A third gyrovirus species in human faeces (original) (raw)
Abstract
Until 2011 the genus Gyrovirus in the family Circoviridae consisted of a single virus (Chicken anemia virus or CAV) causing a common immunosuppressive disease in chickens when a second gyrovirus (HGyV) was reported on the skin of 4 % of healthy humans. HGyV is very closely related to a recently described chicken gyrovirus, AGV2, suggesting that they belong to the same viral species. During a viral metagenomic analysis of 100 human faeces from children with diarrhoea in Chile we identified multiple known human pathogens (adenoviruses, enteroviruses, astroviruses, sapoviruses, noroviruses, parechoviruses and rotaviruses) and a novel gyrovirus species we named GyV3 sharing <63 % similarity with other gyrovirus proteins with evidence of recombination with CAV in its UTR. Gyroviridae consensus PCR revealed a high prevalence of CAV DNA in diarrhoea and normal faeces from Chilean children and faeces of USA cats and dogs, which may reflect consumption of CAV-infected/vaccinated chickens. Whether GyV3 can infect humans and/or chickens requires further studies.
Chicken anemia virus (CAV) is a small non-enveloped virus with a small (2.3 kb) circular ssDNA genome, which until 2011 was the only member of the genus Gyrovirus within the family Circoviridae. CAV was first isolated in 1979 via propagation in 1-day-old chicks in Japan (Schat, 2009). CAV is an economically important pathogen that causes a severe disease in young susceptible chickens and immunosuppression in older chickens. The disease is characterized by anaemia, generalized lymphoid atrophy, bone marrow aplasia and immunosuppression (Adair, 2000). CAV can be transmitted either vertically from hens to their offspring or horizontally between chickens through the oral–faecal route (van Santen et al., 2004), but infection via the respiratory route can also be experimentally performed (Rosenberger & Cloud, 1989). Recently, two gyroviruses, human gyrovirus (HGyV) and avian gyrovirus 2 (AGV2), were identified on the surface of the skin of 4 % of healthy French adults (Sauvage et al., 2011) and in the sera of diseased chicken from Brazil, respectively (Rijsewijk et al., 2011). HGyV and AGV2 are closely related differing by only 3–7 % in their VP1–3 protein sequences.
We report the identification by viral metagenomics of gyroviruses in human faeces including a novel gyrovirus species and investigate their association with unexplained diarrhoea. All studies were approved by the University of California at San Francisco (UCSF) committee on human research. Faecal specimens from Chilean children with acute gastroenteritis were analysed by purifying viral particles, unbiased nucleic acid amplification and high-throughput pyrosequencing (Victoria et al., 2009). A total of 100 faecal samples were pre-tested for rotavirus antigens (IDEIA Rotavirus assay; DAKO) and calicivirus by EIA against baculovirus expressed human calicivirus and RT-PCR and found to be negative (O’Ryan et al., 2007, 2009). The samples were clarified by 15 000 g centrifugation for 10 min and the supernatants filtered through a 0.45 µm filter (Millipore), and then treated with a mixture of DNases [Turbo DNase (Ambion), Baseline-ZERO (Epicentre) and Benzonase (Novagen)] and with RNase (Fermentas) to digest unprotected nucleic acids. Nucleic acids protected from nuclease digestion within viral capsids were then extracted using QIAamp spin-columns (Qiagen). Both RNA and DNA were randomly amplified by using a sequence-independent random RT-PCR where 10 µl of the extracted nucleic acid was used as a template to synthesize cDNA, using SuperScript III reverse transcriptase (Invitrogen) and a primer with an arbitrary sequence with randomized 3′ termini of eight N nucleotides. After reverse transcription followed by heat denaturation and re-annealing of the primer, a single round of primer extension DNA synthesis was performed by using Klenow fragment DNA polymerase (New England Biolabs). PCR amplification was then performed using primers consisting of only the set portion of the 3′ randomized primer. To increase sampling of the viral nucleic acids, the random PCR amplifications were performed in duplicate, pooled and purified by using the QIAquick Purification kit (Qiagen). The purified DNA concentration was determined by Nanodrop (Thermo Scientific) and a DNA library was constructed according to the manufacturer’s protocol for Roche 454 pyrosequencing (GS FLX Titanium General Library Preparation kit; Roche). De novo assembly of the pyrosequence data used the program MIRA (http://chevreux.org/projects_mira.html), with a criterion of at least 95 % identity over 35 bp to merge two fragments. The contig and singlet sequences greater than 100 bp were compared to the GenBank protein databases using blastx.
Thirty-seven infections with known human pathogens associated with diarrhoea were identified: 13 adenovirus (one for type 2, one for type 5, one for type 31, nine for type 41 and one unclassifiable), eight human enteroviruses (three species A: CV-A5, two EV-A7; three species B: E-30, CV-B2, E-16; two species C: two CV-A22), seven astroviruses (three for HAstV1, one for HAstV6, one for HAstV8, one for HAstV-MLB and one for HAstV-HMO-A), five caliciviruses (three for sapoviruses and two for noroviruses), two human bocaviruses (two for HBoV2), one parechovirus (HPeV1) and one rotavirus (RV-A) infections. One adeno-associated virus and numerous instances of anellovirus sequence detection, neither virus associated with diarrhoea, were also detected. One case of rotavirus and five cases of calicivirus shedding were identified despite the negative rotavirus and calicivirus tests using commercial or home-made assays.
Twenty-four sequence reads of >74 000 sequences generated from five faecal samples also showed significant similarities (E value <2×10−30) to gyrovirus proteins. Circular gyrovirus genomes were therefore amplified using inverse PCR (iPCR) with primers designed from the pyrosequenced DNA fragments from four of these samples. iPCR amplicons were directly sequenced by primer walking. Putative ORFs in the gyrovirus genomes were predicted using NCBI ORF finder. A phylogenetic tree with 100 bootstrap resampling of the alignment datasets was generated using the neighbour-joining method (Saitou & Nei, 1987). The genetic distance was calculated using Kimura’s two-parameter method (phylip) (Kimura, 1980). Sequence identity matrix was measured by using BioEdit.
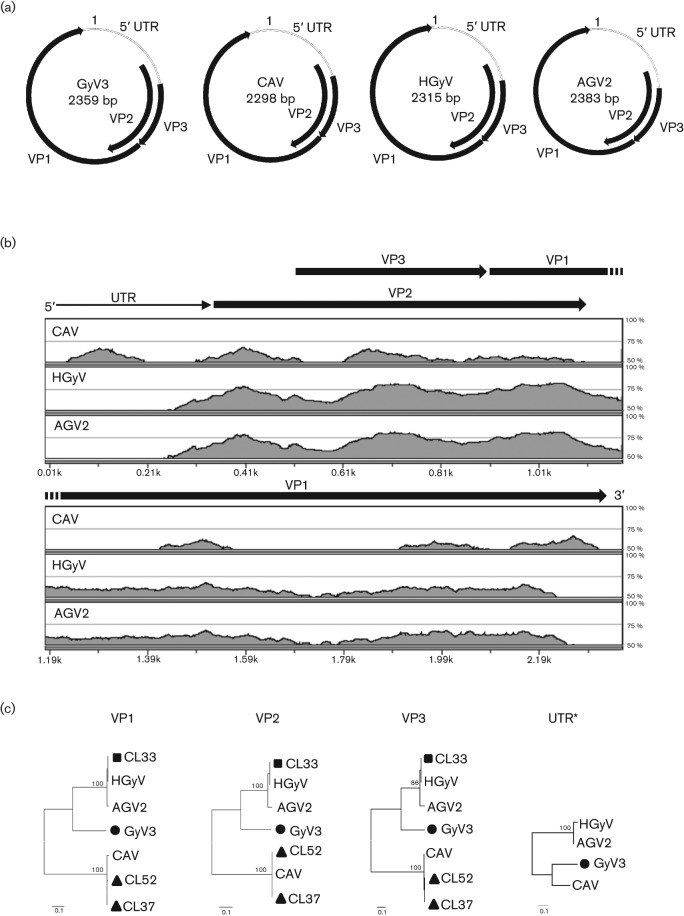
CAV-sequences were found in five faecal specimens; one of which also showed the presence of HGyV/AGV2 as well as novel (divergent) gyrovirus sequences. Complete DNA viral genomes of CAV and HGyV/AGV2 were generated. Two genomes, CL37 and CL52 (GenBank accession nos JQ308213–JQ308214), were 1823 bp in length with 95–99 % nucleotide similarity to CAV. CL37 and CL52 shared 99 % nucleotide similarity with 17 nt mismatches in VP1, resulting in two amino acid changes. These genomes were phylogenetically classified into the CAV genotype D (Eltahir et al., 2011a). Another genome of 1886 bp, CL33 (GenBank accession no. JQ308212), showed high DNA sequence similarities of 99 and 96 % to HGyV and AGV2, respectively. In the same faecal sample where CAV and HGyV/AGV2 were detected, a novel gyrovirus was found which we provisionally named GyV3. We acquired the complete genome (2359 bp) of GyV3 (GenBank accession no. JQ308210) by primer walking the iPCR product (except for the UTR that required plasmid subcloning of a short amplicon and the use of multiple primers to sequence through GC-rich regions). GyV3 had a genomic organization similar to those of CAV and HGyV/AGV2 as shown in Fig. 1(a). Three overlapping ORFs for the VP1 gene (1392 bp), VP2 gene (720 bp) and VP3 gene (378 bp) were detected in GyV3. The intergenic region of VP3 and VP1 was two nucleotides in GyV3 and HGyV/AGV2, but only one nucleotide in CAV. The 5′ UTR (371 bp) contained a polyadenylation signal (AATAAA) and a CAV promoter (TGTACAGGGGGGTACGTCA) with a putative oestrogen-response element (ACGTCA) (Miller et al., 2005). Unlike CAV and HGyV/AGV2 we did not detect multiple copies of this promoter in GyV3. This region also contained two GC-rich sequences GGCGGGGGGCCGGAGGCCCCCCGGTGGCCCCCCGCC and CCCCGCCCACAGGGCGGGC that may form two stem–loops. The first putative stem–loop was also found in CAV, but not in HGyV/AGV2 and the second was restricted to GyV3. The VP1 (463 aa) had a length similar to the major viral structural protein of CAV and HGyV/AGV2. Similar to CAV and HGyV/AGV2A a conserved motif XXTLXXAQ for rolling-circle replication was located near the C terminus of the GyV3 VP1 (Ilyina & Koonin, 1992). GyV3, like HGyV/AVG2, did not contain the rolling-circle replication motifs GQRWHTLVP and TATYALKEPV found in CAV (Ilyina & Koonin, 1992). Pair-wise sequence alignments revealed that GyV3 VP1 only shared 51 and 62 % amino acid similarities to CAV and HGyV, respectively (Table 1). Despite the high divergence in VP1 proteins, direct inspection of the sequence alignment discovered four highly conserved amino acid motifs 59VRLPNP64, 123SKIGGP128, 165WWRWAL170 and 221FSPVASLL228 (GenBank accession no. AF311892) among CAV, HGyV/AGV2 and GyV3. VP2, the second major ORF of GyV3, contained a signature motif CX5R (CNCGGFR) that may be responsible for intracellular signalling in CAV replication (Peters et al., 2002). GyV3 VP2 also shared a conserved motif WX7HX3CXCX5H (WLRECSRTHDAICNCGGFRRH) found in both CAV and torque teno virus (a phylogenetically unrelated and unassigned viral genus also with a small circular ssDNA genome) (Takahashi et al., 2000). VP2 protein in GyV3 shared a low amino acid similarity of 63 % to the genetically closest gyroviruses HGyV/AGV2 (Table 1). In CAV, VP3 protein was named apoptin and reported to induce apoptosis in tumour cells (Pietersen & Noteborn, 2000). The C-terminal region of the VP3 protein of GyV3 showed similarity to the nuclear localization domain of apoptin protein of CAV (Poon et al., 2005a, b). The similarities of VP3 of GyV3 were <57 % at the amino acid level to VP3 of the other gyroviruses. As for CAV (and AGV2) another GC-rich sequence (66 bp) was detected immediately downstream of VP1 in GyV3 that also contains two more potential stem–loops (GGGGGGGGGAAAACCCCCCCCC and CGGGGGGGATCTTCCCCCCCG). Multiple gyrovirus-specific elements were therefore identified in GyV3. The complete genome of GyV3 was aligned with other gyroviruses, showing that except for the UTR region GyV3 similarity was closer to HGyV/AGV2 than CAV in the three ORF coding regions (Fig. 1b). This finding was supported by pair-wise comparison in which GyV3 showed closer similarities to HGyV/AGV2 than to CAV at the nucleotide level for the VP1, VP2 and VP3 (apoptin) proteins (Table 1). The UTR of GyV3 was closer to that of CAV (62 % similar) than to the UTR of HGyV/AGV2 (37–47 % similar). A highly similar region (94 % identity over 109 bases) of high GC content was shared between GyV3 and CAV, indicating that recombination may have brought a CAV UTR region into the GyV3 genome. Phylogenetic analysis confirmed that GyV3 was more related to CAV in the UTR (Fig. 1c). Based on phylogenetic analyses and genetic distances, GyV3 may therefore be considered the third species in the genus Gyrovirus.
Fig. 1.
Human faeces-associated gyroviruses. (a) Genome organizations of a novel genovirus GyV3, CAV, HGyV and AGV2. (b) Global pair-wise sequence alignments of GyV3 with CAV, HGyV and AGV2 reference strains. The sequence similarity (%) is indicated by the height of each point along the y axis. The horizontal axis shows the nucleotide positions in the complete genome. (c) Phylogenetic trees generated with VP1, VP2, VP3 proteins and UTR regions of gyroviruses detected in human faeces and other gyroviruses. The scale indicates amino acid or nucleotide substitutions per position. Note: *The UTR sequences of CL33, CL37 and CL52 were not completed due to GC-rich contents.
Table 1. Pairwise nucleotide (upper right, bold) and amino acid (lower left) sequence similarities (%) between VP1, VP2, VP3 and UTR regions of GyV3 and other gyroviruses.
id, Identical.
| GyV3 | HGyV | AGV2 | CAV | |
|---|---|---|---|---|
| VP1 | ||||
| GyV3 | id | 62 | 63 | 51 |
| HGyV | 62 | id | 93 | 52 |
| AGV2 | 62 | 97 | id | 51 |
| CAV | 43 | 41 | 42 | id |
| VP2 | ||||
| GyV3 | id | 74 | 74 | 53 |
| HGyV | 63 | id | 97 | 57 |
| AGV2 | 63 | 96 | id | 56 |
| CAV | 42 | 43 | 42 | id |
| VP3 | ||||
| GyV3 | id | 70 | 69 | 56 |
| HGyV | 56 | id | 96 | 57 |
| AGV2 | 55 | 93 | id | 56 |
| CAV | 29 | 31 | 30 | id |
| UTR | ||||
| GyV3 | id | 37 | 47 | 62 |
| HGyV | – | id | 81 | 35 |
| AGV2 | – | – | id | 46 |
| CAV | – | – | – | id |
Although numerous molecular epidemiological studies of gyrovirus worldwide have been performed in chickens (Craig et al., 2009; Dos Santos et al., 2012; Eltahir et al., 2011b; Hailemariam et al., 2008; Kim et al., 2010), the prevalence of viruses in this genus in human faeces has not to our knowledge been reported. We used consensus primers based on a VP2 region highly conserved between the three gyrovirus species to test for gyroviruses. Primers ConGyF1 (5′-CGCTGGGGGCAGTGAA-3′) and ConGyR1 (5′-TCTTCGTCGCAGTCK-3′) were used for the first round of PCR, and primers ConGyF1 and ConGyR2 (5′-TCTTGCCATSTTACAGTCTT-3′) for the second round of PCR, resulting in an expected amplicon of ~470 bp. The PCR conditions for the first and second rounds was as follows: denaturation at 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 46 °C for 30 s and 72 °C for 1 min, a final extension at 72 °C for 10 min, and then held at 4 °C. Amplicons were then sequenced directly for species identification. In the faeces of Chilean children with acute gastroenteritis the frequency of CAV was 32 % (32/100), followed by HGyV/AGV2 (1 %; 1/100) and GyV3 (1 %; 1/100) with the HGyV/AGV2 and GyV3 found as co-infections with CAV (i.e. all three viruses in same faecal sample). Faeces from 100 healthy children from the same geographical area in Chile were also tested by consensus PCR for gyrovirus DNA. CAV was detected in 25/100 (25 %) faecal specimens (Table 2). No other gyroviruses were detected in the control group. The proportion of diarrhoeal children containing CAV (32/100 or 32 %) was not significantly different from that found in healthy controls (25/100 or 25 %) (_P_-value of Chi-squared test >0.05), suggesting that CAV was not associated with these Chilean children’s diarrhoea.
Table 2. Frequency of gyrovirus detection in faeces of Chilean children.
| CAV | HGyV/AGV2 | GyV3 | |
|---|---|---|---|
| Diarrhoea | 32/100 (32 %) | 1/100 (1 %) | 1/100 (1 %) |
| Control | 25/100 (25 %) | 0/100 (0 %) | 0/100 (0 %) |
The frequent detection of CAV in the Chilean children’s faeces suggested a possible dietary origin from consumed chicken meat. Consistent with this possibility a high rate of anti-CAV antibody detection was reported in 1994 in Chilean chicken (Toro et al., 1994). A high prevalence of CAV infection in chicken may therefore account for its frequent detection in human faeces. Interestingly, 100 faeces from diarrhoeal children in Burkina Faso in West Africa (Bonkoungou et al., 2010) did not contain any gyrovirus DNA possibly reflecting a lower rate of chicken consumption by children from this West African country and/or a lower rate of local chicken infection with CAV. When muscle tissues from chickens bought in USA stores (San Francisco, CA) (Li et al., 2010a) were tested all 21 (100 %) were positive for CAV. This finding was in line with a previous report of a high rate of CAV-specific antibody in USA chicken sera (Toro et al., 2006) and the frequent use of CAV vaccination in USA chickens using attenuated viral strains. Faecal samples from dogs and cats with diarrhoea from the USA were also tested. Thirty-seven per cent of dog samples (13/35) and 13 % of cat samples (2/15) were positive for CAV. Because chicken meat is frequently a component of prepared pet foods a dietary source of CAV in these companion animals is also possible.
Despite the prior PCR detection of CAV in chickens from Nigeria (Ducatez et al., 2006), DNA (1 µg) extracted from muscle tissues of 36 chickens from Nigeria and from muscle tissues of 54 Nigerian goats and 46 Nigerian cows (Li et al., 2010a) were gyrovirus PCR negative (data not shown). DNA (1 µg) from 13 human muscle and 26 human skin samples from autopsies of 28 USA adults was also extracted and also tested negative for gyrovirus DNA (data not shown).
As is the case for the frequent detection of pig circovirus DNA (Victoria et al., 2010) or nucleic acids from plant viruses (Li et al., 2010b; Phan et al., 2011; Victoria et al., 2009; Zhang et al., 2006) in human or animal faeces without evidence of viral replication it is also possible that the frequent detection of gyrovirus DNA in human and animal stools also reflects the common consumption of CAV-infected chicken meat. The recent report of HGyV on the surface of human skin (Sauvage et al., 2011) and its genetic closeness to AGV2 also suggests the possibility of cross-species transmission and gyrovirus replication in humans. Whether GyV3 can infect chickens and/or humans remains to be determined.
Acknowledgements
We acknowledge NHLBI R01HL083254 and support from BSRI and IDEXX to E. D. We thank Dr Phillip Ursell from the Department of Pathology at UCSF for providing human muscle and skin tissue specimens.
References
- Adair B. M.**(2000).**Immunopathogenesis of chicken anemia virus infection. Dev Comp Immunol 24, 247–255 10.1016/S0145-305X(99)00076-2 [DOI] [PubMed] [Google Scholar]
- Bonkoungou I. J., Sanou I., Bon F., Benon B., Coulibaly S. O., Haukka K., Traoré A. S., Barro N.**(2010).**Epidemiology of rotavirus infection among young children with acute diarrhoea in Burkina Faso. BMC Pediatr 10, 94 10.1186/1471-2431-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M. I., Rimondi A., Delamer M., Sansalone P., König G., Vagnozzi A., Pereda A.**(2009).**Molecular characterization of chicken infectious anemia virus circulating in Argentina during 2007. Avian Dis 53, 331–335 10.1637/8478-100808-Reg.1 [DOI] [PubMed] [Google Scholar]
- Dos Santos H. F., Knak M. B., de Castro F. L., Slongo J., Ritterbusch G. A., Klein T. A., Esteves P. A., Silva A. D., Trevisol I. M.**& other authors (2012).**Variants of the recently discovered avian gyrovirus 2 are detected in Southern Brazil and The Netherlands. Vet Microbiol 155, 230–236 [DOI] [PubMed] [Google Scholar]
- Ducatez M. F., Owoade A. A., Abiola J. O., Muller C. P.**(2006).**Molecular epidemiology of chicken anemia virus in Nigeria. Arch Virol 151, 97–111 10.1007/s00705-005-0609-7 [DOI] [PubMed] [Google Scholar]
- Eltahir Y. M., Qian K., Jin W., Qin A.**(2011a).**Analysis of chicken anemia virus genome: evidence of intersubtype recombination. Virol J 8, 512 10.1186/1743-422X-8-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltahir Y. M., Qian K., Jin W., Wang P., Qin A.**(2011b).**Molecular epidemiology of chicken anemia virus in commercial farms in China. Virol J 8, 145 10.1186/1743-422X-8-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailemariam Z., Omar A. R., Hair-Bejo M., Giap T. C.**(2008).**Detection and characterization of chicken anemia virus from commercial broiler breeder chickens. Virol J 5, 128 10.1186/1743-422X-5-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina T. V., Koonin E. V.**(1992).**Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20, 3279–3285 10.1093/nar/20.13.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. R., Kwon Y. K., Bae Y. C., Oem J. K., Lee O. S.**(2010).**Molecular characterization of chicken infectious anemia viruses detected from breeder and broiler chickens in South Korea. Poult Sci 89, 2426–2431 10.3382/ps.2010-00911 [DOI] [PubMed] [Google Scholar]
- Kimura M.**(1980).**A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Li L., Kapoor A., Slikas B., Bamidele O. S., Wang C., Shaukat S., Masroor M. A., Wilson M. L., Ndjango J. B.**& other authors (2010a).**Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84, 1674–1682 10.1128/JVI.02109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J. G., Wang C., Jones M., Fellers G. M., Kunz T. H., Delwart E.**(2010b).**Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84, 6955–6965 10.1128/JVI.00501-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. M., Jarosinski K. W., Schat K. A.**(2005).**Positive and negative regulation of chicken anemia virus transcription. J Virol 79, 2859–2868 10.1128/JVI.79.5.2859-2868.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Ryan M., Díaz J., Mamani N., Navarrete M., Vallebuono C.**(2007).**Impact of rotavirus infections on outpatient clinic visits in Chile. Pediatr Infect Dis J 26, 41–45 10.1097/01.inf.0000247104.01291.71 [DOI] [PubMed] [Google Scholar]
- O’Ryan M. L., Lucero Y., Prado V., Santolaya M. E., Rabello M., Solis Y., Berríos D., O’Ryan-Soriano M. A., Cortés H., Mamani N.**(2009).**Symptomatic and asymptomatic rotavirus and norovirus infections during infancy in a Chilean birth cohort. Pediatr Infect Dis J 28, 879–884 10.1097/INF.0b013e3181a4bb60 [DOI] [PubMed] [Google Scholar]
- Peters M. A., Jackson D. C., Crabb B. S., Browning G. F.**(2002).**Chicken anemia virus VP2 is a novel dual specificity protein phosphatase. J Biol Chem 277, 39566–39573 10.1074/jbc.M201752200 [DOI] [PubMed] [Google Scholar]
- Phan T. G., Kapusinszky B., Wang C., Rose R. K., Lipton H. L., Delwart E. L.**(2011).**The fecal viral flora of wild rodents. PLoS Pathog 7, e1002218 10.1371/journal.ppat.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen A., Noteborn H. M.**(2000).**Apoptin. Adv Exp Med Biol 465, 153–161 10.1007/0-306-46817-4_14 [DOI] [PubMed] [Google Scholar]
- Poon I. K., Oro C., Dias M. M., Zhang J., Jans D. A.**(2005a).**Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res 65, 7059–7064 10.1158/0008-5472.CAN-05-1370 [DOI] [PubMed] [Google Scholar]
- Poon I. K., Oro C., Dias M. M., Zhang J. P., Jans D. A.**(2005b).**A tumor cell-specific nuclear targeting signal within chicken anemia virus VP3/apoptin. J Virol 79, 1339–1341 10.1128/JVI.79.2.1339-1341.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijsewijk F. A., Dos Santos H. F., Teixeira T. F., Cibulski S. P., Varela A. P., Dezen D., Franco A. C., Roehe P. M.**(2011).**Discovery of a genome of a distant relative of chicken anemia virus reveals a new member of the genus Gyrovirus. Arch Virol 156, 1097–1100 10.1007/s00705-011-0971-6 [DOI] [PubMed] [Google Scholar]
- Rosenberger J. K., Cloud S. S.**(1989).**The isolation and characterization of chicken anemia agent (CAA) from broilers in the United States. Avian Dis 33, 707–713 10.2307/1591148 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M.**(1987).**The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Sauvage V., Cheval J., Foulongne V., Gouilh M. A., Pariente K., Manuguerra J. C., Richardson J., Dereure O., Lecuit M.**& other authors (2011).**Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol 85, 7948–7950 10.1128/JVI.00639-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K. A.**(2009).**Chicken anemia virus. Curr Top Microbiol Immunol 331, 151–183 10.1007/978-3-540-70972-5_10 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Iwasa Y., Hijikata M., Mishiro S.**(2000).**Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch Virol 145, 979–993 10.1007/s007050050689 [DOI] [PubMed] [Google Scholar]
- Toro H., McNulty M. S., Hidalgo H., Rosende S., Connor T. J.**(1994).**Detection of chicken anemia virus antibodies in four poultry operations in Chile. Prev Vet Med 21, 103–106 10.1016/0167-5877(94)90035-3 [DOI] [Google Scholar]
- Toro H., Ewald S., Hoerr F. J.**(2006).**Serological evidence of chicken infectious anemia virus in the United States at least since 1959. Avian Dis 50, 124–126 10.1637/7442-092205R.1 [DOI] [PubMed] [Google Scholar]
- van Santen V. L., Joiner K. S., Murray C., Petrenko N., Hoerr F. J., Toro H.**(2004).**Pathogenesis of chicken anemia virus: comparison of the oral and the intramuscular routes of infection. Avian Dis 48, 494–504 10.1637/7155-010904R [DOI] [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E.**(2009).**Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651 10.1128/JVI.02301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J. G., Wang C., Jones M. S., Jaing C., McLoughlin K., Gardner S., Delwart E. L.**(2010).**Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol 84, 6033–6040 10.1128/JVI.02690-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W. H., Run J. Q., Wei C. L., Soh S. W., Hibberd M. L., Liu E. T., Rohwer F., Ruan Y.**(2006).**RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4, e3 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]