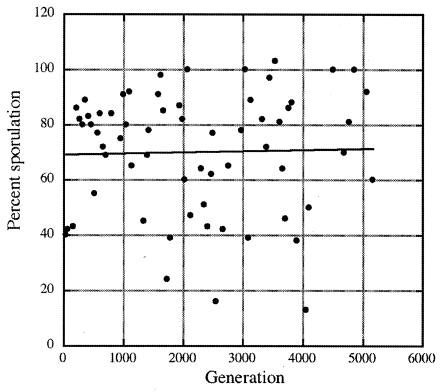
At 50-generation intervals, one of the five experimental populations was chosen on a rotating basis for enumeration of both total viable cells and spores and the percentage of sporulation was calculated. From visual inspection of a plot of the percentage of sporulation versus time from generation 0 to generation >5,000 it appeared that there was no significant increase in the efficiency of sporulation over the course of the experiment (Fig. 1). This observation was supported by regression analysis and analysis of variance (ANOVA) (Fig. 1; best-fit line, _r_2 = 0.011; P = 0.4047). Further analyses done by grouping sporulation frequencies from all cultures in 500-generation intervals also revealed that there was no significant increase in the efficiency of sporulation (data not shown; _r_2 = 0.107; P = 0.5736).
FIG. 1.
Sporulation frequency in all five replicate populations as a function of generations of direct selection for sporulation, including the best-fit regression line through the data (_r_2 = 0.011; ANOVA; P = 0.4047).
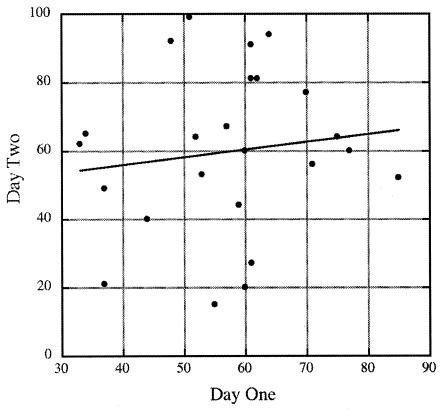
Because fluctuations in the laboratory environment during the >2-year duration of this experiment (e.g., different batches of media, different incubators used for growth, etc.) could possibly have influenced the response to selection for sporulation, we performed the following experiment, in which all environmental factors were strictly controlled. Twenty-four numbered identical 10-ml sporulation cultures were prepared with the same batch of SSM and inoculated at a 1:100 dilution with 24 identical aliquots of heat-shocked spores taken from a single seed culture of strain WN628. All cultures were cultivated for 24 h in the same shaker, and then, as described above, sporulation frequencies were measured at 24 h of growth and heat-shocked spores from each numbered flask were inoculated into a new flask of the same number containing fresh SSM from the same batch as the first set of cultures. After another 24 h of incubation of the second set of 24 flasks, sporulation frequency was again determined. The results of this experiment (Fig. 2), in which sporulation frequency of the first culture is plotted against that of the second culture, again indicated a very low correlation of sporulation efficiency between the two sets of cultures (_r_2 = 0.016; P = 0.5520), again indicating that the decision to sporulate was largely not heritable.
FIG. 2.
Sporulation frequency of cultures on day 2 as a function of their sporulation frequency on day 1, with identical SMM batch and shaker used for growth. The best-fit regression line is indicated (_r_2 = 0.016; ANOVA; P = 0.5520).
While it is clear that none of the five populations responded to direct selection for sporulation by increasing their sporulation efficiency, the hypothesis that this is caused by nonheritable fluctuations in gene expression is not the only possible explanation. Below we discuss three possibilities for the lack of response to selection.
(i) Mutation-limited selection.
Selection can work only on existing genetic variation in populations. Thus if a population exhibits low levels of genetic variation, selection may be ineffective at eliciting a response. Initially, the possibility of mutation limitation in the evolving populations was worrisome, as all populations were started from a single clone of strain WN628. However, we noted that during evolution an increase in the mutation rate to up to an order of magnitude higher than that of the ancestral strain occurred in all five replicate populations (Table 1). With this level of mutational input, coupled with the large effective population sizes of these populations (∼108 cells), it seems very unlikely that mutations are a limiting factor in the response to selection.
TABLE 1.
Mutation rates in sporulating B. subtilis populations.
| Population | Generation | Mutation ratea | Fold increase in mutation rate over ancestor |
|---|---|---|---|
| WN628 (ancestor) | 0 | 8.92 × 10−9 | 1.0 |
| WN628A | 800 | 1.04 × 10−7 | 11.7 |
| WN628B | 800 | 5.63 × 10−8 | 6.3 |
| WN628C | 1600 | 2.81 × 10−8 | 3.2 |
| WN628D | 800 | 6.09 × 10−8 | 6.8 |
| WN628E | 800 | 1.55 × 10−8 | 1.7 |
(ii) Antagonistic pleiotropy.
It is possible that the decision to sporulate could be genetically encoded but that the gene(s) responsible is under another selective constraint in an opposing direction during another period in the life cycle. This seems very unlikely because the majority of sporulation genes are not expressed until growth has ceased and development has been initiated (6) and because all genes required for sporulation have recently been shown to be nonessential for viability and growth (10).
(iii) Sporulation efficiency is already optimized.
It is possible that the process of sporulation is already evolutionarily optimized in this specific strain of B. subtilis such that selection is unable to increase the frequency of sporulation. This does not seem likely because two natural isolates of B. subtilis, one obtained from the interior of Sonoran desert basalt (1) and the other isolated at NASA Jet Propulsion Laboratories (kindly provided by K. Venkateswaran), were tested for their efficiency of sporulation in SSM and showed average sporulation efficiencies of approximately 100 and 98%, respectively (data not shown). Thus, the laboratory strain of B. subtilis used for this study, with an average sporulation efficiency of 58%, is not optimized for the sporulation process, as relatives more recently isolated from the wild exhibit a higher overall sporulation efficiency.
We have demonstrated that B. subtilis populations evolving under strong selection for sporulation do not respond by increasing sporulation efficiency after approximately 5,000 generations of selection, representing more than 800 successive selective events. We propose that this is because the decision to embark on the sporulation pathway is mostly due to intracellular noise. From an evolutionary perspective, this phenomenon seems counterintuitive for the survival of bacterial populations, as they cannot respond to strong selective pressures for such traits. However, a stochastic element in developmental decision making may actually make evolutionary sense in clonal populations of cells capable of a number of response pathways for dealing with environmental stress; phenotypic diversification in this case would increase the probability that a subpopulation of cells would respond to the environment in a manner which increases the likelihood of survival and continued proliferation of the clone. This possible mechanism of adaptation makes sense for B. subtilis, which is normally found in the soil, where environmental conditions are in constant flux. The prevalence and importance of such random responses in nature have yet to be elucidated.
Acknowledgments
We thank C. W. Birky, Jr., B. Walsh, R. E. Lenski, B. Payseur, and L. Reed for helpful discussions about this work; K. Venkateswaran for generous donation of the NASA-SAF B. subtilis isolate; and P. Gerrish for help with fluctuation analysis.
H.M. was supported by an NSF Integrative Graduate Education and Research Traineeship Program Fellowship in Genomics and a research training grant from the Department of Ecology and Evolutionary Biology, University of Arizona.
REFERENCES
- 1.Benardini, J. N., J. Sawyer, K. Venkateswaran, and W. L. Nicholson. 2003. Spore UV and acceleration resistance of endolithic Bacillus pumilus and B. subtilis isolates obtained from Sonoran desert basalt: implications for lithopanspermia. Astrobiology **3:**709-718. [DOI] [PubMed]
- 2.Branda, S. S., J. E. Gonzalez-Pastor, B. Y. Sigal, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98**:**11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung, J. D., G. Stephanopoulos, K. Ireton, and A. D. Grossman. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 176**:**1977-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubnau, D., and C. M. Lovett, Jr. 2002. Transformation and recombination, p. 453-471. In A. L. Sonenshein. J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 5.Elowitz, M. B., A. J. Levine, E. D. Siggia, and P. S. Swain. 2002. Stochastic gene expression in a single cell. Science 297**:**1183-1186. [DOI] [PubMed] [Google Scholar]
- 6.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97**:**8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frederick, K., and J. D. Helmann. 1996. FlgM is a primary regulator of σD activity, and its absence restores motility to a sinR mutant. J. Bacteriol. 178**:**7010-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29**:**477-508. [DOI] [PubMed] [Google Scholar]
- 9.Hadden, C., and E. W. Nester. 1968. Purification of competent cells in the Bacillus subtilis transformation system. J. Bacteriol. 95**:**876-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100**:**4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin, M. D., C. J. Morton-Firth, W. N. Abouhamad, R. B. Bourret, and D. Bray. 1998. Origins of individual swimming behavior in bacteria. Biophys. J. 74**:**175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28**:**491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAdams, H. H., and A. Arkin. 1997. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA 94**:**814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirel, D. B., W. F. Estacio, M. Mathieu, E. Olmsted, J. Ramirez, and L. M. Márquez-Magaña. 2000. Environmental regulation of Bacillus subtilis σD-dependent gene expression. J. Bacteriol. 182**:**3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Msadek, T., F. Kunst, and G. Rappaport. 1993. Two-component regulatory systems, p. 729-745. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 16.Novick, A., and M. Weiner. 1957. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 43**:**553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozbudak, E. M., M. Thattai, I. Kurtser, A. D. Grossman, and A. van Oudenaarden. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31**:**69-73. [DOI] [PubMed] [Google Scholar]
- 18.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 19.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 20.Rao, C. V., D. M. Wolf, and A. P. Arkin. 2002. Control, exploitation and tolerance of intracellular noise. Nature 420**:**231-237. [DOI] [PubMed] [Google Scholar]
- 21.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20**:**4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54**:**704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solomon, J. M., and A. D. Grossman. 1996. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 12**:**150-155. [DOI] [PubMed] [Google Scholar]
- 24.Spudich, J. L., and D. E. Koshland, Jr. 1976. Non-genetic individuality: chance in the single cell. Nature 262**:**467-471. [DOI] [PubMed] [Google Scholar]
- 25.Sung, H. M., and R. E. Yasbin. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J. Bacteriol. 184**:**5641-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]