A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P (original) (raw)
Abstract
The absence of the fragile X mental retardation protein (FMRP), encoded by the FMR1 gene, is responsible for pathologic manifestations in the Fragile X Syndrome, the most frequent cause of inherited mental retardation. FMRP is an RNA-binding protein associated with polysomes as part of a messenger ribonucleoprotein (mRNP) complex. Although its function is poorly understood, various observations suggest a role in local protein translation at neuronal dendrites and in dendritic spine maturation. We present here the identification of CYFIP1/2 (_Cy_toplasmic _F_MRP _I_nteracting _P_roteins) as FMRP interactors. CYFIP1/2 share 88% amino acid sequence identity and represent the two members in humans of a highly conserved protein family. Remarkably, whereas CYFIP2 also interacts with the FMRP-related proteins FXR1P/2P, CYFIP1 interacts exclusively with FMRP. FMRP–CYFIP interaction involves the domain of FMRP also mediating homo- and heteromerization, thus suggesting a competition between interaction among the FXR proteins and interaction with CYFIP. CYFIP1/2 are proteins of unknown function, but CYFIP1 has recently been shown to interact with the small GTPase Rac1, which is implicated in development and maintenance of neuronal structures. Consistent with FMRP and Rac1 localization in dendritic fine structures, CYFIP1/2 are present in synaptosomal extracts.
The absence of the FMR1 gene product FMRP causes the pathogenic manifestations in Fragile X Mental Retardation Syndrome, an X-linked disorder that affects about 1 in 4,000 males and 1 in 7,000 females (1, 2). The underlying molecular mechanism is transcriptional silencing of FMR1 caused by an unstable hypermethylated CGG repeat expansion in the 5′-untranslated region of the gene (for review, see ref. 2). The FMR1 gene codes for a set of 60- to 78-kDa protein isoforms, deriving from alternative mRNA splicing. FMRP is endowed with a nonclassical nuclear localization signal localized at its N-terminal region and a nuclear export signal encoded by exon 14 (3, 4), suggesting that it shuttles between nucleus and cytoplasm (5). FMRP contains two protein K homology (KH) domains and an RGG box, motifs that are known to be autonomously capable of binding RNA. Indeed, FMRP binds RNA homopolymers in vitro (6–8) and its own mRNA in vitro and in vivo (9, 10). In the cytoplasm, FMRP is associated with actively translating ribosomes (polysomes) via messenger mRNP complexes (11, 12). Immunoprecipitation experiments have demonstrated that FMRP is associated with six different proteins in the mRNP particle (10). Three of them have been identified as nucleolin (a known component of mRNP particles) and the FMRP-related proteins FXR1P/2P (10). FMRP, FXR1P, and FXR2P (FXR proteins) share the same functional domains and form homo- and heteromers (13, 14). FMRP is present in many cell types. It is particularly abundant in the cytoplasm of neurons (15) and is also present in distal dendrites, where its local expression was shown to be increased in response to neurotransmitter activation (16).
The properties of FMRP suggest a possible involvement in nuclear export, cytoplasmic transport, and translational control of target mRNAs (2, 17). It has been proposed that FMRP could play a critical role in the regulation of local protein synthesis at the postsynaptic site, indispensable for normal dendritic spine maturation (18, 19). Abnormal dendritic spines have been observed in both fragile X patients and FMR1 knockout mice (18, 19).
To elucidate the function of FMRP, we initiated a search for additional interacting proteins. We screened an embryonal mouse library by using the yeast two-hybrid system and the highly conserved N terminus of FMRP as bait. Recently, we have identified NUFIP1, a novel RNA-binding protein (20). In the present study, we establish and characterize the interaction between FMRP and another protein found by two-hybrid screening: CYFIP1 (_Cy_toplasmic _F_MRP _I_nteracting _P_rotein 1). As for NUFIP1, we detected no interaction between CYFIP1 and the proteins FXR1P or FXR2P, despite their high sequence similarity with FMRP. We identified a close homologue of CYFIP1, CYFIP2, which is also capable of interacting with FMRP, but unlike CYFIP1, it interacts with all three members of the FXR family. Furthermore, we demonstrate that the region of FMRP encoded by exon 7 is involved in the interaction with CYFIP, overlapping with the FMRP site that mediates homo- and heteromerization of FMRP/FXR1P/FXR2P (14). Finally, we show that CYFIP1/2 are present in synaptosomes isolated from mouse brain. They are thus, to our knowledge, the first identified FXR protein partners present also in synaptic terminals of neurons.
CYFIP1/2 are proteins of unknown function lacking any known motif or functional domain. However, CYFIP1 (p140Sra-1) was recently identified as an interactor of the Rac1 small GTPase (21), a key molecule in actin reorganization (22) involved in generation and maintenance of dendritic spines (23, 24).
Materials and Methods
Antibodies.
Rabbit polyclonal antibodies (pAb) were raised and affinity purified against the synthetic peptides LVHPTDKYSNKDCPDSA (CYFIP1 amino acids 416–432; no. 1467), SQEFQRDKQPNAQPQYLH (CYFIP1 amino acids 872–889; no. 1465), and NEVFAILNKYMKSVETDSST (CYFIP2 amino acids 1215–1234; no. 1716). Ab no. 1467 (anti-CYFIP1) crossreacts with CYFIP2, whereas no. 1465 shows little if any crossreaction. Ab no. 1716 is specific for CYFIP2. Other antibodies used in this study are 1C3 (15), 3FX (25), Ab1937 (26), α-nucleolin (27), α-L7a (28), and commercial antibodies against α-CaMKII (Roche Diagnostics), lactate dehydrogenase (Sigma), myc (9E10) (Sigma), and Pax6 [Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA)].
Plasmid Construction.
Cloning protocols of the constructs used in this study are published as supplemental data on the PNAS web site (www.pnas.org).
Recombinant Proteins.
Glutathione _S_-transferase (GST)-FMRP and His6-CYFIP1 were produced with the baculovirus system. GST-FMRP purification has been described (20). SF9 cells infected with recombinant His6-CYFIP1 baculovirus were briefly sonicated in buffer (500 mM NaCl/20 mM Tris⋅HCl, pH 7.5/1% glycerol/10 mM imidazole/protease inhibitor mixture). The 16,000 × g supernatant was incubated with Ni-NTA agarose (Qiagen, Chatsworth, CA) overnight at 4°C. Beads were washed extensively with lysis buffer.
Interaction Assays.
The yeast two-hybrid screening and the quantitative liquid β-galactosidase (β-gal) assay (ONPG assay) were performed as described (20). The β-gal filter lift assay was carried out as recommended by CLONTECH. Expression of constructs was analyzed by Western blot. None of the constructs used in this study showed any self-activation. Coimmunoprecipitation: HeLa cells (about 5 × 107) were resuspended in lysis buffer (200 mM NaCl/20 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/0.3% Triton X-100, protease inhibitor mixture). Lysates were centrifuged 5 min at 2,000 × g to yield the cytoplasmic supernatant. All following steps were carried out in lysis buffer as described (20) or indicated in Fig. 1C legend. Pull-down assays: GST-pull-down (GST-pd) assays were performed as described (20). The histidine6 pull-down assay was performed in 200 mM NaCl/20 mM Tris⋅HCl, pH 7.5/0,5% Triton/10 mM imidazole/protease inhibitor mixture by using Ni-NTA beads (Qiagen) and the GST-pd protocol. Ni-NTA beads that have been incubated with SF9 cell extract infected by wild-type virus and processed in parallel to CYFIP1 beads were used as negative control.
Figure 1.
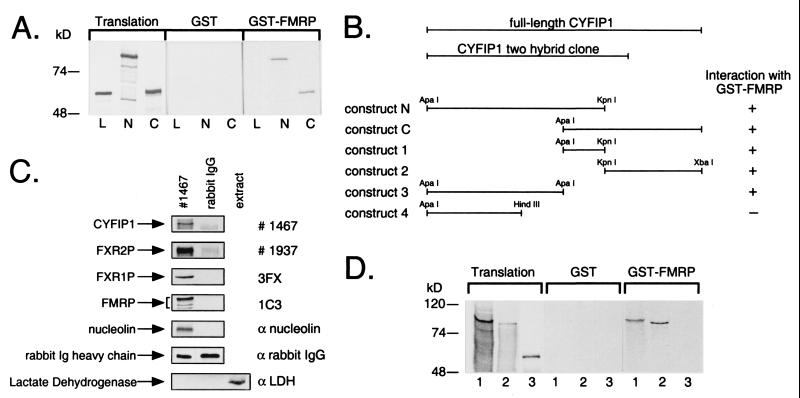
FMRP interacts with CYFIP1/2. (A) Interaction in vitro between GST-tagged full-length FMRP and in vitro translated CYFIP1 N and C terminus (N, C), overlapping by 184 amino acids. GST-pd assays were performed in the presence of 100 mM sodium chloride. In each lane, 10% of the translation product used in each reaction or 40% of the eluate from glutathione beads was loaded. Both overlapping parts of CYFIP1 were retained by GST-FMRP but not by GST alone. Luciferase (L), the negative control, was bound neither by GST-FMRP nor by GST alone. (B) GST-pd assays using additional truncated constructs of CYFIP1 revealed that the N + C overlapping region is not exclusively responsible for interaction with FMRP. No binding affinity to GST-FMRP has been shown by construct 4 (amino acids 1–416). (C) Interaction of CYFIP1 with FMRP in vivo. FMRP, FXR1P, FXR2P, and nucleolin were coimmunoprecipitated from cytoplasmic lysate of HeLa cells by 8 μg of pAb no. 1467 raised against CYFIP1. A functional unrelated protein, lactate dehydrogenase, was not precipitated but was present in the cytoplasmic extract (lane 3). None of the proteins was precipitated by the same amounts of rabbit IgG, demonstrating the specificity of the experiment. One percent of each coimmunoprecipitation reaction was loaded to reveal CYFIP1 and the rabbit IgG heavy chain, eight percent to reveal coprecipitated proteins. (D) CYFIP2 is interacting with GST-FMRP in vitro. CYFIP1 translation product (amino acids 1–959, lane 1) and CYFIP2 translation product (amino acids 1–890, lane 2) were retained by GST-FMRP, but not the negative control (luciferase, lane 3). Assay and loading were performed as described in A. Proteins were separated in 8 or 10% SDS-polyacrylamide gels.
Immunocytochemistry.
Transfection of COS cells and immunostaining were performed as described (3). Cells were fixed 12 h after transfection in 4% paraformaldehyde/1× PBS. Preparations were observed by light or confocal microscopy.
Subcellular Fractionation.
The fractionation procedure has been described by Siomi et al. (14). Western blot signals from three independent experiments were quantified by using the Bio-Rad Imaging Densitometer and the program molecular analyst.
Isolation of Synaptosomes.
Synaptosomes were isolated from mouse brain with an improved two-step purification (29). Animal care was conducted in conformity with institutional guidelines that are in compliance with international laws and policies (29).
Results
To identify novel proteins that interact in vivo with FMRP, we carried out the yeast two-hybrid screening by using the well conserved FMRP N terminus as bait (20). A blast search identified a two-hybrid clone, found multiple times in our screening, as the N-terminal part encoding 881 amino acids of the mouse gene shyc. This gene exhibits an expression pattern in mouse similar to that of fmr1, being very widely expressed during early embryonic development and showing a more restricted expression at later stages (30). In the adult brain, shyc, like fmr1, is strongly expressed in regions rich in neurons (hippocampus, Purkinje cells, cerebral cortex). The shyc orthologous gene in man corresponds to the cDNA clone KIAA0068. The human and mouse genes code for 1,253-aa proteins, which we termed CYFIP1/mCYFIP1, and which lack any known functional domains or motifs.
Interaction Between FMRP and CYFIP1.
To confirm the interaction between FMRP and CYFIP1 observed in the yeast two-hybrid system, we used the GST-pd assay and coimmunoprecipitation experiments. Purified recombinant GST-tagged FMRP full-length protein and GST (20) were incubated with 35S-labeled in vitro translated N- and C-terminal segments of CYFIP1 (constructs N and C; Fig. 1B), overlapping by amino acids 631–815 (we failed to efficiently produce full-length CYFIP1 in vitro, probably because of its high molecular mass of 145 kDa). Both overlapping segments of CYFIP1 were retained by GST-FMRP beads, whereas no nonspecific binding to GST alone was observed (Fig. 1A). Luciferase, the negative control, was bound by neither GST nor GST-FMRP. The same experiment with a set of truncated CYFIP1 constructs (Fig. 1B) showed that indeed a construct corresponding to the N + C overlapping region is capable of interacting with FMRP. Binding also appears to be mediated by neighboring regions, indicating a nonlinear, more complex binding surface for the FMRP interacting site of CYFIP1. A construct spanning amino acids 1–416 of CYFIP1 showed no interaction with FMRP.
A polyclonal anti-CYFIP1 antibody (no. 1467) was used to precipitate endogenous CYFIP1 from the cytoplasmic fraction (see section colocalization) of HeLa cells. By Western blot analysis, we identified FMRP, FXR1P, FXR2P, and nucleolin (Fig. 1C) in the precipitated complex. None of these proteins could be found in the control precipitation with rabbit IgG demonstrating specificity of the precipitation. As a second negative control, we checked that a functionally unrelated protein (lactate dehydrogenase) was absent from the coimmunoprecipitations.
CYFIP2, a CYFIP1 Homologue, Is also Capable of Interacting with FMRP.
While our characterization of CYFIP1 was in progress, we found that it shares high sequence homology (87.7% identity, 94.5% similarity) with the human protein PIR121 (31), whose sequence had been reported in databases. The protein was thus a potential interactor of FMRP. In a GST-pd assay using GST-FMRP, in vitro translated PIR121 N terminus (amino acids 1–890) bound to FMRP with even higher affinity than a comparable in vitro translated CYFIP1 N terminus (amino acids 1–959) (Fig. 1D). Interaction between PIR121 and FMRP was confirmed by yeast two-hybrid assay and again appeared stronger than CYFIP1–FMRP interaction (see Fig. 5). We thus designated PIR121 as CYFIP2.
Figure 5.
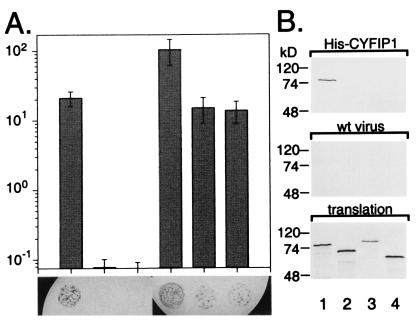
Selective interaction of CYFIP1/2 with FMRP, FXR1P, and FXR2P. (A) Interaction between the CYFIP proteins and the members of the FXR family, determined by yeast two-hybrid system. Diagram illustrating activation of the β-gal reporter in yeast coexpressing the following proteins (from Left to Right): FMRP + CYFIP1, FXR1P + CYFIP1, FXR2P + CYFIP1, FMRP + CYFIP2, FXR1P + CYFIP2, and FXR2P + CYFIP2, measured by liquid ONPG assay. At the foot of each bar, a X-gal filter lift assay is shown. Blue (black) color indicates β-gal activity induced by interaction of the two transformed fusion proteins. CYFIP2 interacts also with the FMRP-related proteins FXR1P/2P, whereas CYFIP1 interaction is restricted to FMRP. (B) The His6 pull-down assay (His-pd) assay using His6-tagged full-length CYFIP1 and in vitro translated proteins FMRP iso7 (lane 1), FXR1P iso a/P70 (lane 2), FXR2P (lane 3), and luciferase (lane 4). His-pd assays were performed in a buffer containing 200 mM sodium chloride. Loadings were performed as indicated in the legend to Fig. 1. FMRP bound specifically to CYFIP1, whereas FXR1P, FXR2P, and the negative control luciferase were not retained. None of the proteins is retained by beads incubated with extract from SF9 cells infected by wild-type baculovirus.
CYFIP1/2 Are Members of a Widely Expressed, Highly Conserved Protein Family.
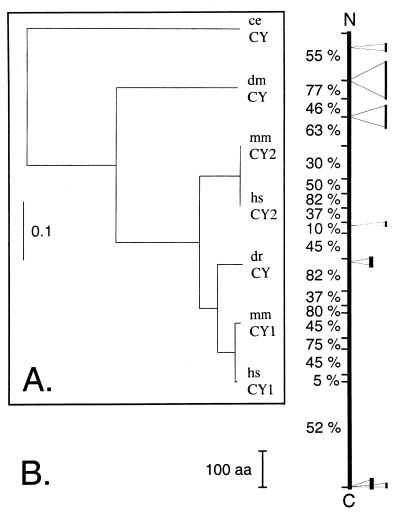
The cDNA sequences of human CYFIP1, CYFIP2, and mouse CYFIP1 are known. In addition, genomic sequences of single CYFIP orthologues in worm (Caenorhabditis elegans) and fly (Drosophila melanogaster) and partial expressed sequence tags (ESTs) for mouse CYFIP2, for the fly, and a zebrafish orthologue are also available. We completed the sequence of mouse CYFIP2 and of zebrafish CYFIP by sequencing of EST clones and reverse transcription–PCR products and confirmed the sequence of fruit fly CYFIP cDNA predicted by the genomic sequence. A phylogenetic study of the CYFIP protein family is shown in Fig. 2A. Human CYFIP1/2 share 98.7% and 99.9% amino acid sequence identity, respectively, with their mouse orthologues. Their homologues in fly and worm share about 67% and 51% amino acid identity with the human proteins, respectively. Although fly and worm homologues exhibit about the same distance to both human proteins, the zebrafish protein appears to be closer related to human CYFIP1 (93% identity) than to human CYFIP2 (86% identity) (Fig. 2A). A second yet-unidentified member of the CYFIP family may thus exist in zebrafish. Our data show that the CYFIP family is even more highly conserved in evolution than the FXR family. The schematic depiction (Fig. 2B) of the CYFIP proteins indicates that conservation between human CYFIP1/2 and their fly and worm orthologues is manifested almost throughout the entire protein, suggesting that it contains numerous functionally and/or structurally indispensable domains. Like FMRP and its related proteins, CYFIP1/2 are widely expressed [Unigene of the National Center for Biotechnology Information, the database of Human unidentified gene-encoded large proteins (HUGE) of the Kazusa DNA Research Institute, Chiba, Japan].
Figure 2.
The CYFIP proteins are highly conserved in evolution. (A) Phylogenetic tree of the CYFIP protein family comprising two members in human (hsCY1/2) and mouse (mmCY1/2) and one member in zebrafish (drCY), Drosophila (dmCY), and C. elegans (ceCY). The multiple protein sequence alignment performed by clustalx is provided as supplementary material (www.pnas.org); the tree was generated by using the program phylowin and the neighbor-joining algorithm. (B) Schematic depiction of CYFIP conservation along the protein. Percentage to the left indicate the portion of residues conserved (similar) among human CYFIP1, CYFIP2, and CYFIP proteins of D. melanogaster and C. elegans. The fly and worm proteins exhibit some inserted sequences that are indicated on the right (thick bar for D. melanogaster, thin for C. elegans). The C. elegans protein sequence is predicted by the genomic sequence and has not been verified.
Colocalization of CYFIP1/2 with FMRP.
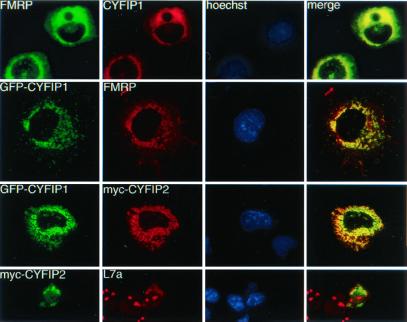
Myc-tagged CYFIP2 (PIR121) was reported to be cytoplasmic (31). We studied subcellular distribution of CYFIP1 and its relation to cytoplasmic FMRP as well as CYFIP1-CYFIP2 colocalization in transfected Cos cells. We cloned the CYFIP1 ORF in the eukaryotic expression vector pEGFP harboring an N-terminal green fluorescent protein (GFP) tag (GFP-CYFIP1) as well as in the vector pTL1 (CYFIP1) to allow expression of nontagged CYFIP1. Both CYFIP1 constructs colocalized with FMRP iso7 (Fig. 3, rows 1 and 2) and appear in a characteristic cytoplasmic pattern with strong labeling of the perinuclear region. A similar distribution of CYFIP1 has been observed in cells overexpressing exclusively CYFIP1 (not shown), indicating that its subcellular localization is not, at least not exclusively, determined by interaction with FMRP. As a first attempt to get information about similar and/or distinct functions of the two CYFIP proteins, we performed a colocalization study. We cotransfected Cos cells with the GFP-CYFIP1/pEGFP construct and a myc-tagged CYFIP2/pTL1 construct. The localization of CYFIP1/2 was exactly the same (Fig. 3, row 3) in all cells and confocal sections analyzed. In Cos cells transfected only with myc-tagged CYFIP2, CYFIP2 was distributed in a way similar to cytoplasmic ribosomes (Fig. 3, row 4), stained here with an antibody against the ribosomal subunit L7a. L7a is also strongly present in the nucleolus (Fig. 3, row 4, panel 2), the locus of ribosome assembly. In conclusion, CYFIP1/2 appear mostly colocalized with FMRP and ribosomes in the cytoplasm.
Figure 3.
Coimmunolocalization studies in transiently transfected Cos cells. Row 1: cells transfected with CYFIP1 and FMRP iso7. FMRP was revealed by mAb 1C3, CYFIP1 by pAb no. 1465. Row 2: cells cotransfected with GFP-CYFIP1 and FMRP iso7, revealed by mAb 1C3. Row 3: cells cotransfected with GFP-tagged CYFIP1 and a myc-tagged CYFIP2 construct, revealed by mAb α-myc 9E10. Row 4: Cos transfected with myc-CYFIP2 (observed by 9E10) and compared with localization of endogenous ribosomes, revealed by a pAb raised against L7a, a protein of the ribosomal large subunit. Anti-L7a also stains nucleoli, the loci of ribosome assembly. Column 3: Hoechst staining. Column 4: merging images (yellow) from series 1 and 2, respectively, indicate colocalization.
CYFIP1/2 Show No RNA-Binding Properties.
All protein components of the FMRP-containing mRNP complex known so far (FMRP, FXR1P, FXR2P, and nucleolin), as well as the nuclear FMRP interactor NUFIP1, exhibit RNA-binding properties. Although no putative RNA-binding domains have been detected within CYFIP1/2, we decided to test RNA binding of the proteins. We carried out a Northwestern experiment by using purified His6-tagged recombinant CYFIP1 and a labeled riboprobe transcribed from the polylinker of the pBluescript vector. No binding activity of CYFIP1 was detected. Furthermore, RNA–homopolymer-binding assays, by using in vitro synthesized proteins, and an Oligo-dT cellulose mRNA copurification experiment indicated no RNA-binding abilities by CYFIP1/2 (data not shown).
CYFIP1/2 Cofractionate Partially with FMRP and Are also Present in Synaptosomal Extracts.
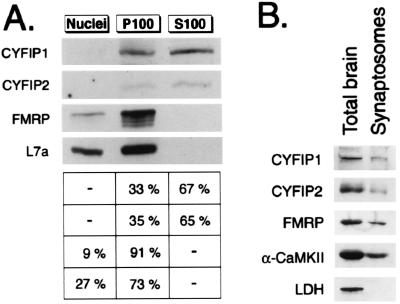
To investigate whether CYFIP1/2 are associated with heavy sedimenting structures (that include polysomes and FMRP) (14), we first fractionated HeLa cells into a nuclear fraction (N), a cytoplasmic insoluble/heavy fraction (P100), and a cytoplasmic soluble fraction (S100). We compared the endogenous CYFIP content in each fraction with that of FMRP and ribosomal subunit L7a (Fig. 4A). Nine percent and ninety-one percent of FMRP were present in the N and P100 fractions, and no FMRP was detected in S100. As expected, CYFIP1/2 were found in cytoplasmic fractions. Sixty-seven percent of CYFIP1 was localized in the S100 fraction and 33% of CYFIP1 pelleted with FMRP in the P100 fraction. CYFIP2 distribution appeared similar to that of CYFIP1. The finding that the largest part of CYFIP1/2 is not sedimenting together with FMRP strongly suggests that the CYFIP proteins also carry out FMRP-independent functions in the cell.
Figure 4.
Subcellular fractionations. (A) CYFIP1/2, FMRP, and L7a content in three subcellular fractions of HeLa cells analyzed by Western blot by using pAb no. 1465, pAb no. 1716, mAb 1C3, and anti-L7a. Fractions represent nuclei (N), insoluble cytoplasm (P100), and soluble cytoplasm (S100). Signals from three independent experiments were quantified. Percentage of total cellular proteins in each fraction is indicated on the right. (B) CYFIP1/2 are present in synaptosomes. Western blot analysis of proteins in total brain extract (15 μg loaded) and synaptosomal extract (14 μg loaded).
With regard to the mental retardation phenotype of Fragile X Syndrome, one of the most interesting features is certainly the presence of FMRP at the postsynaptic site of neurons and its increased dendritic synthesis after neurotransmitter activation (16). To address the question whether CYFIP1/2 are also localized in distal dendrites, we isolated synaptosomes consisting of pre- and postsynaptic terminals from mouse brain (29). By Western blot, CYFIP1/2 were detected in the synaptosomal extracts (Fig. 4B). The ratio of the two proteins in the synaptic fraction and the total brain extract is similar to that seen for FMRP and for the α-subunit of the well described synaptic protein CaMKII (for review, see ref. 32), revealing the presence of significant amounts of CYFIP1/2 at the synapses. Nonsynaptic controls, cytoplasmic proteins lactate dehydrogenase (Fig. 4B), and nuclear protein PAX6 (data not shown) were not present in the synaptic preparation.
Selective Interaction of CYFIP1/2 with FXR1P/2P.
The FXR1P/2P proteins show high homology (69% identity) with FMRP in the N-terminal region. Despite this homology, NUFIP1 was recently found to interact only with FMRP but not with FXR1P/2P (20). The ability of CYFIP1/2 to interact with the FMRP-related proteins was examined by yeast two-hybrid assay (Fig. 5A). CYFIP1 did not interact with either FXR1P/2P, in both 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) filter lift assay and quantitative β-gal (ONPG) assay. In contrast, CYFIP2 showed strong interaction with FXR1P/2P in both assays.
A pull-down assay by using His6-tagged CYFIP1 produced in the baculovirus system and in vitro translated FXR proteins [FMRP (iso7), FXR1P (iso a/P70), and FXR2P] verified the lack of interaction of CYFIP1 with FXR1P/2P, while confirming interaction with FMRP (Fig. 5B).
FMR1 Exon 7 Is Implicated in Interaction with CYFIP1.
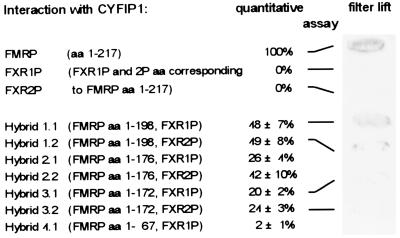
To define the site within the FMRP N terminus responsible for interaction with CYFIP1, we generated a set of chimeric constructs (Fig. 6) that span the region encoded by FMR1 exon 1–7/8 (amino acids 1–217) but are partially substituted by the corresponding FXR1 or FXR2 sequences. This approach minimizes the risk of changes in protein structure that might occur in truncated constructs. By testing the chimeric constructs in the yeast two-hybrid system, we found that the region encoded by FMR1 exon 7 is implicated in the interaction (Fig. 6). Replacement of that particular region (amino acids 173–217, hybrids 3.1 and 3.2) with the corresponding FXR1/2 sequences resulted in a dramatic 80% reduction of interaction, determined by quantitative ONPG assay. In this assay, interaction with CYFIP1 appears completely abolished when exons 4–7 were replaced. In the X-gal filter lift assay, substitution of exon 7 appears sufficient to suppress almost completely detectable interaction with CYFIP1 (Fig. 6). We conclude that FMRP exon 7, containing the homo- and heteromerization domain of the FXR family (14), is also the major site for interaction of FMRP with CYFIP1.
Figure 6.
The FMRP sequence encoded by FMR1 exon 7 is implicated in the interaction with CYFIP1. Yeast two-hybrid assay between CYFIP1 and chimeric FMRP/FXR1P and FMRP/FXR2P proteins. In hybrid constructs, 1.1–2.2 increasing parts of FMRP exon 7 were replaced by FXR1P/2P, in hybrids 3.1 and 3.2 the entire exon 7. In construct 4.1, exon 4–7/8 of FMR1 is substituted by the FXR1 sequence. To the right of each construct, the strength of interaction with CYFIP1 in relation to interaction FMRP-CYFIP1 is indicated. For constructs 1.1, 1.2, 3.1, and 3.2, also an X-gal filter lift assay is shown. A more detailed representation of the hybrid constructs, the FMRP exon 7 region, and the corresponding FXR1P/2P sequences is published as supplemental data on the PNAS web site, www.pnas.org.
Discussion
We identified two proteins that interact with FMRP. These proteins, termed CYFIP1/2, represent the two members in humans from a protein family highly conserved in evolution. CYFIP1/2 are distributed in an identical pattern in the cytoplasm, showing colocalization with FMRP and ribosomes. Both proteins harbor no known motifs, which would indicate a possible function. However, it has been shown that CYFIP1 (p140Sra-1) interacts with the small GTPase Rac1 (21). The authors identified CYFIP1 by microsequencing of copurified material. The presence of CYFIP2, still unknown at that time, was not reported, but it is notable that CYFIP1/2 share 100% sequence conservation among the identified peptide sequences. Indeed, cotransfection experiments showed changed localization of both CYFIPs when transfected together with Rac1V12, a constitutive active form of Rac1 (ref. 21; unpublished results), suggesting that both homologues are partners of Rac1.
The members of the Rho family of GTPases play well established roles in dynamic reorganization of the actin cytoskeleton (22). In neurons, actin is a major component of the neuronal cytoskeleton and appears highly enriched in dendritic spines (33). Consistently, Rho family small GTPases have been found to be involved in establishment of neuronal cell polarity, in morphogenesis and migration of growth cones (for review see refs. 34 and 35). Purkinje cells of transgenic mice overexpressing Rac1V12 showed reduced axon terminals and prevalent abnormal structure of dendritic spines (23). Furthermore, it was recently demonstrated that Rac1, in addition to its role during spine maturation, is also essential for maintenance of these structures, whereas other small GTPases show effects on other subneuronal structures (24). The only consistent abnormality observed in both the brain of fragile X patients and of the fragile X mouse knockout model is abnormal dendritic spines (18, 19) that are too long, thin, and slightly more numerous. The identification of CYFIP1 as a FMRP-interacting protein provides the first link to pathways directly involved in development and maintenance of dendritic spines and neuronal plasticity. Because FMRP and Rac1 have both been observed in dendritic spines (16, 24), we checked also for the presence of CYFIP proteins. Indeed, we found CYFIP1/2 represented in synaptosomal extracts, which are free from cell soma contaminations, in similar ratios as for the synaptic protein CaMKII and FMRP itself. Moreover, we have preliminary results suggesting that CYFIP1/2 mRNAs are also translocated to the distal portion of dendrites. The finding of mRNA presence at the postsynaptic site of neurons indicates that a protein is locally translated and thus probably needed for fast response to extracellular stimuli.
There is a growing indication that FMRP inhibits the translation of mRNAs (36) and that an altered regulation could be responsible for neuronal abnormalities in fragile X patients. Taking together our data and current knowledge on Rac1 GTPase, we propose that Rac1 and CYFIP1/2 are part of the pathway regulating translational control (or other functions executed) by FMRP, possibly in a neurotransmitter activation-dependent manner. Rho GTPases have been suggested to link surface receptors to the organization of the actin cytoskeleton (22). It is tempting to speculate that in dendrites, cytoskeleton reorganization and activity of the local protein machinery required for developing spine compartments could be coregulated via this pathway.
CYFIP2 is capable of interacting with FMRP and its related proteins FXR1P/2P, whereas CYFIP1 interaction is restricted to FMRP. As FMRP/FXR1P/FXR2P (at least in their N terminus) as well as CYFIP1/2 differ only in less than 30 and 13% of their amino acid residues, respectively, this selectivity of interaction is surprising and indicates highly specific recognition sites. Although CYFIP1 is not interacting directly with FXR1P/2P, we found the two proteins coimmunoprecipitated by CYFIP1 pAb no. 1467. This was expected because FXR1P/2P are part of the same FMRP-containing mRNP complex (10) and the presence of nucleolin further indicates coprecipitation of the entire mRNP particle. Partial coprecipitation of the FXR proteins might also occur via CYFIP2, because pAb no. 1467 shows crossreaction with the CYFIP1 homologue. However, in addition to the putative role of CYFIP1/2 in a regulating pathway, the two proteins may be important for determination of common and distinct function of FMRP and its two related proteins. A specific function for CYFIP1, showing no interaction with FXR1P/2P, could be a key for understanding also the unique function(s) of FMRP, not replaceable by the FXR1P/2P proteins.
Our interaction studies indicate that the sequence encoded by FMR1 exon 7 is causally involved in binding of CYFIP1 (thus probably also in binding CYFIP2). That the hetero- and homomerization site of the FXR family has been ascribed to that region (14) raises the question—even in the case that the FMRP interaction sites are overlapping and not identical—whether binding of CYFIP1/2 and homo/heteromerization can occur simultaneously. If FMRP binding to a FXR molecule and to a CYFIP molecule will turn out to be mutually exclusive, one could imagine that this is part of a mechanism modulating FMRP properties, as RNA-binding affinity/specificity or translational control.
Supplementary Material
Supplemental Data
Acknowledgments
We thank T. Nagase and R. Iggo for providing the full-length cDNAs of CYFIP1 (KIAA0068) and CYFIP2 (PIR121, tagged and nontagged), P. Blader for the zebrafish cDNA library, and A. Hoogeveen, P. Bouvet, A. Ziemiecki, and the Developmental Studies Hybridoma Bank for providing FXR2P, nucleolin, L7a, and PAX6 antibodies, respectively. We are grateful to F. Amaldi, F. Blondeau, E. Lalli, A. Lunkes, T. Riedl, and N. Roy for discussions and comments on the manuscript. We further acknowledge L. Bianchetti, M. Boegler, and the Institut de Génétique et de Biologie Moléculaire et Cellulaire core facilities for their help and support. This study was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg, and the FRAXA Foundation. A.S. is an Ernst Schering Research Foundation fellow.
Abbreviations
mRNP
messenger ribonucleoprotein
FMR1
fragile X mental retardation gene
FMRP
fragile X mental retardation protein
FXR1 and -2
_FMR1-_related genes 1 and 2
FXR1P/2P
FXR1 and -2 proteins
β-gal
β-galactosidase
GST-pd
glutathione _S_-transferase pull-down
GFP
green fluorescent protein
X-gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
pAb
polyclonal antibodies
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. D38549 (KIAA0068/human CYFIP1), AF160973 (PIR121/human CYFIP2), AF072697 (shyc/mouse CYFIP1), AF334144 (mouse CYFIP2), AY017342 (zebrafish CYFIP), AY017343 (fly CYFIP), and AF038619 (worm CYFIP)].
References
- 1.Hagerman R J. In: Fragile X Syndrome: Diagnosis, Treatment and Research. Hagerman R J, Cronister-Silverman A, editors. Baltimore: Johns Hopkins Univ. Press; 1996. pp. 3–68. [Google Scholar]
- 2.Imbert G, Feng Y, Nelson D, Warren S T, Mandel J-L. In: Genetic Instabilities and Hereditary Neurological Diseases. Warren S T, Wells R D, editors. San Diego: Academic; 1998. pp. 27–53. [Google Scholar]
- 3.Sittler A, Devys D, Weber C, Mandel J-L. Hum Mol Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 4.Eberhart D E, Malter H E, Feng Y, Warren S T. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 5.Tamanini F, Bontekoe C, Bakker C E, van Unen L, Anar B, Willemsen R, Yoshida M, Galjaard H, Oostra B A, Hoogeveen A T. Hum Mol Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- 6.Ashley C T, Jr, Wilkinson K D, Reines D, Warren S T. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 7.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 8.Adinolfi S, Bagni C, Musco G, Gibson T, Mazzarella L, Pastore A. RNA. 1999;5:1248–1258. doi: 10.1017/s1355838299990647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson K D, Warren S T. J Biol Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 10.Ceman S, Brown V, Warren S T. Mol Cell Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian E W. Hum Mol Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, Absher D, Eberhart D E, Brown V, Malter H E, Warren S T. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, O'Connor J P, Siomi M C, Srinivasan S, Dutra A, Nussbaum R L, Dreyfuss R. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devys D, Lutz Y, Rouyer N, Bellocq J P, Mandel J L. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 16.Weiler I J, Irwin S A, Klintsova A Y, Spencer C M, Brazelton A D, Miyashiro K, Comery T A, Patel B, Eberwine J, Greenough W T. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin P, Warren S T. Hum Mol Genet. 2000;9:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 18.Comery T A, Harris J B, Willems P J, Oostra B A, Irwin S A, Weiler I J, Greenough W T. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irwin S A, Galvez R, Greenough W T. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 20.Bardoni B, Schenck A, Mandel J-L. Hum Mol Genet. 1999;8:2557–2566. doi: 10.1093/hmg/8.13.2557. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura Y, Iwamatsu A, Kaibuchi K. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- 22.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 23.Luo L, Hensch T K, Ackerman L, Barbel S, Jan L Y, Jan Y N. Nature (London) 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama A Y, Harms M B, Luo L. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khandjian E W, Bardoni B, Corbin F, Sittler A, Giroux S, Heitz D, Tremblay S, Pinset C, Montarras D, Rousseau F, Mandel J-L. Hum Mol Genet. 1998;7:2121–2128. doi: 10.1093/hmg/7.13.2121. [DOI] [PubMed] [Google Scholar]
- 26.Bakker C E, de Diego Otero Y, Bontekoe C, Raghoe P, Luteijn T, Hoogeveen A T, Oostra B A, Willemsen R. Exp Cell Res. 2000;258:162–170. doi: 10.1006/excr.2000.4932. [DOI] [PubMed] [Google Scholar]
- 27.Bouvet P, Diaz J J, Kindbeiter K, Madjar J J, Amalric F. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 28.Ziemiecki A, Muller R G, Fu X C, Hynes N E, Kozma S. EMBO J. 1990;9:191–196. doi: 10.1002/j.1460-2075.1990.tb08095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagni C, Mannucci L, Dotti C G, Amaldi F. J Neurosci. 2000;20:RC76. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koster F, Schinke B, Niemann S, Hermans-Borgmeyer I. Neurosci Lett. 1998;252:69–71. doi: 10.1016/s0304-3940(98)00531-x. [DOI] [PubMed] [Google Scholar]
- 31.Saller E, Tom E, Brunori M, Otter M, Estreicher A, Mack D H, Iggo R. EMBO J. 1999;18:4424–4437. doi: 10.1093/emboj/18.16.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soderling T R. Curr Opin Neurobiol. 2000;10:375–380. doi: 10.1016/s0959-4388(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 33.Fischer M, Kaech S, Knutti D, Matus A. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 34.Drubin D G, Nelson W J. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 35.Luo L, Jan L Y, Jan Y. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- 36.Laggerbauer B, Ostareck D, Keidel E, Ostareck-Lederer A, Fischer U. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data