Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma (original) (raw)
. Author manuscript; available in PMC: 2014 Aug 15.
Abstract
A comprehensive description of genomic alterations in lung squamous cell carcinoma (lung SqCC) has recently been reported, enabling the identification of genomic events that contribute to the oncogenesis of this disease. In lung SqCC, one of the most frequently altered receptor tyrosine kinase families is the fibroblast growth factor receptor (FGFR) family, with amplification or mutation observed in all four family members. Here, we describe the oncogenic nature of mutations observed in FGFR2 and FGFR3, which are each observed in 3% of samples, for a mutation rate of 6% across both genes. Using cell culture and xenograft models, we show that several of these mutations drive cellular transformation. Transformation can be reversed by small molecule FGFR inhibitors currently being developed for clinical use. We also show that mutations in the extracellular domains of FGFR2 lead to constitutive FGFR dimerization. Additionally, we report a patient with an _FGFR2_-mutated oral squamous cell carcinoma who responded to the multi-targeted tyrosine kinase inhibitor pazopanib. These findings provide new insights into driving oncogenic events in a subset of lung squamous cancers, and recommend future clinical studies with FGFR inhibitors in patients with lung and head and neck SqCC.
Keywords: Squamous cell lung cancer, Fibroblast growth factor receptors, tyrosine kinase inhibitors, lung cancer genomics
Introduction
Two goals of comprehensive next-generation sequencing of cancers are to discover novel, targetable somatic alterations, and to identify new targets for which therapies already exist. Genome-scale analyses of tumors representing many cancer types have recently been completed (1–6), enabling discoveries consistent with both goals.
Historically, targetable oncogenic alterations in cancer were discovered on an individual gene basis. This was the case for cancer-causing alterations observed in several tyrosine kinases, including EGFR and ALK in lung adenocarcinoma (7–9), FGFR2 in endometrial carcinoma (10, 11), and FGFR3 in urothelial carcinoma (12). These studies and others have led to demonstrations of the successful application of targeted therapeutic agents and their superiority to conventional chemotherapy (13, 14).
Lung squamous cell carcinoma (lung SqCC) is a prevalent and deadly disease for which no targeted therapies are approved. Recent data reported by The Cancer Genome Atlas (TCGA) lung SqCC project (4) demonstrated that the Fibroblast Growth Factor Receptor (FGFR) tyrosine kinases are one of the most frequently altered kinase families in this disease. Amplification of FGFR1 was observed, in agreement with prior reports (15, 16). Furthermore, mutations in FGFR2 and FGFR3 were reported. While the frequency of these mutations did not reach statistical significance at the cohort size examined by TCGA, several features including recurrence, prior observation in other cancer types and congenital syndromes, and lack of other dominant oncogenic alterations in tumors with FGFR mutations, suggested they might be driving, targetable events in a subset of patients presenting with this disease.
Germline mutations in the FGFR tyrosine kinase family were first described in craniofacial and skeletal syndromes (17). Somatic point mutations identical to those germline events have also been observed in malignancies (18). The FGFR family is made up of four active members that each contain an extracellular domain (ECD) and a cytoplasmic kinase domain. Activation is stimulated by binding fibroblast growth factor (FGF) and heparan sulfate proteoglycan (HSPG) in the ECD, and subsequent dimerization of two receptor-ligand complexes, leading to transphosphorylation of the kinase domains. This leads to phosphorylation of binding partner FRS2 and downstream activation of Ras/MAPK and PI3K/AKT pathways (19).
The FGF family is made up of more than 20 members, all of which retain specificities for both different FGFR family members and different isoforms of each receptor (20). In addition, tissue types vary in which receptors, isoforms, and ligands are expressed, adding further levels of complexity to the system. Dysregulation can lead to oncogenesis, as has been shown with altered expression of receptors (15, 16, 21), altered isoform expression (22, 23), and altered ligand specificity (24) driven by somatic genomic events.
Aberrant FGFR signaling has been implicated in the development of several cancer types. In addition to lung SqCC, FGFR1 amplification is observed in 10% of breast cancers (21). Point mutations in FGFR2 are observed in 12% of endometrial carcinomas (10) and mutations in FGFR3 are observed in more than 30% of urothelial carcinomas (12). Cell lines harboring these events have demonstrated sensitivity to inhibition by FGFR small molecule inhibitors, and clinical trials are now testing FGFR inhibitors in patients harboring somatic events in FGFRs (18).
Here, we characterize FGFR2 and FGFR3 mutations observed in lung SqCC and demonstrate the oncogenic potential of these mutations using models of transformation and dependency. We demonstrate that cells harboring these mutations are sensitive to inhibition by several FGFR and multi-kinase inhibitors. In addition, we report a case of a patient with an _FGFR2_-mutated oral squamous cell carcinoma, who responded to pazopanib, an inhibitor of multiple tyrosine kinases including the FGFR family. Together, these data identify a promising new therapeutic target for patients with lung SqCC and other squamous epithelial tumors.
Methods
Patient samples and genomic analysis
We manually reviewed FGFR2 and FGFR3 exome sequencing data generated by the TCGA research network. Additionally, we queried publically available sequencing data generated from 18 samples that were excluded from the initial TCGA report. All data were de-identified and obtained in accordance with patient protection standards set by the TCGA and were obtained from the TCGA Data Portal.
For the individual with a clinical response to pazopanib, total RNA was extracted using the AllPrep DNA/RNA Mini Kit (Qiagen #80204). Poly-adenylated mRNA was enriched using the Ambion MicroPoly(A)Purist kit starting from 30 μg of total RNA as an input according to the manufacturer’s protocol.
Illumina transcriptome sequencing libraries were prepared as previously described (25) from mRNA and from total RNA and were subjected to 76 bp paired-end sequencing on a single lane of an Illumina GAIIx sequencer. Sequencing reads were first aligned to all curated protein-coding transcripts and were mapped back to reference human genome, hg18 as previously described (25). Potential mutations were called using the Unified genotyper from the GATK tool (26).
This individual was consented for the analysis according to Institutional Protocol 94138 at the Dana-Farber Cancer Institute. The FGFR2 P253R mutation was found in both the total RNA-seq data and mRNA-seq data, and it was confirmed from genomic DNA by Sanger sequencing in a CLIA-certified laboratory.
Cell lines, antibodies, ligands, and inhibitors
NIH-3T3 cells and Ba/F3 cells were obtained from the American Type Culture Collection and maintained as described previously (10, 20). Antibodies against FGFR2 (C-8) and FRS2 (H-91) were purchased from Santa Cruz Biotechnology, Inc. Antibodies against FGFR3 (C51F2), p-FGFR, p-FRS2 (Y436), AKT (C67E7), p-AKT (T308, 244F9), Erk 1/2 (137F5), p-Erk 1/2 (E10), and beta-actin (8H10D10) were obtained from Cell Signaling Technology, Inc.
For FGFR stimulation experiments, the FGF1 ligand was obtained from Abcam. FGF7 and FGF9 were obtained from Life Technologies. Interleukin-3 (IL-3) was purchased from VWR and heparin from StemCell Technologies, Inc.
Ponatinib (AP24534), dovitinib (TKI258), and cediranib (AZD2171) were obtained from Selleck Chemicals. Brivanib alaninate (BMS-582664) was obtained from Fischer Scientific. Pazopanib (GW786034) was obtained from Axon Medchem. AZD4547 was obtained from Active Biochem. E7080 was obtained from American Custom Chemicals Corporation. BGJ398 was a gift from Novartis Pharmaceuticals Corporation (Basel, CH).
Mutagenesis and cellular transfection and infection
Mutagenesis primers developed for each mutation were generated using the Agilent QuikChange Primer Design tool. FGFR2 isoforms IIIb and IIIc, and FGFR3 isoform IIIc were cloned into pDONR223 and mutated by site-directed mutagenesis with the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies. Sequence-verified constructs were cloned into pBabe-puro and transfected into HEK-293T cells with Fugene-6 (Promega) as described previously (10). NIH-3T3 and Ba/F3 cells were infected with the resulting virus and after two days and cells were selected with 2 μg/mL puromycin.
Western blot analysis and visualization of unreduced dimers
Cells were lysed in buffer containing 0.5% NP-40, 50 mM Tris pH 8, 150 mM MgCl2, and phosphatase and protease inhibitors, and proteins were separated by SDS/PAGE and transferred to nitrocellulose membranes via the iBlot dry transfer system (Invitrogen). Antibody binding was detected using the LI-COR Odyssey IR imaging system (LI-COR Biosciences).
To visualize receptor dimers formed by extracellular domain mutations to cysteine residues, NIH-3T3 cells expressing the appropriate mutations were serum starved for eight hours in the presence of PBS or FGF1 and heparin, washed with PBS containing 10mM iodoacetamide, and lysed in lysis buffer containing 1% Triton, 10% glycerol, 50mM Tris pH 7.4, and 10 mM iodoacetamide. Two 100 μg aliquots of each protein sample were prepared, one with reducing agent and one without. Electrophoresis was performed using 4–12% Tris-glycine SDS/PAGE gels (Invitrogen)
To confirm loss of phosphorylation of relevant kinases in the presence of inhibitor, NIH-3T3 cells expressing mutated FGFR2 or FGFR3 were washed with PBS, serum starved for four hours in the presence of indicated concentrations of inhibitor, and ligand stimulated with FGF1 for 30 minutes before lysis.
Soft agar colony formation assays
Two mL of 0.5% Select agar (Gibco) and media were plated to each well of a non-tissue culture-treated 6-well plate and allowed to solidify. 5×104 cells were suspended in 330 μL media and mixed with 770 μL 0.5% Select agar and media and plated onto the solidified bottom layer in triplicate. Plates were incubated for three weeks, photographed using QuickCapture (Logitech), and quantified via ImageJ for colony formation. Statistical comparison was performed using the Student’s t-test.
To evaluate the effect of clinical inhibitors on soft agar colony formation, the above protocol was performed with the following alteration: 5×104 cells were suspended in 330 μL media plus relevant inhibitor prior to addition of 0.5% agar solution and plating.
Xenograft studies
All animal experiments were performed according to institutional guidelines regarding animal safety. Immuno-compromised mice were injected with NIH-3T3 cells stably expressing exogenous FGFR2-IIIb WT, W290C, S320C or K660N mutant isoforms. Cohorts of 7 mice were injected at 3 sites for each cell type with two million cells per site, and mice were observed until tumor volume reached 200–300 mm3. Mice were then treated with BGJ-398 at 15 mg/kg or vehicle (PEG-300) control daily for 2 weeks, and tumor size was measured during the treatment period.
Ba/F3 dependency and inhibitor studies
Ba/F3 cells expressing each mutation construct were selected in media containing IL-3 and puromycin. To establish cells dependent on FGFR signaling, three million cells were washed twice with PBS and seeded into 2 mL of media containing FGF7 (for FGFR2 IIIb) or FGF9 (for FGFR2 IIIc) and heparin. These cells were maintained until IL-3 independent cells emerged. 5000 FGFR-dependent cells per well were seeded into 96-well plates in 100 μL media containing FGF and heparin. 10 μL drug was added in quadruplicate for final concentrations of 0.3nM-10 μM in half logs, with two DMSO controls, and incubated for three or four days. 50 μL Cell Titer Glo (Promega) was added to each well and luminescence was measured on the SpectroMax 5 imager. Percent survival compared to DMSO controls was calculated and plotted in Prism (GraphPad Software, Inc).
Results
FGFR2 and FGFR3 are recurrently mutated in lung squamous cell carcinoma
We analyzed whole-exome sequencing data generated by The Cancer Genome Atlas (4), for mutations in the FGFR2 and FGFR3 genes. We identified 5 FGFR2 and 6 FGFR3 mutations in analysis of exome sequencing data of 178 tumor/normal pairs, as well as an FGFR2 K660N mutation in a sample that was excluded from the TCGA report due to poor RNA quality (TCGA-21-1083), for a total of 12 mutations.
Patients in the reported TCGA cohort with FGFR mutations (n=10, as one subject had two FGFR mutations) ranged from 58 to 81 years old with a median age of 73. All patients were current or former smokers with a pack year history of 9 to 63 pack years (median 49). Tumors were obtained from resected specimens with a T stage of 1 (n=3) or 2 (n=7) and N stage of 0 (n=8) or 1 (n=2). More extensive patient data are available in Table S1.
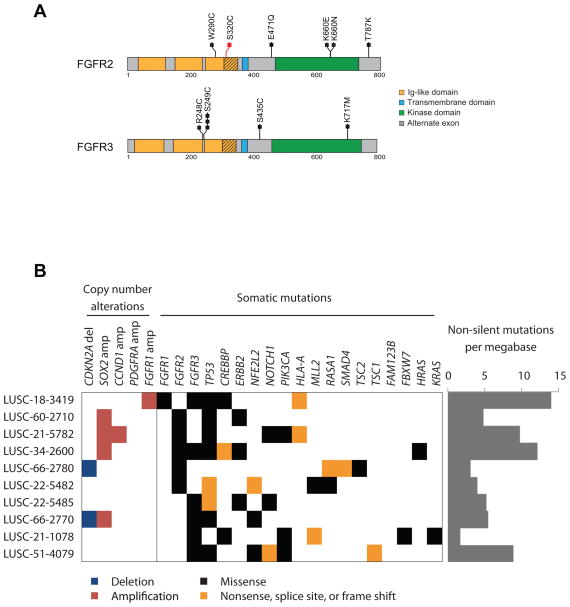
The observed mutations fell in both the extracellular and kinase domains of FGFR2 and FGFR3, both in codons in which mutations have been previously reported in endometrial carcinoma (10, 11) and urothelial carcinoma (12), and at novel residues (Figure 1A). In the samples containing FGFR2 or FGFR3 mutations, the IIIb isoforms of each protein were overexpressed compared to the IIIc isoforms (Figure S1). FGFR kinase alterations were significantly enriched in the basal expression subtype (27) (Fisher’s Exact test; _p_=0.016, Figure S1).
Figure 1.
Recurrent mutations in FGFR2 and FGFR3 are observed in lung squamous cell carcinoma. (A) Sequencing data from TCGA were analyzed and recurrent mutations were observed in FGFR2 and FGFR3. The mutation S320C in FGFR2, in red, is located in the alternatively spliced exon in the IG-3 domain of FGFR2 IIIb; the remaining mutations are annotated to the IIIc isoform. FGFR3 mutations are annotated in the IIIc isoform. (B) Co-occurring somatic copy number alterations and mutations in samples with mutation.
FGFR mutations co-occurred with mutations in known oncogenes in only three cases. LUSC-21-1078 had a high somatic mutation rate and harbored mutations in HRAS at codon 61 and PDGFRA at codon 842, both previously reported to be sites of oncogenic mutation, as well as a novel ERBB2 E1021Q mutation (Figure 1B). LUSC-21-1078 contained a non-canonical KRAS mutation G118S and LUSC-21-5485 had a previously unreported ERBB2 mutation G1075V. Other samples contained no known oncogenic somatic mutations, except that FGFR2 and FGFR3 mutations commonly co-occurred with mutations in TP53 (8/10) and PIK3CA (3/10), the latter a gene with mutations that commonly co-occur with driving oncogenes. Four of ten samples with FGFR mutation harbored 3q amplification of SOX2 and two samples CDKN2A homozygous deletion (Figure 1B). The presence of these events may suggest that FGFR mutations are not solely driving oncogenesis; however, due to the high heterogeneity observed in the lung SqCC samples, even the presence of other known oncogenic events does not guarantee that events co-occur in cells or that subsets of tumor cells would not be sensitive to FGFR targeted therapy.
FGFR2 and FGFR3 mutations drive anchorage-independent growth of NIH-3T3 cells
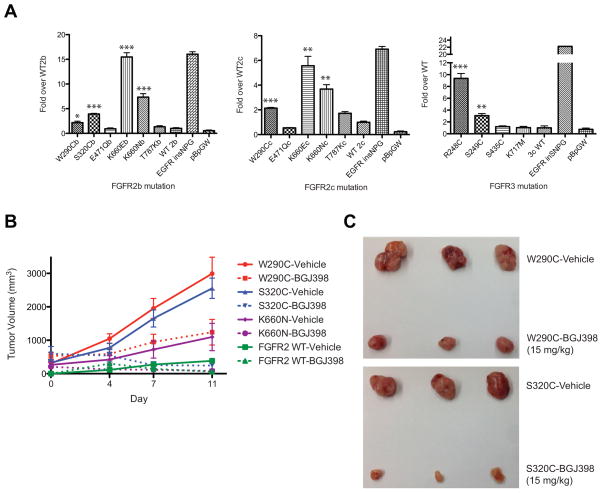
To determine whether the mutations identified in lung SqCC were oncogenic, we established NIH-3T3 cells stably expressing each mutation to assess anchorage-independent growth in soft agar. We observed colony formation in cells expressing the majority of observed FGFR2 and FGFR3 mutations (Figure 2A). We determined that extracellular domain mutations W290C and S320C in FGFR2, and R248C and S249C in FGFR3, significantly increased colony formation compared to wild type FGFR2 or FGFR3, as did kinase domain mutations K660E and K660N in FGFR2 (p<0.05 by Student’s t-test). In contrast, FGFR2 mutations E471Q and T787K, and FGFR3 mutations S435C and K717M did not form colonies above wild type. Robust formation of colonies was observed in NIH-3T3 cells expressing an activating EGFR insertion mutation. FGFR2 mutations were generated in both common isoforms of FGFR2 with similar results obtained for all assayed mutations with the exception of FGFR2 T787K, which was very modestly transforming only in isoform IIIc (Figure 2A).
Figure 2.
A subset of lung SqCC mutations in FGFR2 and FGFR3 are transforming in anchorage independent growth assays and xenograft assays. (A) Colony formation compared to wild type in NIH-3T3 cells expressing FGFR mutations was calculated for each isoform and graphed. EGFR insNPG was included as a positive control, and the pBabe-puro Gateway empty vector (pBp GW) was included as a negative control. P-values were calculated with the student’s t-test and significance is indicated by asterisks; * < 0.05, ** < 0.01, *** < 0.001. (B) Nude mice injected with transforming FGFR2 mutant cells from (A) developed tumors, which were treated with BGJ398 (dashed lines) or vehicle (solid lines). (C) Tumors were dissected from the mice for visual inspection comparing treatment with vehicle or drug. Top panel, FGFR2-W290C tumors; bottom panel, FGFR2-S320C. Tumor images corresponding to FGFR2-K660N and FGFR2-WT tumors are shown in Figure S2.
FGFR2 and FGFR3 mutations drive tumor formation in xenograft models
NIH-3T3 cells expressing transforming FGFR2 mutations or wild type were injected into nude mice. Tumors reached approximately 200–300 mm3 in all mice injected with mutant cells by day 13 and began treatment with a pan-FGFR inhibitor, BGJ398 (28), or vehicle, with ECD mutations driving particularly strong tumor formation (Figure 2B, solid lines). Tumors formed by cells expressing wild type FGFR2 grew more slowly, and began treatment on day 16 (Figure 2B).
Tumors treated with BGJ398 slowed or reversed their growth compared to vehicle (Figure 2B, dashed lines), so that by the end of the study, tumor burden in vehicle-treated versus BGJ398-treated mice was noticeably distinct (Figure 2C, Figure S2).
Extracellular domain mutations form ligand sensitive intermolecular disulfide bonds
A common mechanism of activation of the FGFR2 and FGFR3 kinases is through the formation of covalently bound receptor dimers (29, 30). While wild type RTKs maintain extracellular structure required for ligand binding and receptor dimerization through intramolecular disulfide bonds, mutant receptors can form intermolecular disulfide dimers due through a novel cysteine residue created by the mutation itself, or through instability created by a mutated residue near a structural intramolecular disulfide bond (29). This mechanism was previously established for FGFR3 mutations that we have observed in lung SqCC, R248C and S249C (30).
To assess whether mutations in the extracellular domain of FGFR2 and FGFR3 lead to covalent dimerization, and whether dimerization could be increased by ligand stimulation, we serum starved cells in the presence of PBS or 5 nM FGF1 and 2 μg/mL heparin for eight hours, or 5 nM FGF1 and 2 μg/mL heparin for 30 minutes, followed by washing with PBS and serum starving in the presence of PBS for the remaining 7.5 hours followed by electrophoresis in both reducing and non-reducing conditions. FGFR2 ECD mutations were sufficient to drive covalent dimerization in the absence of ligand, but dimerization was increased in the presence of even 30 minutes of ligand stimulation (Figure S3A). In FGFR3 mutations, on the other hand, dimerization was observed but not increased under ligand-stimulation conditions (Figure S3B). As has been demonstrated previously (31), FGFR proteins typically form highly glycosylated folded protein products. While FGFR2 W290C appears to undergo a glycosylation defect contributing to its lower molecular weight, this mutant form still retains the capacity to dimerize.
We then seeded the same cells into soft agar in the presence of PBS, 2 μg/mL heparin alone, or 5 nM FGF1 and 2 μg/mL heparin. After three weeks, we observed greater colony formation in response to FGF1 and heparin treatment than in heparin alone or PBS treated cells (Figure S3C).
FGFR2 and FGFR3-driven cellular transformation is blocked by clinically relevant FGFR inhibitors
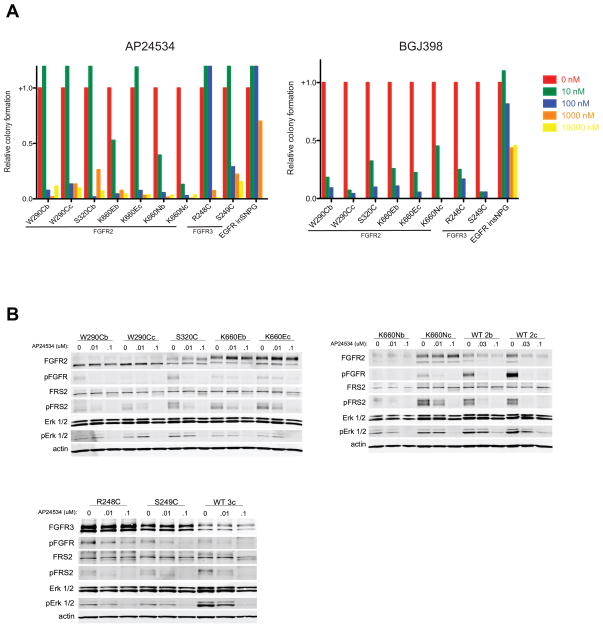
Having established that FGFR2 and FGFR3 mutations in lung SqCC drive anchorage-independent growth in NIH-3T3 cells, we asked whether this transformation could be blocked by small molecule inhibitors of FGFRs. NIH-3T3 cells were seeded into soft agar in the presence or absence of the multi-kinase inhibitor AP24534 (ponatinib), which targets imatinib-resistant BCR-ABL (32), and has activity against FGFR family members (33). Colony formation was inhibited in the presence of ponatinib in cells harboring activating FGFR2 or FGFR3 mutations, but not in cells harboring an activating EGFR insertion (Figure 3A, left panel). All extracellular domain mutations in FGFR2 and S249C in FGFR3 lost colony forming potential when exposed to 100 nM of drug, whereas kinase domain mutations lost colony forming potential at 10 nM of drug. Exceptions were FGFR2 K660E expressed in the IIIc isoform, which behaved similarly to the FGFR2 ECD mutations, and FGFR3 R248C, which had a ten-fold higher inhibitory concentration than any other mutation, at 1 μM. Colony formation driven by EGFR was not lost until cells were exposed to 10 μM of drug.
Figure 3.
Anchorage independent colony formation is abrogated in the presence of anti-FGFR inhibitors. (A) NIH-3T3 cells expressing each transforming mutation were seeded in the presence of increasing concentrations of ponatinib (AP24534) (left panel) and BJG398 (right panel). (B) Cells were serum starved and exposed to the indicated concentrations of ponatinib for four hours and then ligand stimulated for 30 minutes with FGF1, after which cells were lysed and probed via immunoblot. These experiments were performed with several other clinical inhibitors; those results are documented in Figure S4.
To determine whether ponatinib was inhibiting colony formation driven by mutant FGFR2 and FGFR3, we assessed phosphorylation of several proteins in the FGFR signaling pathway. Levels of phospho-FGFR, phospho-FRS2, and phospho-Erk all decreased in response to increasing concentrations of ponatinib (Figure 3B), suggesting that colony formation was lost due to a decrease in FGFR-mediated signaling.
To evaluate whether ponatinib was acting by specific inhibition of FGFR kinases, these assays were also performed with BGJ398, a selective FGFR kinase inhibitor (28) as well as pazopanib (GW786034) (34) and dovitinib (TKI-258) (35), two multi-kinase inhibitors with specificity for FGFR family members. Colony formation was inhibited by at least 50% in the presence of 10 nM BGJ398 for all cells expressing FGFR mutations, while cells expressing the activating EGFR insertion did not lose the capacity for colony formation until 1 μM BGJ398 (Figure 3A, right panel), and wild type phosphorylation was lost at 10 nM under ligand stimulation conditions (Figure S4A). Dovitinib also inhibited colony formation in cells expressing mutant FGFR compared to activated EGFR, but with less uniformity across mutations. FGFR2 ECD mutations lost 50% colony formation between 100 nM and 1 μM dovitinib. In contrast, colony formation was inhibited by 50% between 10 nM and 100 nM for FGFR2 kinase domain mutations excluding K660E IIIc, which behaved similarly to the FGFR2 ECD mutations. Cells expressing FGFR3 R248C and S249C were sensitive between 10 nM and 100 nM. Again, cells transformed by mutant EGFR did not lose colony formation until exposed to 10 μM drug (Figure S4B, left panel). Mutant EGFR-expressing cells had sustained phosphorylation at AKT T308 up to 10 μM dovitinib, as detected by immunoblot, while detectable AKT phosphorylation was lost by 100 nM to 1 μM dovitinib in cells expressing FGFR mutations (Figure S4C). Pazopanib similarly inhibited colony formation in cells expressing all FGFR2 and FGFR3 mutations at concentrations of 100 nM-1 μM drug, while cells expressing mutant EGFR formed colonies even in the presence of 10 μM drug (Figure S4B, right panel). Consistently, biochemical studies revealed sustained AKT T308 phosphorylation in mutant EGFR cells exposed to 10 μM pazopanib, while detectable AKT T308 phosphorylation was lost in mutant FGFR cells at 100 nM to 1 μM pazopanib (Figure S4D).
In NIH-3T3 cells expressing the extracellular domain mutations of both FGFR2 and FGFR3 and in the kinase domain mutation FGFR2 K660E IIIc, we observed that low concentrations of ponatinib (10 nM) conferred a growth promoting phenotype above control, which was abrogated at higher concentrations (Figure 3A, left panel). This could be due to the multi-kinase inhibitory properties of ponatinib, which may inhibit a second kinase that could impact FGFR2 or FGFR3 signaling. This phenomenon was also observed when these experiments were performed with the two other multi-kinase inhibitors with anti-FGFR activity, pazopanib and dovitinib (Figure S4B), but not with BGJ398, a more selective FGFR kinase inhibitor (Figure 3A, right panel).
Analysis of FGFR2 and FGFR3 inhibition in IL-3 independent Ba/F3 cells
To test whether cellular transformation driven by mutated FGFR2 could be abrogated in a second system by small molecule FGFR inhibitors and to test the relative efficacy of these compounds, we generated Ba/F3 cells expressing the FGFR2 mutations that had demonstrated significant colony formation in the NIH-3T3 anchorage-independence assay. These cell lines were dependent on FGFR signaling in the presence of FGF and heparin, and in the absence of IL-3. Phosphorylation of the FGFR kinase domain and FRS2 were measured by immunoblot, and interestingly, cells expressing FGFR2 K660E IIIc showed a greater degree of phosphorylation of both molecules despite similar expression levels as compared to cells expressing other mutations (Figure 4A).
Figure 4.
Ba/F3 cells dependent on FGFR signaling are sensitive to FGFR inhibitors. (A) Ba/F3 cells dependent on FGFR signaling were isolated by exchanging IL-3 with FGF-7 or FGF-9 and heparin. These cells were lysed and probed for FGFR2 or FGFR3 expression, phospho-FGFR, FRS2, and phospho-FRS2 Y436. Actin was used as a loading control. (B) Ba/F3 cells expressing each mutation construct were seeded into 96-well plates, in the presence of increasing concentrations of ponatinib (left panel) or BGJ398 (right panel). After four days, proliferation was measured with Cell Titer Glo. (C) IC50 values were calculated for each mutation. These experiments were performed with other FGFR inhibitors; those results are documented Figure S5.
Ba/F3 cells expressing wild-type and mutated FGFR2 transgenes were first seeded into media containing increasing concentrations of ponatinib. We observed that ponatinib inhibited IL-3 independent proliferation of Ba/F3 cells expressing the FGFR mutations at about 10 nM of drug treatment, but cells expressing an EGFR activating insertion or parental Ba/F3 cells grown in the presence of IL-3 were only inhibited by 10 μM of drug (Figure 4B, left panel). IC50 values for Ba/F3 cells expressing each mutant were also calculated and plotted (Figure 4C, left panel). These assays were also performed on cells seeded into media containing BGJ398, and similarly, cells expressing FGFR mutations, but not the EGFR insertion or parental Ba/F3 cells, were inhibited at about 10 nM inhibitory concentrations of drug (Figure 4B, right panel and Figure 4C, right panel). Interestingly, insensitive controls in the presence of ponatinib appeared to gain a growth advantage in the presence of drug at concentrations in the range of 10–100 nM (Figure 4B), similar to our observations in the anchorage independence colony formation assay (Figure 3A, Figure S4B).
To further assess the potency of small molecule FGFR kinase inhibitors in the Ba/F3 system, we assembled a panel of FGFR kinase inhibitors described in the literature ((28, 32–39), Table S2) and tested the Ba/F3 inhibitory response in the presence of each. Each of these inhibitors demonstrated similar trends to those seen for ponatinib and BGJ398: a multi-log increase in drug sensitivity in cells expressing FGFR mutations compared to controls (Figure S5). IC50 values for each mutation in the presence of each drug were also calculated (Figure S5). Strikingly, FGFR2 K660E expressed in the IIIc isoform (in yellow) repeatedly exhibited a 5–10 fold higher IC50 concentration as compared to the IIIb isoform and either isoform of the K660N mutation in the FGFR2 kinase domain (Figure S5). This observation was consistent with the concentrations at which anchorage independent growth observed for FGFR2 K660E IIIc was lost in the presence of several inhibitors (Figure 3A and Figure S4B).
Case report of a head and neck SqCC patient responding to an FGFR inhibitor
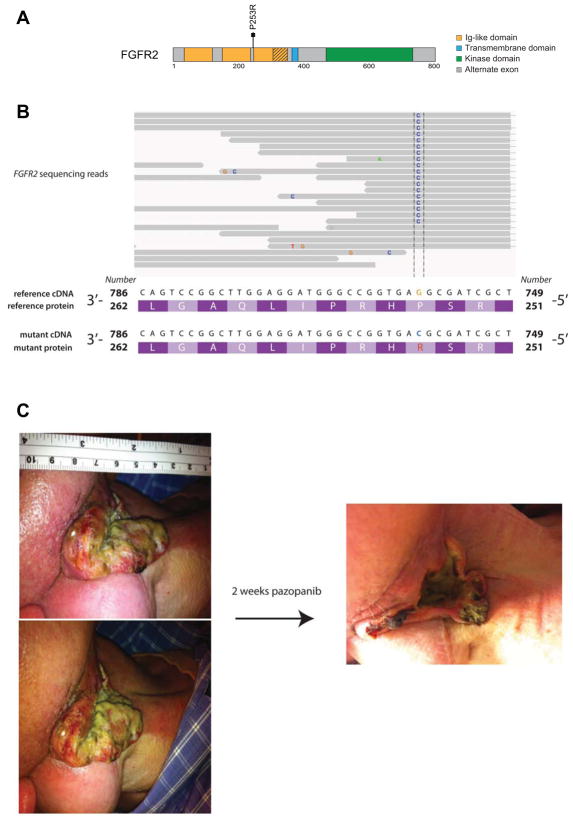
We identified an individual with squamous cell carcinoma of the head and neck who was found to harbor an extracellular FGFR2 mutation (p.P253R) in a biopsy specimen (Figure 5A). This mutation was initially identified in RNA sequencing data and then confirmed by Sanger sequencing in a CLIA-certified laboratory (Figure 5B). FGFR2 mutations have previously been observed at low frequencies in head and neck cancer (40, 41), and confirmed by initial reports from TCGA where seven mutations were observed in exome sequencing data of 279 individuals as of October 1, 2012 (data obtained from the TCGA Data Coordinating Center). FGFR2 P253R has previously been observed in endometrial carcinoma (10). Cellular and biochemical analysis of the FGFR2 P253R mutation suggest that this event is transforming and sensitive to targeted therapies in our assays, similar to the events observed in lung SqCC (Figure S6).
Figure 5.
An oral squamous cell carcinoma patient harboring a somatic FGFR2 P253R mutation demonstrates a partial response to an FGFR inhibitor. (A) A schematic shows the P253R mutation in the FGFR2 extracellular domain. (B) mRNA sequencing was performed and a somatic mutation in FGFR2 was identified, shown in the IGV viewer. (C) Pre- and post-treatment images from the patient.
The patient was diagnosed with locally advanced (T2N1M0, stage III) squamous cell carcinoma of the right tongue in 2008 at the age of 52. He had no history of tobacco use or alcohol abuse and was treated with a right hemiglossectomy and post-operative radiation therapy. He subsequently developed recurrences in the right and left neck over a period of three years and was treated with surgery, two additional courses of radiation therapy and multiple courses of chemotherapy including carboplatin, paclitaxel, cisplatin and cetuximab. In 2012, he had further progression in the right neck and left axilla. He began daily treatment with 800 mg pazopanib starting on April 12, 2012. At this time, he had gross disease in the right neck (Figure 5C, left panels). A follow up visit 12 days later showed a marked reduction in tumor size (Figure 5C, right panel). He continued on pazopanib for two months, when he presented with a right carotid hemorrhage. Pazopanib was discontinued at that time, and the patient remains alive as of March 15, 2013 under hospice care. This correlative observation does not definitively identify FGFR2 as the target of pazopanib, but we believe that this result provides compelling rationale to continue to pursue treatment of _FGFR2_-mutated tumors with anti-FGFR targeted therapies.
Discussion
Lung squamous cell carcinoma is a poorly characterized disease responsible for 40,000 new deaths per year in the US. One of the most provocative findings from genomic analysis is that of recurrent FGFR2 and FGFR3 mutations, which are significant given that germline FGFR mutations are known to be pathogenic (17), that somatic mutations have been described in other malignancies (18), and that focal FGFR1 amplification is known to occur in lung SqCC and appears to be a therapeutic target (15, 16).
We have confirmed that a subset of observed mutations drive transformation in NIH-3T3 cells in an anchorage independent growth assay and xenograft assays, and that this is reversible by pan-FGFR and multi-kinase inhibitors. Some mutations were not transforming, but given the very high somatic mutation rate in lung SqCC, this observation is not surprising. We found that extracellular domain mutations in FGFR2 are able to form ligand sensitive covalent receptor-dimers, as has been observed in other FGFR2 ECD mutations (29) and in FGFR3 mutations that have been described previously in urothelial carcinoma, and which we also observe here in the lung SqCC data (30). This finding is especially relevant given that the FGFR2 W290C mutation has been observed independently in lung SqCC sequencing on two previous occasions (10, 42). It is also possible that the glycosylation deficiency that we observed in the expressed protein harboring this mutation impacts protein function, a phenomenon with precedence in this receptor family (31).
We found that the FGFR mutations also exhibited sensitivity to inhibition by FGFR inhibitors in the Ba/F3 system, which models dependency on oncogenic pathways. Many drugs in the panel of inhibitors that we tested are already approved for clinical use in other malignancies, and clinical trials are underway to test sensitivity to FGFR inhibitors in patients harboring FGFR events (NCT01004224, NCT01457846, NCT00979134). While we cannot infer in vivo sensitivity to these inhibitors from our models, we believe that this study provides a compelling rationale for extending trials of FGFR kinase inhibitors to patients with lung and oral SqCC harboring FGFR2 or FGFR3 mutations.
This study represents one of the first functionally validated novel recurrent targets to emerge from analysis of the systematic genomic profiling of lung SqCC by the TCGA Research Network. It is our expectation that these findings will continue with the publication of more genomic studies of malignancies, and that this will lead to improved treatment options for patients with this disease.
Supplementary Material
1
2
3
4
5
6
7
8
9
Significance.
FGFR2 and FGFR3 mutations, each found in 3% of lung squamous cell carcinomas, drive cellular transformation and are associated with response to FGFR kinase inhibitors currently in clinical development. These findings provide a rationale for therapeutic targeting of FGFR2 and FGFR3 mutations in lung and head and neck squamous cell carcinoma.
Acknowledgments
The authors thank Ami S. Bhatt, member of the Meyerson laboratory, Pamela M. Pollock, investigator at Queensland Institute of Technology (AUS), and David M. Ornitz, investigator at Washington University in St. Louis, for helpful discussion and technical support.
Grant Support
P.S.H. is a recipient of a Young Investigator Grant from the National Lung Cancer Partnership and is supported by NCI grants 1K08CA163677, and the Stephen D. and Alice Cutler Investigator Fund. M.M. is supported by Uniting Against Lung Cancer, the Lung Cancer Research Foundation, the American Lung Association, Novartis Pharmaceuticals, and NCI grant P50CA090578. M.D.W. is supported by a Ruth L. Kirschstein National Research Service Award Individual Fellowship from the National Cancer Institute (NIH F32CA142039). T.J.P. is supported by a Canadian Institutes of Health Research Fellowship.
Footnotes
Financial disclosure: P.S.H. is a recipient of a Young Investigator Grant from the National Lung Cancer Partnership and is supported by NCI grants 1K08CA163677, and the Stephen D. and Alice Cutler Investigator Fund. M.M. is supported by Uniting Against Lung Cancer, the Lung Cancer Research Foundation, the American Lung Association, Novartis Pharmaceuticals, and NCI grant P50CA090578. M.D.W. is supported by a Ruth L. Kirschstein National Research Service Award Individual Fellowship from the National Cancer Institute (NIH F32CA142039). T.J.P. is supported by a Canadian Institutes of Health Research Fellowship.
Conflict of interest: M.M. is a consultant to Novartis and receives research support from Novartis, and is a founding advisor and consultant to, and an equity holder in, Foundation Medicine. P.S.H. reports consulting fees from ARIAD. R.I.H. reports consulting fees from Aveo and is an advisor to Boehringer-Ingelheim. M.D.W. consulted for GeneCentric Diagnostics.
References
- 1.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. Epub 2012/07/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012 doi: 10.1038/ng.2405. Epub 2012/09/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012 doi: 10.1038/ng.2396. Epub 2012/09/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 doi: 10.1038/nature11404. Epub 2012/09/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–34. doi: 10.1016/j.cell.2012.08.024. Epub 2012/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–20. doi: 10.1016/j.cell.2012.08.029. Epub 2012/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. Epub 2004/05/01. [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. Epub 2004/05/01. [DOI] [PubMed] [Google Scholar]
- 9.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. Epub 2007/07/13. [DOI] [PubMed] [Google Scholar]
- 10.Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(25):8713–7. doi: 10.1073/pnas.0803379105. Epub 2008/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H, et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26(50):7158–62. doi: 10.1038/sj.onc.1210529. Epub 2007/05/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23(1):18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 13.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine. 2010;362(25):2380–8. doi: 10.1056/NEJMoa0909530. Epub 2010/06/25. [DOI] [PubMed] [Google Scholar]
- 14.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. Epub 2009/08/21. [DOI] [PubMed] [Google Scholar]
- 15.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Science translational medicine. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. Epub 2010/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE. 2011;6(6):e20351. doi: 10.1371/journal.pone.0020351. Epub 2011/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes & development. 2002;16(12):1446–65. doi: 10.1101/gad.990702. Epub 2002/06/25. [DOI] [PubMed] [Google Scholar]
- 18.Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends in molecular medicine. 2011;17(5):283–92. doi: 10.1016/j.molmed.2011.01.012. Epub 2011/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–29. doi: 10.1038/nrc2780. Epub 2010/01/23. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. The Journal of biological chemistry. 2006;281(23):15694–700. doi: 10.1074/jbc.M601252200. Epub 2006/04/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast cancer research: BCR. 2007;9(2):R23. doi: 10.1186/bcr1665. Epub 2007/04/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwabi-Addo B, Ropiquet F, Giri D, Ittmann M. Alternative splicing of fibroblast growth factor receptors in human prostate cancer. The Prostate. 2001;46(2):163–72. doi: 10.1002/1097-0045(20010201)46:2<163::aid-pros1020>3.0.co;2-t. Epub 2001/02/15. [DOI] [PubMed] [Google Scholar]
- 23.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan WL. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Molecular and cellular biology. 1993;13(8):4513–22. doi: 10.1128/mcb.13.8.4513. Epub 1993/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K, Herr AB, Waksman G, Ornitz DM. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14536–41. doi: 10.1073/pnas.97.26.14536. Epub 2000/12/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin JZ, Berger MF, Adiconis X, Rogov P, Melnikov A, Fennell T, et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome biology. 2009;10(10):R115. doi: 10.1186/gb-2009-10-10-r115. Epub 2009/10/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. Epub 2011/04/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(19):4864–75. doi: 10.1158/1078-0432.CCR-10-0199. Epub 2010/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem. 2011;54(20):7066–83. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 29.Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell. 2000;101(4):413–24. doi: 10.1016/s0092-8674(00)80851-x. Epub 2000/06/01. [DOI] [PubMed] [Google Scholar]
- 30.d’Avis PY, Robertson SC, Meyer AN, Bardwell WM, Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by mutations responsible for the lethal skeletal dysplasia thanatophoric dysplasia type I. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 1998;9(1):71–8. Epub 1998/01/23. [PubMed] [Google Scholar]
- 31.Gartside MG, Chen H, Ibrahimi OA, Byron SA, Curtis AV, Wellens CL, et al. Loss-of-function fibroblast growth factor receptor-2 mutations in melanoma. Molecular cancer research: MCR. 2009;7(1):41–54. doi: 10.1158/1541-7786.MCR-08-0021. Epub 2009/01/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer cell. 2009;16(5):401–12. doi: 10.1016/j.ccr.2009.09.028. Epub 2009/11/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Molecular cancer therapeutics. 2012;11(3):690–9. doi: 10.1158/1535-7163.MCT-11-0450. Epub 2012/01/13. [DOI] [PubMed] [Google Scholar]
- 34.Sonpavde G, Hutson TE. Pazopanib: a novel multitargeted tyrosine kinase inhibitor. Current oncology reports. 2007;9(2):115–9. doi: 10.1007/s11912-007-0007-2. Epub 2007/02/10. [DOI] [PubMed] [Google Scholar]
- 35.Renhowe PA, Pecchi S, Shafer CM, Machajewski TD, Jazan EM, Taylor C, et al. Design, structure-activity relationships and in vivo characterization of 4-amino-3-benzimidazol-2-ylhydroquinolin-2-ones: a novel class of receptor tyrosine kinase inhibitors. J Med Chem. 2009;52(2):278–92. doi: 10.1021/jm800790t. Epub 2008/12/31. [DOI] [PubMed] [Google Scholar]
- 36.Cai Z-w, Zhang Y, Borzilleri RM, Qian L, Barbosa S, Wei D, et al. Discovery of brivanib alaninate ((S)-((R)-1-(4-(4-fluoro-2-methyl-1H-indol-5-yloxy)-5-methylpyrrolo[2,1-f][1,2,4]triazin-6-yloxy)propan-2-yl)2-aminopropanoate), a novel prodrug of dual vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1 kinase inhibitor (BMS-540215) J Med Chem. 2008;51(6):1976–80. doi: 10.1021/jm7013309. [DOI] [PubMed] [Google Scholar]
- 37.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, et al. AZD4547: An orally bioavailable, potent and selective inhibitor of the Fibroblast Growth Factor Receptor tyrosine kinase family. Cancer Res. 2012:1–41. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 38.Matsui J, Yamamoto Y, Funahashi Y, Tsuruoka A, Watanabe T, Wakabayashi T, et al. E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition. International journal of cancer Journal international du cancer. 2008;122(3):664–71. doi: 10.1002/ijc.23131. Epub 2007/10/19. [DOI] [PubMed] [Google Scholar]
- 39.Wedge SR, Kendrew J, Hennequin LF, Valentine PJ, Barry ST, Brave SR, et al. AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res. 2005;65(10):4389–400. doi: 10.1158/0008-5472.CAN-04-4409. Epub 2005/05/19. [DOI] [PubMed] [Google Scholar]
- 40.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–60. doi: 10.1126/science.1208130. Epub 2011/07/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–7. doi: 10.1126/science.1206923. Epub 2011/07/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65(17):7591–5. doi: 10.1158/0008-5472.CAN-05-1855. Epub 2005/09/06. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8
9