Mobile genetic elements of the human gastrointestinal tract: Potential for spread of antibiotic resistance genes (original) (raw)
Abstract
The human intestine is an important location for horizontal gene transfer (HGT) due to the presence of a densely populated community of microorganisms which are essential to the health of the human superorganism. HGT in this niche has the potential to influence the evolution of members of this microbial community and to mediate the spread of antibiotic resistance genes from commensal organisms to potential pathogens. Recent culture-independent techniques and metagenomic studies have provided an insight into the distribution of mobile genetic elements (MGEs) and the extent of HGT in the human gastrointestinal tract. In this mini-review, we explore the current knowledge of mobile genetic elements in the gastrointestinal tract, the progress of research into the distribution of antibiotic resistance genes in the gut and the potential role of MGEs in the spread of antibiotic resistance. In the face of reduced treatment options for many clinical infections, understanding environmental and commensal antibiotic resistance and spread is critical to the future development of meaningful and long lasting anti-microbial therapies.
Keywords: antibiotics resistance, mobile genetic elements, gut, gene transfer, microbiome
Introduction
It has long been recognized that the human intestine is a candidate location for horizontal gene transfer (HGT) between members of the gut microbial community with potential evolutionary and therapeutic implications.1-6 Culture-independent techniques which include metagenomic analysis and next generation sequencing approaches, combined with novel methods such as transposon-aided capture (TRACA),7 have provided a glimpse of the distribution of mobile genetic elements (MGEs) and the extent of HGT in the human gastrointestinal tract (GIT). The microbiota of the gut has been described as the equivalent of a virtual organ for the metabolic potential it provides to the host.8 In the years ahead, deeper sequencing of greater numbers of individuals will reveal further insights into the evolutionary contributions of MGEs in shaping the functional properties of the gut microbiota. The purpose of this review is to explore the current knowledge of MGEs in the GIT and to examine their potential role in the spread of antibiotic resistance.
Mobile Genetic Elements in the Gut
MGEs have been fundamental to the evolution and genetic variation of many living things including plants,9,10 animals and humans.11,12 On sequencing the human genome, over 200 genes were found to have significant homology to those encoding bacterial proteins indicating significant horizontal acquisition of bacterial genes during human evolution.13 In addition, the relevance of trans-kingdom HGT to the gut microbiota has been highlighted recently with the detection of eukaryotic genes for starch breakdown in various gut bacterial genomes.14 The significance of intra-Kingdom (bacterial) HGT in shaping the evolution of microbial communities in the gut was underlined recently through studies of gut bacteria isolated from Japanese populations consuming seaweed. In the GIT of some Japanese individuals, horizontally acquired genes from marine bacteria are utilized by gut Bacteroides for the breakdown of porphyrans from seaweed.15 Such bacteria are absent from the gut microbial communities of North American individuals and therefore indicate the adaptation of gut bacteria to host diet.15
In recognition of their ecological and evolutionary significance in nature, the database “A CLAssification of Mobile genetic Elements” (ACLAME) has been dedicated to the collection, classification and analysis of sequenced MGEs, particularly phage and plasmids,16 and should help toward the expansion of our knowledge of this elusive yet important group. An overview of the environmental and clinical relevance of some MGEs and their contributions in the gut microbial community follows.
Plasmids
Plasmid-mediated transfer of antibiotic resistance between bacteria is well established and plasmids have also been linked with resistance to toxic heavy metals such as mercury, cadmium, silver and pollutant breakdown.17 They are also known to encode genes for bacteriocin production, for utilization of additional nutrients such as sucrose, lactose and urea,18 for DNA repair and for proteins that govern the symbiotic relationship between prokaryotes and eukaryotes. A well-defined example of a role for plasmids in establishing symbiosis and infection exists in the Agrobacterium model where plasmid encoded traits influence survival and mediate inter-kingdom transfer of DNA from the bacteria to plant cells.19,20 Recent publications highlight known functions of plasmids in gut associated bacteria to include cell wall biosynthesis, redox balance,21 nutrient and carbohydrate utilization and bacteriocin production.4 Functions for establishment and survival such as adherence to epithelial cells and antibiotic resistance have been noted on plasmids in both pathogenic22 and commensal23 Escherichia coli in the gut. Such findings highlight two polar aspects of plasmid carriage in the gut. Furthermore, a recent study demonstrated in the murine model that pathogen-induced gut inflammation can cause population increases in Salmonella spp and E. coli which result in an increase in transfer of colicin-plasmid p2 from Salmonella spp to commensal E. coli. In the absence of inflammation, HGT was inhibited by the commensal microbiota. The authors suggest that inflammation could drive the HGT and re-assortment of plasmids between pathogens and commensals in the gut promoting the spread of antibiotic resistance, fitness and virulence associated traits.24
Plasmids play a central part in the clinical dissemination of antibiotic resistance genes between pathogens and have allowed for the rapid development of multi-antibiotic resistance. For most classes of antibiotics in clinical use, plasmid encoded resistance exists.25Multiple resistances have been spread in Vietnamese Salmonella spp by a conjugative plasmid carrying an integron26 and carbapenem resistance in Acinetobacter baumannii has been linked to a putatively conjugative plasmid.27 Additionally, recent outbreaks of resistant E. coli in Canada have suggested the possible conjugative transfer of carbapenem resistance between Klebsiella pneumonia and E. coli in hospital patients.28,29 It is clear that plasmids play a role in contributing to antibiotic resistance in the environment and in the clinical setting. However, the extent of their role in conferring antibiotic resistance to populations in the healthy GIT remains to be fully established. Continued research is needed to enhance knowledge of plasmid borne resistances in the healthy gut and the future implications of their transfer.
Conjugative Transposons (CTns)
In lactic acid bacteria, CTns are known to confer resistance to the antibiotics tetracycline, chloramphenicol, kanamycin and erythromycin, as well as genes for sucrose metabolism and nisin resistance.30 A classic example of antibiotic resistance being spread by mobile genetic elements is the CTn contribution to the loss of tetracycline as a treatment for opportunistic Bacteroides spp infections, as is their continued spread of resistance to other drugs for the treatment of such infections.31,32
The broad host range of some families of CTns (Tn_916_ and Tn_1549_) as well as the potential of those of Bacteroides spp to mobilise other MGEs in their proximity make them particularly relevant to HGT and adaptation of communities to life in the healthy gut. The suggestion that an enriched set of CTn-like genes, CTnRINT (CTn Rich in Intestine) is involved in HGT in the gut33 speaks to the evolutionary significance of CTns in the gut. Furthermore, a recent study uncovered a Tn_916_-likeCTn, Tn_6079_in the gut of a healthy infant. Tn_6079_ carries tetracycline (tetM and tetL) and erythromycin (ermT) resistance genes indicating the presence of antibiotic resistance genes and significant potential for HGT in early gut development.6 Additionally, reports by Brouwer and colleagues highlight the role of CTns in bacterial adaptation to the gut environment as they describe CTns of Clostridium difficile which carry antibiotic resistance and other genes thought to aid its adaptation to life in the gut.34,35
Bacteriophage
Metagenomic analysis of the Sargasso sea microbiome demonstrated the significant influence of viral (bacteriophage) horizontal gene transfer on marine microbial evolution and diversity.36 Similarly, upon examination of conserved functions in the gut microbial community, a small, but significant proportion (5%) of these functions involved pro-phage related proteins indicating that these are an essential part of the gut ecosystem.37 These may present structural and functional ecological determinants previously unknown to us.38 The gut specificity of certain bacteriophage isolated from human fecal material has been shown and reflected in their absence in the general environment as well as fecal samples from horses, sheep, pigs, cattle, rabbits and poultry.39 A similar scenario has been noted with plasmids from healthy feces.40 Indeed very recent analyses of Bacteroides spp specific bacteriophage φB124–14 and φB40–8 revealed that these are confined to the human gut. Ogilvie and colleagues (2012) found a variation in the distribution of these bacteriophage across individuals raising the possibility that these and other phage may influence inter-individual differences in the composition and function of the gut microbiota.41 The influence of bacteriophage in shaping the gut microbiota has been reviewed by Mills and coworkers.42
Transposons and Integrons
Transposase genes flanking putative antibacterial resistance genes in the genomes of bacteria such as Bordetella pertussis and Mycobacterium smegmatis suggest the HGT of the genes by transposons before speciation of the bacteria.43Transposons are commonly linked to clinical resistance in Enterobacteriaceae, indeed the suspected origin of the blaNDM-1 gene in pathogens is a composite transposon44,45.Recent experimental evidence has also suggested natural transformation to be a meaningful player in the dissemination of transposons and integrons within and between bacterial species.46 Integrons were first discovered in association with the spread of antibiotic resistance genes,47 are found associated with transposons, conjugative transposons and plasmids and have significant potential to spread antibiotic resistance genes. Approximately 100 resistance gene cassettes are known and these are related to many classes of antibiotics as well as antiseptics/disinfectants such as quaternary ammonium compounds.48 Additionally, the role of integrons has been extended with the discovery of chromosomal integron (CI) structures or superintegrons in the genomes of hundreds of bacterial species25,49.CIs harbouring multiple antibiotic resistance gene cassettes have been identified in Vibrio cholerae,49 highlighting how significant these elements can be to the evolution and adaptation of species to changing environmental conditions. This topic was recently reviewed by Hall (2012) in more depth50.
Antibiotic Resistance and Its Spread in the Gut
Naturally occurring antibiotic resistance
The recent discovery of antibiotic resistance genes in DNA recovered from 30,000 y old permafrost samples verified that antibiotic resistance is an ancient phenomenon that existed prior to the widespread clinical use of antibiotics.51 With this widespread antibiotic use came the challenge of addressing the spread of antibiotic resistance to clinical strains. Two years prior to FDA approval, a report of resistance to penicillin was published.52 Shortly after the first use of penicillin, resistant strains of Staphylococcus aureus emerged clinically. The situation was similar when second generation antibiotics were developed, with the rapid emergence of methicillin-resistant S. aureus (MRSA) and other resistant pathogens. Since the 1980s MRSA populations have risen, staphylococcal nosocomial blood infections have increased and community acquired MRSA as well as resistant Streptococcus pneumoniae are very real public health concerns.53 Within the last decade, vancomycin resistance has also emerged in S. aureus.54-56 We are facing the possible loss of treatment for some clinical infections with the onset of multi drug resistances in A. baumannii and K. pneumoniae rendering them almost untreatable.43,57 The recent emergence of Totally Drug Resistant strains of M. tuberculosis is also significant in this regard.58
During an investigation into the antibiotic resistome of the cultivable spore-forming Actinomycetes spp in soil, resistance to every class of antibiotics clinically available, synthetic and natural, was outlined.59 Included was resistance to daptomycin, an antibiotic that had just received FDA clearance. All isolates detected were multi-drug resistant. Such findings highlight the broad reservoirs of antibiotic resistance genes currently available in the natural environment and the problems they could cause should they become mobile.59 Indeed a recent publication has provided strong evidence that this scenario is already in play with identical mobile resistance determinants seen in commensal gut bacteria, soil microbes and clinical pathogens.60
Resilience of the human gut microbiome following antibiotic administration
Interesting findings around the resilience of human fecal communities following antibiotic treatment have emerged. Observations following ciprofloxacin administration indicated that the community structure of the feces returned to that measured pre-treatment. However, several taxa, most notably members of the Firmicutes did not return.61 Analysis of the effects of an antibiotic treatment commonly used for H. pylori therapy indicated that perturbation of the microbiota could persist for up to four years following antibiotic treatment. Significantly, at the four year time point in this study the authors could detect the persistence of the macrolide resistance gene _erm_B.62 Furthermore, a recent study demonstrated that infants administered ampicillin and gentamicin within 48hrs of birth exhibit a dramatically altered gut microbiome at 8 weeks of life with an over-representation of members of the Proteobacteria and a reduction in Bifidobacterium species.63 The study indicates the significant impact of early antibiotic treatment upon development of the gut microbiota. Although antibiotic resistance genes are not highly represented in the recently reported minimal genome or metagenome of the gut,37 it is clear from other reports that upon selection, a functional resistome is present in the healthy gut.64 In particular, the previously unknown resistance genes noted in the non-cultivable fraction of feces may account for the resilience of resident commensal populations in the wake of antibiotic treatment.
Influence of the Agricultural Use of Antibiotics on Antibiotic Resistance and Microbial Populations
There is evidence to support the link between widespread use of antibiotics in agriculture and the emergence of resistant microbes in humans and the environment.65 Half of US antibiotic use is in agriculture, animal husbandry and food production.66 Use of antibiotics in food production has resulted in the appearance of rising antibiotic resistance in food pathogens. For example, avoparcin was used as a feed additive in the poultry and pig industry during the 1980s. Such use is suspected to have led to the emergence of vancomycin resistant enterococci (VRE) in the guts of animals67and humans alike in Denmark.68 Lateral vanA gene transfer between animal and human enterococci has been suggested as contributory to clinical VRE.69 Multi-drug resistant Salmonellae spp are also suspected to have spread through food.70 It is thought that the use of tetracycline in the poultry industry may have led to the appearance of tetracycline resistant Campylobacter spp.71,72
Furthermore, a recent study by Looft and colleagues73 provided significant insight into the impact of antibiotic-supplemented feed upon the population dynamics of existing bacteria in the swine gut as well as the development of antibiotic resistance phenotypes in the porcine intestinal microbiota. A group of pigs were fed a diet containing performance-enhancing antibiotics [chlortetracycline, sulfamethazine and penicillin (known as ASP250)] and compared with a control group fed the same diet without antibiotics. Administration of antibiotics resulted in a substantial increase in members of the Proteobacteria in general and E. coli in particular with a general increase in genes associated with energy extraction. Interestingly, significant levels of antibiotic resistance genes were detected in the gut microbiota of control fed animals, in particular ermA, ermB, mefA, _tet_32 and aadA. However, antibiotic resistance genes were increased further in both richness and abundance through the administration of antibiotic-containing feed. Indeed antibiotic administration also enhanced the spread of resistance against agents that were not administered in the feed possibly indicating selection of genes that are adjacent to those conferring resistance to ASP250 antibiotics on transferrable elements.73 This study is key to our understanding of the effects agricultural antibiotic use is having on resistance gene spread.
Though antibiotic levels in foods are closely regulated, consuming foods containing low levels of antibiotics theoretically has the potential to impact the gut microbiota. Sub-therapeutic levels of antibiotics have also recently been shown to alter the colonic microbiome in nascent mice toward a population that enhances fat deposition in adult animals.74 The work raises the possibility that exposure to sub-therapeutic levels of antibiotics may have implications for the metabolic capacity of the host, influencing adiposity and other metabolic parameters. Indeed the same group performed a longitudinal study following the impact of early life exposure to antibiotics upon subsequent weight gain in humans.75 They found that infant exposure to antibiotics during the first 6 months of life (but not at later stages of infancy) was associated with increases in body mass at 10 to 38 months of age.75
Another recent study demonstrated the outgrowth of an antibiotic-resistant E. coli pathobiont in antibiotic-treated mice that gave rise to inflammation and sepsis in these animals.76 These studies suggest that low levels of antibiotics appearing via the food chain may influence metabolic and disease parameters in subjects via changes in the microbiota. While the data are compelling, more work is required to analyze exact levels of antibiotic exposure from natural sources and to focus upon larger cohorts in human studies. Furthermore, the role of HGT and antibiotic resistance genes in influencing surviving bacterial populations needs to be established. Guidelines for modern food production approaches which encourage lower antibiotic use in agriculture, animal husbandry and fish farming are to be welcomed.
Progress in the Study of Antibiotic Resistance and Its Spread in the Healthy Gut
Historically, studies into antibiotic resistance genes, mechanisms and spread were initially limited to clinical isolates. Subsequently work has progressed via the analysis of culturable populations of bacteria from the gut. More modern approaches have utilized functional and sequence-based metagenomic techniques to analyze the antibiotic resistome. In addition comparative sequence analyses of whole bacterial genomes enables identification of genes spread by HGT. If gene homologs exist on two very different bacterial genomes and are over 95% similar they may be deemed to have been acquired horizontally.77
Early studies of commensal gut antibiotic resistance investigated the resistances of the culturable facultative anaerobic fraction of the fecal microbiota,78-83 which accounts for 1% of the gut microbiota. In particular, a 1996 report focused on the resilience of antibiotic resistant Enterobacteriaceae in pediatric fecal samples. Over 13 weeks, samples were monitored for resistant Shigella spp and Klebsiella spp and E. coli. Trimethoprim, ampicillin and tetracycline resistant E. coli isolates were detected in the majority of donors throughout the duration of the study. E. coli isolates with other resistances were also detected, but in fewer donors and with seemingly transient colonisation. Klebsiella spp with resistance to trimethoprim, tetracycline, chloramphenicol, gentamicin and nitrofurnatoin were briefly observed. Resistant Shigella spp were noted in a minority of the children, one exhibiting tetracycline resistance, a second displaying trimethoprim resistance and a third with resistances to ampicillin, trimethoprim and tetracycline. The study confirmed commensal colonization with multi-drug resistant E. coli in healthy children.84
A study in 2000, from Finland, investigated the antimicrobial resistances of fecal Enterobacteriaceae in three groups of subjects; healthy people, short-term hospital patients and long-term hospital patients. Again in this study, E. coli were the most highly represented carriers of resistance in all three groups tested. In healthy people and short-term hospital patients, a general trend of resistance to more than two antibiotics was noted in E. coli and the levels of resistance detected were comparable to resistance levels linked to MGE’s. Significant resistances to streptomycin, sulfamethoxazole, streptomycin, ampicillin and trimethoprim were noted in E. coli isolated from the healthy group. Multidrug resistance was presumed transferable in this study and found to the same extent in the healthy and short-term hospital stay groups and to a higher degree in the long-term stay patients.85
A study in 2001, by Millar and coworkers, explored antibiotic resistances in the GIT of healthy children between 7 to 8 y and revealed pediatric oral carriage of Staphylococcus spp with resistance to chloramphenicol, tetracycline and methicillin as well as Hemophilus spp with resistance to ampicillin and erythromycin.86 Fecal carriage of Gram negative bacteria with resistance to chloramphenicol, ampicillin, spectinomycin and streptomycin was also detected. Notably, bacterial resistance to antibiotics that the children would not have had previous exposure to were detected, echoing the findings of previous studies.78,84 It was suggested that these bacteria may have been acquired through family, friends, pets or food.86 The study did not examine whether the source of the detected resistance was chromosomal or of mobile origin.
Deeper Insights into the Spread of Antibiotic Resistance in the Healthy Gut
A study in 2000, by Scott and colleagues, focused on tetracycline resistance and described the presence of the resistance gene teeth in two human gut commensals Faecalibacterium prausnitzii and Bifidobacterium longum.87 The teeth gene had been described previously in the rumen isolate, Butyrivibrio fibrisolvens, and had been shown to transfer between strains.88 Facultative anaerobes common to animals and man were the suggested transfer source of teeth between the rumen and human gut populations. The first report of mobile antibiotic resistance gene transfer from a gut commensal to a rumen isolate was furnished by Melville and coworkers in 2001.89 The group identified a new tetracycline resistance gene tet32 in the same anaerobic commensal gut isolate, F. prausnitzii mentioned above, and its transfer to the rumen isolate B. fibrisolvens by a possible MGE; however, the authors could not identify the element within the scope of the study. Emphasis was placed on the fact that although no solid evidence was available for the transfer between such commensals and pathogens, the same resistance genes can be found in commensal and pathogenic bacteria and could be horizontally spread.89
Culture independent techniques revealed a link between horizontal gene transfer and increased antibiotic resistance in gut Bacteroides spp over 30 years.90 Horizontally transferred alleles of tetQ were identified in different species of Bacteroides spp and it was confirmed that these alleles were transferred by a CTn among Bacteroides spp species in the gut. It was determined that ermF and ermG seem to be spreading in a similar manner, potentially through CTnDOT and CTn_7853_. It was concluded that the human colon is the only possible place such an increase in the spread of resistance could occur. Transfer of CTns is stimulated by tetracycline and it was suggested by the authors that increases in resistance seen in previous decades may have been triggered by environmental exposure to tetracycline. Nevertheless, lateral transfer of tetQ had already occurred in up to 30% of isolates before 1970.90
The healthy gut is both a source of antibiotic resistance genes and a location for their spread by CTns, as identical macrolide and tetracycline resistance genes can be seen in both Gram negative and Gram positive gut bacteria.1,77,91 Further evidence was furnished in 2008 by Scott and colleagues when they described the transfer of CTns carrying tetracycline resistance from rumen and human isolates to the gut commensal Roseburia inulinivorans. The study demonstrated that antibiotic conjugal gene transfer occurs between important human and gut microbes under conditions in vitro.92
Metagenomics Analyses of Antibiotic Resistance Elements in the Healthy Gut
The most comprehensive analysis to date of antibiotic resistance in the healthy gut was performed by Sommer and coworkers.93 In 2009 the group reported characterization of the antibiotic resistome in the human gut. Samples were obtained from two healthy human volunteers who had not taken antibiotics in the previous year. Employing metagenomic analysis alongside aerobic functional screens of saliva and fecal samples, a striking catalog of antibiotic resistance genes was compiled. Ninety five unique inserts conferring functional resistance to all 13 antibiotics used in the study were identified from a clone library constructed in E. coli. In general, sequence homology in GenBank was not above 70% for nucleotides and 65% for amino acids. One fifth of sequences did show high amino acid homology to the tetQ product mentioned above and the CTX-M-15 enzyme, a common broad spectrum β lactamase. Primarily the closest related homologs to inserts were from innocuous commensals such as Bifidobacterium longum, as well as those with a penchant for opportunistic pathogenesis such as Bacteroides fragilis and Bacteroides uniformis, including a gene from the latter that had not previously been characterized as a broad spectrum resistance gene. The majority of resistance genes identified in the healthy gut were related to those currently known to be carried in pathogens. However, 78 novel genes did not exhibit high homology to known proteins for resistance in pathogens, suggesting that these resistance genes are inaccessible to or rarely transferred to pathogens. The authors infer that given the functionality seen in E. coli, there must be a hindrance to the gene transfer itself or processes other than functional compatibility. Phylogenetic analysis of the inserts indicated that the sequences were from Bacteroidet_e_s and Firmicutes as would be expected from the gut community indicating that a good insight into antibiotic resistance in this population was gained.93
The authors also examined the facultative culturable fraction during this study. Phylogenetic analysis placed the 572 bacterial strains mainly into the Proteobacteria with a scattering of Firmicutes and Actinobacteria. Facultative isolates did not show resistance to all the antibiotics tested. Chloramphenicol and minocycline were the most common resistances in this group. An interesting finding was that 95% of 115 unique inserts with transferable resistance in the culturable fraction were over 90% similar at the nucleotide level to known pathogens, indicating an evolutionary relationship between the facultative and cultivable fraction of the healthy gut and clinical pathogens. A group of resistance genes with 100% homology to those of pathogens were a class of tetracycline efflux pumps (tetA), two classes of aminoglycoside-modifying enzymes and three classes of β lactamases (TEM, ampC and CTX-M).93 Additionally, in a recent report from Forsberg and colleagues outlining evidence for the horizontal transfer of antibiotic resistance genes between environmental soil bacteria and human pathogens, a link was made between _tet_A in the human GIT, soil bacteria and human pathogens.60
Recent comparative analysis of gut metagenomic data has reported enrichment of conjugative transposons,33 pro-phage related proteins37 and a plasmid gene subset40 in the healthy gut. Further studies have detailed HGT of specific genes. For example a study by Thornton and colleagues found two C10 cysteine protease genes located in the B. fragilis genome to be associated with a conjugative transposon-like element and a bacteriophage-like element, one of which showed potential for excision. Homologs of these genes were detected in both clinical isolates of B. fragilis and normal human fecal microbiota.94 Another example is teeth, a gene located on conjugative transposons of the Bacteroides species in the gut, but on an insertion sequence 21 (IS_21_) family transposon in the Prevotella species in the oral cavity.95 This observation not only indicates HGT, but also different routes of dissemination of the same resistance gene by HGT in two locations of the GIT.
Insights from the Human Microbiome Project and Antibiotic Resistance Genes Database (ARDB)
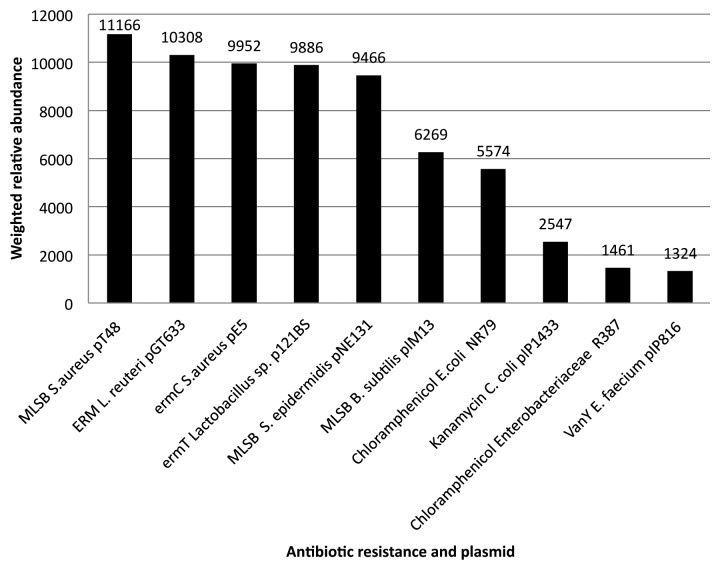
In June of 2012 we conducted comparative analysis of stool data sets from the Human Microbiome Project96 and antibiotic resistance genes annotated in the ARDB.97 Among proteins found to have the highest weighted relative abundance within all these data sets were; proteins for resistance to tetracycline in an uncultured bacterium and Prevotella copra and beta-lactam antibiotics in Bacteroides spa (Table 1). Furthermore, tetracycline resistance proteins of the TetX family related to Pseudomonas aeruginosa, Bacteroides thetaiotaomicron, Sphingobacterium spp, Bacteroides fragile and uncultured bacteria had high weighted relative abundance within 80% or more of these datasets (Table 2). MGE associated resistance determinants were detected in all HMP stool data sets with individual variations in carriage. A range of plasmid associated resistances were among these (Fig. 1). Macrolide resistance associated with plasmids of Staphylococcus, Lactobacillus and Bacillus plasmids scored highest in weighted relative abundance. Chloramphenicol resistance linked to Enterobacteriaceae plasmids, kanamycin resistance on a Campylobacter plasmid and vancomycin on an Enterococcus plasmid also scored high in weighted relative abundance in this group (Fig. 2). Conjugative transposons in Clostridium and Streptococcus species were linked to tetracycline and chloramphenicol carriage (authors` analysis of data from http://www.hmpdacc.org/HMGOI/ June 2012). These links suggest HGT has contributed to antibiotic resistance gene spread in the GIT.
Table 1. Most abundant resistance determinants represented in all data sets.
| Resistance determinant | Bacteria | Weighted relative abundance1 |
|---|---|---|
| Hypothetical protein PREVCOP_01173 | Prevotella copri DSM 18205 | 874781 |
| Tetracycline resistance protein Tet37 | uncultured bacterium | 410954 |
| Hypothetical protein BACOVA_05313 | Bacteroides ovatus ATCC 8483 | 393166 |
| Beta-lactamase | Bacteroides fragilis NCTC 9343 | 205868 |
| Beta-lactamase | Bacteroides fragilis | 205213 |
| Cephalosporinase | Bacteroides fragilis | 205213 |
| Beta-lactamase | Bacteroides fragilis YCH46 | 199943 |
| Class A β-lactamase CFXA3 | Bacteroides sp D1 | 160332 |
Table 2. Weighted relative abundance and bacterial associations of TetX, the most abundant resistance determinant in 80% or more HMP data sets.
| Resistance determinant | Bacteria | Weighted relative abundance1 |
|---|---|---|
| Tetracycline inactivating enzyme | Pseudomonas aeruginosa | 177876 |
| TetX2 protein | Bacteroides thetaiotaomicron | 177935 |
| Tetracycline resistance protein | Sphingobacterium sp PM2-P1–29 | 177674 |
| TetX transposon Tn4351/Tn4400. | Bacteroides fragilis | 177674 |
| TetX | uncultured bacterium HH1107 | 163932 |
Figure 1. Distribution of most abundant resistance plasmids across HMP stool data sets. E.coli NR79 (Chloramphenicol), Enterobacteriaceae R387 (Chloramphenicol), E. faecium pIP816 (vanY) were represented in all of the 146 stool data sets. Lactobacillus sp P121BS (ermT) and L. reuteri pGT633 (ERM) were noted in 90% or more of the stool data sets. S. aureus pT48 (MLSB), S. aureus pE5 (ermC) and B. subtilis pIM13 (MLSB) S. epidermidis pNE131 (MLSB) were detected in over 80% of the data sets.
Figure 2. Plasmids of highest weighted relative abundance in HMP stool data sets. The resistance plasmid with the most weighted relative abundance in the HMP stool data sets was found to be S.aureus pT48 (MLSB) and this was followed by L. reuteri pGT633 (ERM), S. aureus pE5 (ermC), Lactobacillus sp p121BS (ermT), S. epidermidis pNE131 (MLSB), B. subtilis pIM13 (MLSB), E.coli NR79 (Chloramphenicol), C. coli pIP1433 (Kanamycin), Enterobacteriaceae R387 (Chloramphenicol), E. faecium pIP816 (vanY).
Analysis of HGT in Maternal and Infant Subjects
Another metagenomic study recently published by de Vries and colleagues described tetracycline resistance in the feces of a mother and her one month old infant. In the mother’s gut, more genes were detected (tetO, tetW and tetX) in a greater variety of bacteria (Firmicutes and Bacteroidetes). Possible HGT was noted in the mothers’ gut as the same resistance genes were detected in unrelated bacteria. Transferable resistance was noted in the infant metagenomic library with the detection of a streptococcal CTn carrying tetM and tetL and ermT. Tetracycline resistance genes, tetO and tetW, were confirmed to be common to mother and infant by PCR analysis of infant total metagenomic fecal DNA. The study confirms the presence and potential for spread of antibiotic resistances in the human gut from its early development.6
A recent study by Morowitz and colleagues explored the composition of a pre-term infant gut community. Detection of phage and plasmids in the major populations, Citrobacter and Enterococcus, of the infant gut suggests that such MGEs are part of the community even from its earliest development. An interesting observation was that numerous mobile elements had corresponding dynamics to the minor populations of Klebsiella and Enterobacter. The finding indicates that the presence of MGE may not be limited to the most dominant populations in the early gut community.98 The study highlights that with deep enough sequencing the mobile metagenome of the gut can be accessed and annotated and should be considered as a tool for the furthering of knowledge in the field.
Conclusions
The emergence and spread of antibiotic resistance in clinically important pathogens has encouraged significant research into MGEs and the mechanisms of HGT which drive the spread of antibiotic resistance genes. Over the past 20 y, research into the human gut and the associated microbiota has demonstrated significant potential for the spread of MGEs in this important niche. HGT has been linked to adaptation to the effect of host diet and MGEs have been noted in the infant GIT. Furthermore, investigations by Sommer and colleagues in 2009,64 Forsberg and coworkers 60 and our own analysis of stool data from the Human Microbiome Project outlined here, link MGEs and HGT to antibiotic resistance determinants and their potential to spread from the mobile metagenome in the large intestine through the mechanisms of conjugation, transformation and natural transduction. Insights gained from these recent metagenomic studies enhance our understanding of the contribution made by HGT to the evolution of antibiotic resistance in the GIT. As we face the loss of viable treatments for some clinical infections, understanding commensal and environmental antibiotic resistances and their spread is critical to the future development of meaningful and long lasting anti-microbial therapies.
Acknowledgments
This work was supported by Science Foundation Ireland through the funding of the Alimentary Pharmabiotic Centre under the Centres for Engineering Science and Technology (CSET) program.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
References
- 1.Scott KP. The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell Mol Life Sci. 2002;59:2071–82. doi: 10.1007/s000180200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–6. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Jones BV. The human gut mobile metagenome: a metazoan perspective. Gut Microbes. 2010;1:415–31. doi: 10.4161/gmic.1.6.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schjørring S, Krogfelt KA. Assessment of bacterial antibiotic resistance transfer in the gut. Int J Microbiol. 2011;2011:312956. doi: 10.1155/2011/312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries LE, Vallès Y, Agersø Y, Vaishampayan PA, García-Montaner A, Kuehl JV, et al. The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS One. 2011;6:e21644. doi: 10.1371/journal.pone.0021644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 8.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, et al. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- 11.Iyer LM, Aravind L, Coon SL, Klein DC, Koonin EV. Evolution of cell-cell signaling in animals: did late horizontal gene transfer from bacteria have a role? Trends Genet. 2004;20:292–9. doi: 10.1016/j.tig.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Chénais B, Caruso A, Hiard S, Casse N. The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene. 2012;509:7–15. doi: 10.1016/j.gene.2012.07.042. [DOI] [PubMed] [Google Scholar]
- 13.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 14.Arias MC, Danchin EG, Coutinho P, Henrissat B, Ball S. Eukaryote to gut bacteria transfer of a glycoside hydrolase gene essential for starch breakdown in plants. Mob Genet Elements. 2012;2:81–7. doi: 10.4161/mge.20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 16.Leplae R, Lima-Mendez G, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 2010;38(Database issue):D57–61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas CM. Paradigms of plasmid organization. Mol Microbiol. 2000;37:485–91. doi: 10.1046/j.1365-2958.2000.02006.x. [DOI] [PubMed] [Google Scholar]
- 18.Mobley HLT, Chippendale GR, Fraiman MH, Tenney JH, Warren JW. Variable phenotypes of Providencia stuartii due to plasmid-encoded traits. J Clin Microbiol. 1985;22:851–3. doi: 10.1128/jcm.22.5.851-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridé M, Ridé S, Petit A, Bollet C, Dessaux Y, Gardan L. Characterization of plasmid-borne and chromosome-encoded traits of Agrobacterium biovar 1, 2, and 3 strains from France. Appl Environ Microbiol. 2000;66:1818–25. doi: 10.1128/AEM.66.5.1818-1825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-de los Santos A, Brom S, Romero D. Rhizobium plasmids in bacteria-legume interactions. World J Microbiol Biotechnol. 1996;12:119–25. doi: 10.1007/BF00364676. [DOI] [PubMed] [Google Scholar]
- 21.Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren JP, Cerdeño-Tárraga AM, et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc Natl Acad Sci U S A. 2006;103:6718–23. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weintraub A. Enteroaggregative Escherichia coli: epidemiology, virulence and detection. J Med Microbiol. 2007;56:4–8. doi: 10.1099/jmm.0.46930-0. [DOI] [PubMed] [Google Scholar]
- 23.Oshima K, Toh H, Ogura Y, Sasamoto H, Morita H, Park SH, et al. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 2008;15:375–86. doi: 10.1093/dnares/dsn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109:1269–74. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008;153(Suppl 1):S347–57. doi: 10.1038/sj.bjp.0707607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vo AT, van Duijkeren E, Gaastra W, Fluit AC. Antimicrobial resistance, class 1 integrons, and genomic island 1 in Salmonella isolates from Vietnam. PLoS One. 2010;5:e9440. doi: 10.1371/journal.pone.0009440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Yang ZL, Wu XM, Wang Y, Liu YJ, Luo H, et al. Complete genome sequence of Acinetobacter baumannii MDR-TJ and insights into its mechanism of antibiotic resistance. J Antimicrob Chemother. 2012;67:2825–32. doi: 10.1093/jac/dks327. [DOI] [PubMed] [Google Scholar]
- 28.Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg Infect Dis. 2011;17:103–6. doi: 10.3201/eid1701.101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borgia S, Lastovetska O, Richardson D, Eshaghi A, Xiong J, Chung C, et al. Outbreak of carbapenem-resistant enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin Infect Dis. 2012;55:e109–17. doi: 10.1093/cid/cis737. [DOI] [PubMed] [Google Scholar]
- 30.Mathur S, Singh R. Antibiotic resistance in food lactic acid bacteria--a review. Int J Food Microbiol. 2005;105:281–95. doi: 10.1016/j.ijfoodmicro.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Salyers AA, Shoemaker NB, Stevens AM, Li LY. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–90. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–10. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 33.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–81. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One. 2011;6:e23014. doi: 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brouwer MS, Roberts AP, Mullany P, Allan E. In silico analysis of sequenced strains of Clostridium difficile reveals a related set of conjugative transposons carrying a variety of accessory genes. Mob Genet Elements. 2012;2:8–12. doi: 10.4161/mge.19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, et al. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vacharaksa A, Finlay BB. Gut microbiota: metagenomics to study complex ecology. Curr Biol. 2010;20:R569–71. doi: 10.1016/j.cub.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Ebdon J, Muniesa M, Taylor H. The application of a recently isolated strain of Bacteroides (GB-124) to identify human sources of faecal pollution in a temperate river catchment. Water Res. 2007;41:3683–90. doi: 10.1016/j.watres.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 40.Jones BV, Sun F, Marchesi JR. Comparative metagenomic analysis of plasmid encoded functions in the human gut microbiome. BMC Genomics. 2010;11:46. doi: 10.1186/1471-2164-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogilvie LA, Caplin J, Dedi C, Diston D, Cheek E, Bowler L, et al. Comparative (meta)genomic analysis and ecological profiling of human gut-specific bacteriophage φB124-14. PLoS One. 2012;7:e35053. doi: 10.1371/journal.pone.0035053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–86. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 44.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18:263–72. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 46.Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012;8:e1002837. doi: 10.1371/journal.ppat.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, et al. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol. 2008;190:5095–100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–84. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 49.Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003;13:428–42. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall RM. Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci. 2012;1267:71–8. doi: 10.1111/j.1749-6632.2012.06588.x. [DOI] [PubMed] [Google Scholar]
- 51.D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 52.Abraham E, Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;146:837. doi: 10.1038/146837a0. [DOI] [PubMed] [Google Scholar]
- 53.Cars O, Nordberg P. Antibiotic resistance – The faceless threat. International journal of risk and safety in medicine. 2005;17:103–10. [Google Scholar]
- 54.Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, et al. Vancomycin-Resistant Staphylococcus aureus Investigative Team Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003;348:1342–7. doi: 10.1056/NEJMoa025025. [DOI] [PubMed] [Google Scholar]
- 55.Sievert DM, Rudrik JT, Patel JB, McDonald LC, Wilkins MJ, Hageman JC. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin Infect Dis. 2008;46:668–74. doi: 10.1086/527392. [DOI] [PubMed] [Google Scholar]
- 56.Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;3:e00112–12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D, Participants of the 3rd World Healthcare-Associated Infections Forum Ready for a world without antibiotics? The Pensières Antibiotic Resistance Call to Action. Antimicrob Resist Infect Control. 2012;1:11. doi: 10.1186/2047-2994-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller B, Borrell S, Rose G, Gagneux S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 2012;29:160–9. doi: 10.1016/j.tig.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–7. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 60.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–11. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. 2012;56:5811–20. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazimierczak KA, Rincon MT, Patterson AJ, Martin JC, Young P, Flint HJ, et al. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob Agents Chemother. 2008;52:4001–9. doi: 10.1128/AAC.00308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lipsitch M, Singer RS, Levin BR. Antibiotics in agriculture: when is it time to close the barn door? Proc Natl Acad Sci U S A. 2002;99:5752–4. doi: 10.1073/pnas.092142499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heuer OE, Pedersen K, Jensen LB, Madsen M, Olsen JE. Persistence of vancomycin-resistant enterococci (VRE) in broiler houses after the avoparcin ban. Microb Drug Resist. 2002;8:355–61. doi: 10.1089/10766290260469624. [DOI] [PubMed] [Google Scholar]
- 68.Hammerum AM, Lester CH, Neimann J, Porsbo LJ, Olsen KE, Jensen LB, et al. A vancomycin-resistant Enterococcus faecium isolate from a Danish healthy volunteer, detected 7 years after the ban of avoparcin, is possibly related to pig isolates. J Antimicrob Chemother. 2004;53:547–9. doi: 10.1093/jac/dkh101. [DOI] [PubMed] [Google Scholar]
- 69.Kühn I, Iversen A, Finn M, Greko C, Burman LG, Blanch AR, et al. Occurrence and relatedness of vancomycin-resistant enterococci in animals, humans, and the environment in different European regions. Appl Environ Microbiol. 2005;71:5383–90. doi: 10.1128/AEM.71.9.5383-5390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo FJ. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–8. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 71.Wardak S, Szych J, Zasada AA, Gierczynski R. Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrob Agents Chemother. 2007;51:1123–5. doi: 10.1128/AAC.01187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rozynek E, Dzierzanowska-Fangrat K, Korsak D, Konieczny P, Wardak S, Szych J, et al. Comparison of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from humans and chicken carcasses in Poland. J Food Prot. 2008;71:602–7. doi: 10.4315/0362-028x-71.3.602. [DOI] [PubMed] [Google Scholar]
- 73.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A. 2012;109:1691–6. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trasande L, Blustein J, Liu M, Corwin E, Cox LM, Blaser MJ. Infant antibiotic exposures and early-life body mass. Int J Obes (Lond) 2013;37:16–23. doi: 10.1038/ijo.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–6. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Moorhouse EC. Transferable drug resistance in enterobacteria isolated from urban infants. Br Med J. 1969;2:405–7. doi: 10.1136/bmj.2.5654.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Linton KB, Lee PA, Richmond MH, Gillespie WA, Rowland AJ, Baker VN. Antibiotic resistance and transmissible R-factors in the intestinal coliform flora of healthy adults and children in an urban and a rural community. J Hyg (Lond) 1972;70:99–104. doi: 10.1017/S0022172400022130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levy SB, Marshall B, Schluederberg S, Rowse D, Davis J. High frequency of antimicrobial resistance in human fecal flora. Antimicrob Agents Chemother. 1988;32:1801–6. doi: 10.1128/AAC.32.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lester SC, del Pilar Pla M, Wang F, Perez Schael I, Jiang H, O’Brien TF. The carriage of Escherichia coli resistant to antimicrobial agents by healthy children in Boston, in Caracas, Venezuela, and in Qin Pu, China. N Engl J Med. 1990;323:285–9. doi: 10.1056/NEJM199008023230501. [DOI] [PubMed] [Google Scholar]
- 82.Bonten M, Stobberingh E, Philips J, Houben A. Antibiotic resistance of Escherichia coli in fecal samples of healthy people in two different areas in an industrialized country. Infection. 1992;20:258–62. doi: 10.1007/BF01710790. [DOI] [PubMed] [Google Scholar]
- 83.London N, Nijsten R, van der Bogaard A, Stobberingh E. Carriage of antibiotic-resistant Escherichia coli by healthy volunteers during a 15-week period. Infection. 1994;22:187–92. doi: 10.1007/BF01716700. [DOI] [PubMed] [Google Scholar]
- 84.Calva JJ, Sifuentes-Osornio J, Cerón C. Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob Agents Chemother. 1996;40:1699–702. doi: 10.1128/aac.40.7.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Österblad M, Hakanen A, Manninen R, Leistevuo T, Peltonen R, Meurman O, et al. A between-species comparison of antimicrobial resistance in enterobacteria in fecal flora. Antimicrob Agents Chemother. 2000;44:1479–84. doi: 10.1128/AAC.44.6.1479-1484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Millar MR, Walsh TR, Linton CJ, Zhang S, Leeming JP, Bennett PM, ALSPAC Study Team. Avon Longitudinal Study of Pregnancy and Childhood Carriage of antibiotic-resistant bacteria by healthy children. J Antimicrob Chemother. 2001;47:605–10. doi: 10.1093/jac/47.5.605. [DOI] [PubMed] [Google Scholar]
- 87.Scott KP, Melville CM, Barbosa TM, Flint HJ. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother. 2000;44:775–7. doi: 10.1128/AAC.44.3.775-777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scott KP, Barbosa TM, Forbes KJ, Flint HJ. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1997;63:3405–11. doi: 10.1128/aem.63.9.3405-3411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melville CM, Scott KP, Mercer DK, Flint HJ. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob Agents Chemother. 2001;45:3246–9. doi: 10.1128/AAC.45.11.3246-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol. 2001;67:561–8. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whittle G, Shoemaker NB, Salyers AA. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell Mol Life Sci. 2002;59:2044–54. doi: 10.1007/s000180200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scott KP, Martin JC, Mrazek J, Flint HJ. Transfer of conjugative elements from rumen and human Firmicutes bacteria to Roseburia inulinivorans. Appl Environ Microbiol. 2008;74:3915–7. doi: 10.1128/AEM.02807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thornton RF, Kagawa TF, O’Toole PW, Cooney JC. The dissemination of C10 cysteine protease genes in Bacteroides fragilis by mobile genetic elements. BMC Microbiol. 2010;10:122. doi: 10.1186/1471-2180-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tribble GD, Garza JJ, Yeung VL, Rigney TW, Dao DH, Rodrigues PH, et al. Genetic analysis of mobile tetQ elements in oral Prevotella species. Anaerobe. 2010;16:604–9. doi: 10.1016/j.anaerobe.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Consortium HMP, Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu B, Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37(Database issue):D443–7. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, Relman DA, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci U S A. 2011;108:1128–33. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]