Inertial Focusing for Tumor Antigen–Dependent and –Independent Sorting of Rare Circulating Tumor Cells (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 3.
Abstract
Circulating tumor cells (CTCs) are shed into the bloodstream from primary and metastatic tumor deposits. Their isolation and analysis hold great promise for the early detection of invasive cancer and the management of advanced disease, but technological hurdles have limited their broad clinical utility. We describe an inertial focusing–enhanced microfluidic CTC capture platform, termed “CTC-iChip,” that is capable of sorting rare CTCs from whole blood at 107 cells/s. Most importantly, the iChip is capable of isolating CTCs using strategies that are either dependent or independent of tumor membrane epitopes, and thus applicable to virtually all cancers. We specifically demonstrate the use of the iChip in an expanded set of both epithelial and nonepithelial cancers including lung, prostate, pancreas, breast, and melanoma. The sorting of CTCs as unfixed cells in solution allows for the application of high-quality clinically standardized morphological and immunohistochemical analyses, as well as RNA-based single-cell molecular characterization. The combination of an unbiased, broadly applicable, high-throughput, and automatable rare cell sorting technology with generally accepted molecular assays and cytology standards will enable the integration of CTC-based diagnostics into the clinical management of cancer.
INTRODUCTION
The rarity of circulating tumor cells (CTCs) in the blood of cancer patients has required development of highly specialized technologies for their isolation (1, 2). Once detected, enumeration and molecular characterization of CTCs have been applied to prognostic classifications of breast, prostate, and colon cancers (3), and to predictive markers of targeted drug therapy in lung cancer (4). However, the limited sensitivity of commercially available approaches combined with the complexity and heterogeneity of the disease has restricted the broad acceptance and dissemination of CTC-based diagnostics (5).
Several strategies have been used to process blood for analysis of CTCs, including platforms for rapid scanning of unpurified cell populations (6–8). The most common enrichment approaches have used antibodies against the cell surface protein epithelial cell adhesion molecule (EpCAM). Labeling CTCs with anti-EpCAM–coated beads, followed by bulk magnetic enrichment methods (9–11), has been tested. The U.S. Food and Drug Administration (FDA)–approved Veridex system, CellSearch, immunomagnetically labels CTCs and then enriches the cells by bulk purification across a magnetic field. Conceptually, EpCAM-based CTC capture may have limited ability to identify tumor cells with reduced expression of this epithelial marker as a result of the epithelial-mesenchymal transition (EMT) (12). However, tumor antigen–independent CTC enrichment, such as bulk depletion of hematopoietic cells, suffers from poor yields and low purity (13, 14). Together, CTC isolation approaches have traditionally involved multiple batch processing steps, resulting in substantial loss of CTCs (14).
Recently, we introduced microfluidic methods to improve the sensitivity of CTC isolation (15), a strategy that is particularly attractive because it can lead to efficient purification of viable CTCs from unprocessed whole blood (16–21). The micropost CTC-Chip (μpCTC-Chip) relies on laminar flow of blood cells through anti-EpCAM antibody-coated microposts (15), whereas the herringbone CTC-Chip (HbCTC-Chip) uses microvortices generated by herringbone-shaped grooves to direct cells toward antibody-coated surfaces (16). Although promising, these methods require surface functionalization to bind to tumor antigens on CTCs and thus yield CTCs that are immobilized within a micro-fluidic chamber and are not readily subjected to either standard clinical cytopathological imaging or single-cell molecular characterization.
To address the shortcomings of the current approaches, we developed a strategy that combines the strengths of microfluidics for rare cell handling while incorporating the benefits of magnetic-based cell sorting. After the magnetic labeling of cells in whole blood, this capture platform integrates three sequential microfluidic technologies within a single automated system: (i) debulking by separation of nucleated cells, including CTCs and white blood cells (WBCs), from red blood cells (RBCs) and platelets using deterministic lateral displacement (22); (ii) alignment of nucleated cells within a microfluidic channel using inertial focusing (23); and (iii) deflection of magnetically tagged cells into a collection channel. In essence, these three integrated microfluidic functions replace bulk RBC lysis and/or centrifugation, hydrodynamic sheath flow in flow cytometry, and magnetic-activated cell sorting (MACS). We call this integrated microfluidic system the CTC-iChip, based on the inertial focusing strategy, which allows positioning of cells in a near-single file line, such that they can be precisely deflected using minimal magnetic force. This integrated microfluidic platform, with its ability to isolate CTCs in suspension using both tumor antigen–dependent and tumor antigen–independent modes, is compatible with high-definition imaging and single-cell molecular analyses, as well as standard clinical cytopathology. We demonstrate its capabilities for diverse cancer diagnostic applications in both epithelial and nonepithelial cancers.
RESULTS
CTC-iChip design and function
The overall CTC-iChip isolation strategy is depicted in Fig. 1 and fig. S1. We explored two modes of immunomagnetic sorting to isolate CTCs: a positive selection mode (posCTC-iChip), whereby CTCs are identified and sorted on the basis of their expression of EpCAM, and a negative selection mode (negCTC-iChip), in which the blood sample is depleted of leukocytes by immunomagnetically targeting both the common leukocyte antigen CD45 and the granulocyte marker CD15.
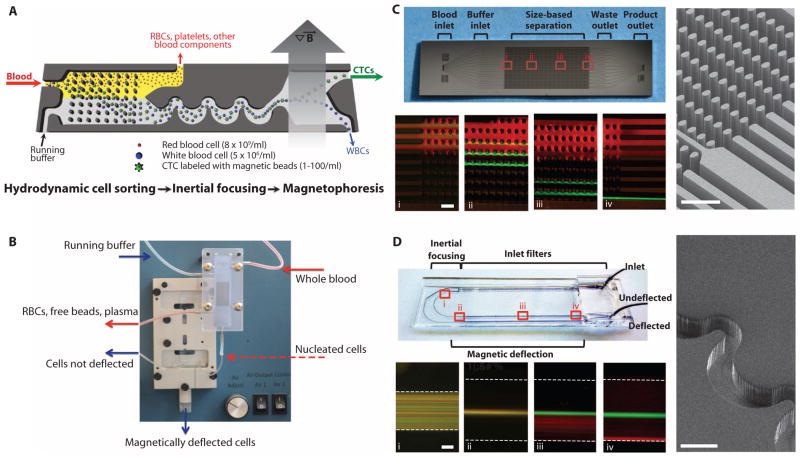
Fig. 1.
The CTC-iChip system. (A) Three microfluidic components of the CTC-iChip are shown schematically. Whole blood premixed with immunomagnetic beads and buffer comprises the inputs. The figure demonstrates the positive isolation method; however, the system can be operated in negative depletion mode. (B) Integrated microfluidic system. The debulking array sits in a custom polycarbonate manifold that enables fluidic connections to the inputs, waste line, and second-stage microfluidic channels. The inertial focusing and magnetophoresis chip is placed in an aluminum manifold that houses the quadrupole magnetic circuit. Magnetically deflected cells are collected in a vial. (C) Hydrodynamic size–based sorting. A mixture of 2-μm (red) and 10-μm (green) beads enters the channel (i). Whereas the 2-μm beads remain in laminar flow and follow the fluid streamlines, the 10-μm spheres interact with the post-array (ii and iii) as shown in the scanning electron microscope (SEM) image (right panel). Larger beads are fully deflected into the coincident running buffer stream by the end of the array (iv). Scale bars, 100 μm. (D) Cell focusing and magnetophoretic sorting. Magnetically labeled SKBR3 (red) and unlabeled PC3-9 (green) cell populations are mixed and enter the channel in random distribution (i). After passing through 60 asymmetric focusing units (pictured in the SEM, right panel), the cells align in a single central stream (ii). Magnetically tagged cells are then deflected (iii) using an external magnetic field, and separation is achieved by the end of the channel (iv). Scale bars, 100 μm.
Target cell labeling was developed and characterized for both operational modes (fig. S2). After labeling, the first stage within the CTC-iChip used hydrodynamic size–based sorting to achieve low shear microfluidic debulking of whole blood (22, 24). RBCs, platelets, plasma proteins, and free magnetic beads were discarded, whereas nucleated cells (WBCs and CTCs) were retained and presented to the second stage for inertial focusing. The efficient removal of free beads is critical because these may accumulate in the magnetophoresis channel and significantly reduce the sensitivity and specificity of the approach. The operational principle of microfluidic debulking is based on hydrodynamic size–dependent deterministic lateral displacement (22, 24), in which coincident flow of cell-containing and cell-free solutions through an array of microposts leads to rapid size-based separation (Fig. 1C and fig. S3). We tested two different array configurations with gaps between microposts of 20 or 32 μm. An array with 20-μm gaps retains virtually all nucleated cells with minimal contaminating RBCs but has a cutoff for cells larger than 21 μm and may therefore lose large CTCs or CTC clusters. In contrast, an array with 32-μm gaps has an extended operating range for cells between 8 and 30 μm but retains only 60% of WBCs. Because the cells lost in the 32-μm gap array are granulocytes and lymphocytes that are smaller than the reported CTC sizes (16), we selected this array for the CTC-iChip.
The second CTC-iChip component orders nucleated cells within the microfluidic channel, both laterally and longitudinally, so they can be precisely deflected into a collection channel with minimal magnetic moment. The rationale underlying the inertial focusing of cells in microchannels is based on the principles of pipe flow (23, 25); essentially, a cellular fluid entering asymmetric, curved channels emerges as a tight row of individual cells traveling within a defined streamline position (Fig. 1D). We tested variable cell suspensions for focusing performance; WBCs as well as cancer cell lines were well focused within the operational parameters (hematocrit less than 0.4%; flow rate between 50 and 150 μl/min; nucleated cell concentration less than 3 × 106/ml) (fig. S4). Inertial focusing operational parameters were matched to output of the preceding debulking array, and the in-line integration of these complex microfluidic structures within the CTC-iChip thus avoided cell losses associated with commonly used batch processing strategies.
In the final CTC-iChip component, magnetically labeled cells are separated from unlabeled cells within a deflection channel. The precise control over cell position provided by inertial focusing prevents cellular collisions during magnetophoresis; therefore, cell displacement occurs as a predictable function of magnetic load. We modeled the forces exerted on cells labeled with 1-μm beads using a quadrupole magnetic circuit (fig. S5) and predicted deflection patterns under different flow and magnetic load conditions (Fig. 2A). This model was tested using magnetically labeled PC3-9 human prostate cancer cells. The measured deflection distance, plotted as a function of magnetic load, matched the prediction (Fig. 2B).
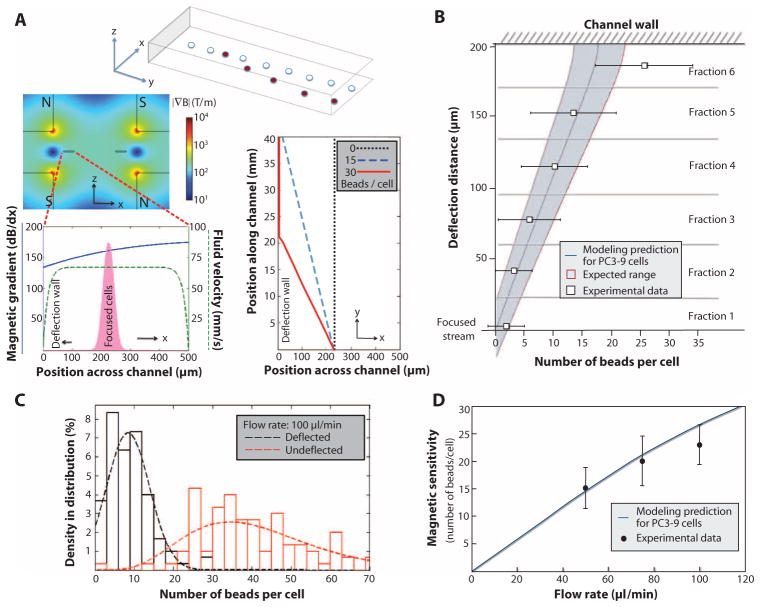
Fig. 2.
Modeling and magnetic sensitivity of the system. (A) A mathematical model describes the deflection of labeled cells (red) from a focused stream (white). Finite element method analysis of the quadrupole magnetic circuit and fluid flow in the channel provided estimates of the magnetic gradient (blue) and flow rate (green) across the deflection channel (left panel). This information, in conjunction with our experimental understanding of cell position in the focused stream (pink), was used to construct an overall model to predict the trajectories of focused cells with varying magnetic loads (right panel). (B) High sensitivity of inertial focusing enhanced magnetophoresis. Human PC3-9 cells were labeled with varying numbers of magnetic beads and collected in separate exit streams after traveling in the 4-cm-long magnetic deflection channel, fractionating the cells based on magnetic deflection distance. The beads on a representative population of cells were counted in each fraction. The deflection distance was measured from focused stream position to the channel wall. Fraction 6 included cells that deflected all the way and traveled at the wall; therefore, this data point did not match the simulation. The expected variations in cell sizes and the initial distribution of cells in the focused stream contribute to a variation in the deflection pattern that is reflected by shading the expected range around the model prediction. (C) The experimental “magnetic sensitivity” was determined by plotting histograms of bead loading density for deflected and undeflected cells for a given flow rate. The intersection of curve fits of these data represents the minimum number of beads required to deflect a cell. (D) The minimum required magnetic load increases with higher flow rates, as expected, and is accurately predicted by the model.
To demonstrate the dependence of sensitivity on flow speed, we processed labeled cells at various flow rates and quantified the number of beads per cell for deflected and nondeflected outputs (Fig. 2C and fig. S6). The improvement in sensitivity with increasing magnetic residence time (by reducing flow speed) correlated with the predictive model (Fig. 2D), indicating high magnetic sensitivity for the overall system (5 to 20 beads per cell, depending on cell size). The process parameters characterized for the posCTC-iChip applied similarly to the negCTC-iChip.
Evaluating the CTC-iChip using cells spiked into whole blood
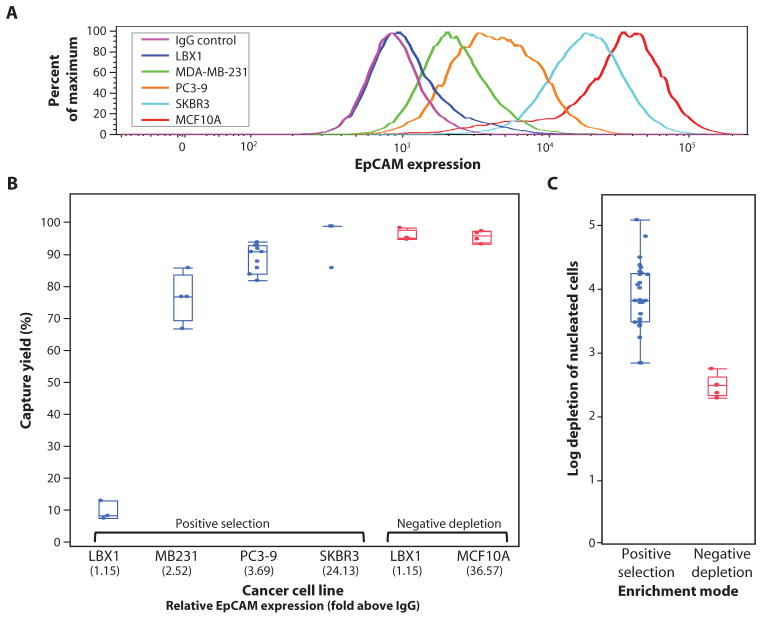
To evaluate the efficiency of the CTC-iChip, we spiked five cell lines spanning a broad range of EpCAM expression into healthy whole blood and isolated using posCTC-iChip or negCTC-iChip modes. The EpCAM expression of each cell line was quantified by comparing the anti-EpCAM signal to that of a matched irrelevant antibody (Fig. 3A). Recovery of SKBR3 human breast cancer cells [24-fold EpCAM signal over control immunoglobulin G (IgG)] was 98.6 ± 4.3% (mean ± SD), and capture of human prostate PC3-9 cancer cells (3.7-fold EpCAM signal) was 89.7 ± 4.5% (Fig. 3B). Even cells with minimal EpCAM expression, such as MDA-MB-231 (26), a “triple-negative” mesenchymal breast cancer cell line (only 2.5-fold EpCAM signal over control), were recovered with 77.8 ± 7.8% capture efficiency by posCTC-iChip. Virtually complete abrogation of EpCAM expression, achieved by ectopic expression of the EMT master regulator LBX1 in MCF10A human breast cancer cells (MCF10A-LBX1) (27), resulted in 10.9 ± 3.0% capture efficiency.
Fig. 3.
Evaluation of overall system performance using cancer cell lines spiked into whole blood. (A) Quantitation of variable EpCAM expression in five cell lines using flow cytometry. (B) Capture yield of positive selection and negative depletion modes using various cell lines expressing different levels of EpCAM. (C) Background in posCTC-iChip product is measured, achieving >3.5-log depletion of WBCs. In contrast, negCTC-iChip has an order of magnitude lower purification. In both (B) and (C), each data point is an experimental result. Upper and lower bounds of the boxes signify the 75th and 25th quantiles, respectively. Perpendicular line in the box represents median value, and data points left above or below the error bars are outliers.
Switching to the negCTC-iChip, both the epithelial parental MCF10A cells and their highly mesenchymal MCF10A-LBX1 derivatives were captured at equal efficiency (96.7 ± 1.9% for MCF10As and 97.0 ± 1.7% for the MCF10A-LBX1 derivatives) (Fig. 3B). Together, these two modes demonstrate the flexibility of the CTC-iChip to isolate a broad spectrum of rare cells with high efficiency in both tumor antigen–dependent and tumor antigen–independent modes.
Sample purity was analyzed for both operating modes. Using EpCAM-based positive selection, we achieved an average >3.5-log purification (mean, 1500 WBCs/ml of whole blood; range, 67 to 2537 WBCs/ml). In the leukocyte depletion mode, purification was 2.5 log (mean, 32,000 WBCs/ml; range, 17,264 to 39,172 WBCs/ml) (Fig. 3C). In the posCTC-iChip, the vast majority of contaminating WBCs carried magnetic beads, suggesting that nonspecific interactions between WBCs and either the anti-EpCAM antibody or the beads themselves caused the contamination. In the negCTC-iChip, contaminating WBCs were free of beads, suggesting that they comprise a population of leukocytes with reduced CD45 or CD15 expression, as confirmed by flow cytometry (table S1).
posCTC-iChip isolation of CTCs
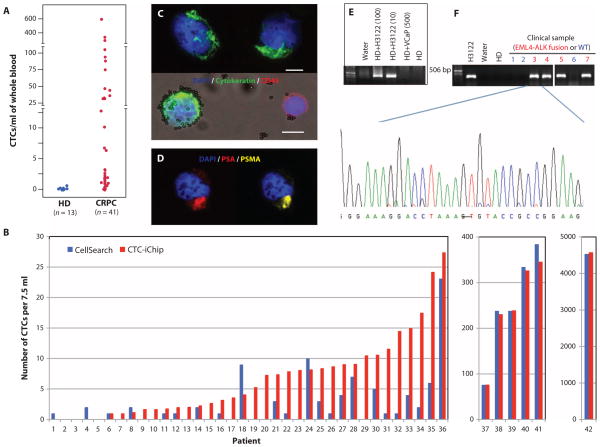
We tested the posCTC-iChip in patients with prostate cancer, a disease in which metastatic lesions primarily affect bone, and hence, CTC analysis is key to analyzing recurrences after resection of the primary tumor. On average, 10 ml (range, 6 to 12 ml) of whole blood was analyzed from these patients. Using triple staining for cytokeratins (CKs) (epithelial marker), CD45 (leukocytes), and 4′,6-diamidino-2-phenylindole (DAPI) (nuclear marker), we identified ≥0.5 CTC/ml in 37 of 41 (90%) prostate patients with recurrent (castration-resistant) disease (mean, 50.3/ml; range, 0.5 to 610/ml; median, 3.2/ml) (Fig. 4A). The detection cutoff of 0.5 CTC/ml was more than 2 SDs above the mean number of CK+ cells detected in 13 healthy donors (excluding an outlier with 0.7/ml; mean ± SD, 0.17 ± 0.12/ml; median, 0.19/ml; range, 0 to 0.33/ml). WBC contamination in the posCTC-iChip product was low (mean, 1188/ml; median, 352/ml; range, 58 to 9249/ml), resulting in high sample purities (mean, 7.8%; median, 0.8%; range for samples with ≥0.5 CTC/ml, 0.02 to 43%) (fig. S7).
Fig. 4.
CTC isolation by posCTC-iChip in cancer patients. (A) CTCs isolated from castrate-resistant prostate cancer (CRPC) patients were enumerated and compared with blood specimens processed from healthy donors. (B) EpCAM-based isolation using posCTC-iChip was compared with the Cell-Search system. Clinical samples were metastatic cancer patients of prostate (n = 19), breast (n = 12), pancreas (n = 6), colorectal (n = 2), and lung (n = 2). All counts were normalized to 7.5 ml. (C) For enumeration of CTCs from CRPC patients, CK8/18/19 staining was used (green). CD45 antigen (red) was used to identify contaminating leukocytes. Scale bars, 10 μm. (D) A CTC from a CRPC patient was stained for prostate-specific antigen (PSA) (red), prostate-specific membrane antigen (PSMA) (yellow), and DAPI (blue) to demonstrate dual immunofluorescence staining for PSAs. (E) Validation of EML4-ALK RT-PCR assay was completed with cell lines. posCTC-iChip products of whole blood from a healthy donor (HD) spiked with 0, 10, and 100 H3122 cells (expressing EML4-ALK variant 1) per 10 ml were subjected to RT-PCR for detection of the EML4-ALK fusion. Product isolated from healthy donor blood spiked with 500 VCaP cells/ml was processed as a negative control. (F) posCTC-iChip products from patient samples known to harbor the EML4-ALK translocation by FISH were similarly processed as in (E), and the bands were sequenced to confirm the presence of the fusion transcript. A representative sequence trace from patient 3 shows the translocation breakpoint between exon 13 of EML4 and exon 20 of ALK. CTC analysis of three patients whose cancer lacks the translocation was used to establish specificity: a prostate cancer patient (lane 1), an EGFR mutant lung cancer patient (lane 2), and a _HER2_-amplified lung cancer patient (lane 6).
We performed a detailed comparison of the posCTC-iChip with the FDA-approved CellSearch system (Fig. 4B). To minimize reagent variability between platforms, we used anti-EpCAM capture as well as CK and CD45 staining antibodies from the same source, and consistent criteria were used to evaluate putative CTCs. CTCs were defined as DAPI+/CD45−/CK+, and WBCs were defined as DAPI+ or DAPI+/CD45+ events. Specimens from prostate (n = 19) and other cancers (breast, n = 12; pancreas, n = 6; colorectal, n = 2; lung, n = 2) were compared. Although both assays performed well with high CTC loads (>30 CTCs per 7.5 ml), at lower CTC numbers, there was a marked differential in capture efficiency. Among the 86% (36 of 42) of metastatic cancer patients with fewer than 30 CTCs/7.5 ml, the number of CK+ CTCs isolated with the posCTC-iChip was significantly higher in 22 cases (P < 0.001, paired t test analysis). The remaining 14 cases had CTCs below detection limits for both systems (Fig. 4B and table S2). Thus, the sensitivity of the CTC-iChip is particularly critical in patients with a lower CTC burden.
In addition to capturing more CTCs in patients with lower CTC burdens, the iChip isolates these cells in suspension, which in turn enables their immobilization on a standard glass slide for high-resolution imaging and standard clinical cytopathological examination (fig. S8), as well as simultaneous staining for multiple biomarkers (Fig. 4, C and D). Beyond imaging, molecular genetic tools are increasingly applied to the characterization of CTCs. Nowhere is this more evident than in non–small cell lung cancer (NSCLC), where targeted therapies can provide marked clinical benefit (28). Among the most challenging assays is detection of the EML4-ALK translocation in about 3% of cases, which marks those responsive to the selective targeted inhibitor crizotinib. Detection of this intrachromosomal translocation by fluorescence in situ hybridization (FISH) is difficult, and, at the molecular level, the variability of chromosomal breakpoints necessitates RNA-based detection of the fusion transcript, which cannot be readily achieved using either fixed CTCs or free plasma nucleic acids.
We established a reverse transcription polymerase chain reaction (RT-PCR) assay capable of detecting the EML4-ALK translocation in H3122 lung cancer cells spiked into WBCs at a purity of 0.1% or introduced into whole blood (10 cells/10 ml) and processed through the posCTC-iChip (Fig. 4E). In patient specimens, the EML4-ALK transcript was detected in CTCs from four cases known to have this chromosome rearrangement by FISH analysis of the primary tumor. It was absent in CTCs from two NSCLC patients and one patient with prostate cancer whose tumors were all known to lack this abnormality. In cases where CTC-based RNA analysis identified the expected product, nucleotide sequencing confirmed the breakpoint in the fusion transcript (Fig. 4F). Thus, the posCTC-iChip allowed purification of CTCs for RNA-based molecular genotyping.
negCTC-iChip to isolate CTCs
Given the heterogeneity of circulating cancer cells, including the subset thought to undergo EMT, depletion of normal blood cells from clinical specimens should allow characterization of unlabeled nonhematopoietic cells. We analyzed CTCs from 10 patients with metastatic breast cancer, including luminal (ER+/PR+, n = 6), triple-negative (ER−/PR−/HER2−, n = 2), and HER2+ (n = 2) subtypes. Triple-negative breast cancers are noteworthy in that they express primarily mesenchymal markers and are unlikely to be captured efficiently using positive selection for EpCAM+ cells (20).
We stained the enriched CTC specimens using the Papanicolaou (Pap) stain, which is used for cytopathology analysis in clinical laboratories. In selected cases, the hematoxylin and eosin (H&E)–stained primary tumor tissue was compared with Pap-stained fine needle aspirates (FNAs) of the tumor or pleural effusions from the same patient. A remarkably similar morphological appearance was evident between cancer cells in the primary breast tumors and the isolated CTCs, as shown for three different patients in Fig. 5A. An ER+ breast cancer patient revealed small and regularly shaped cells in H&E, cytology, and CTC samples. Similarly, larger and more irregular tumor cells were found in a HER2+ primary breast cancer by H&E cytology and CTC analysis. In another example from a triple-negative high-grade breast cancer patient, pleomorphic CTCs similar to the patient’s previously sampled cytology specimen were seen.
Fig. 5.
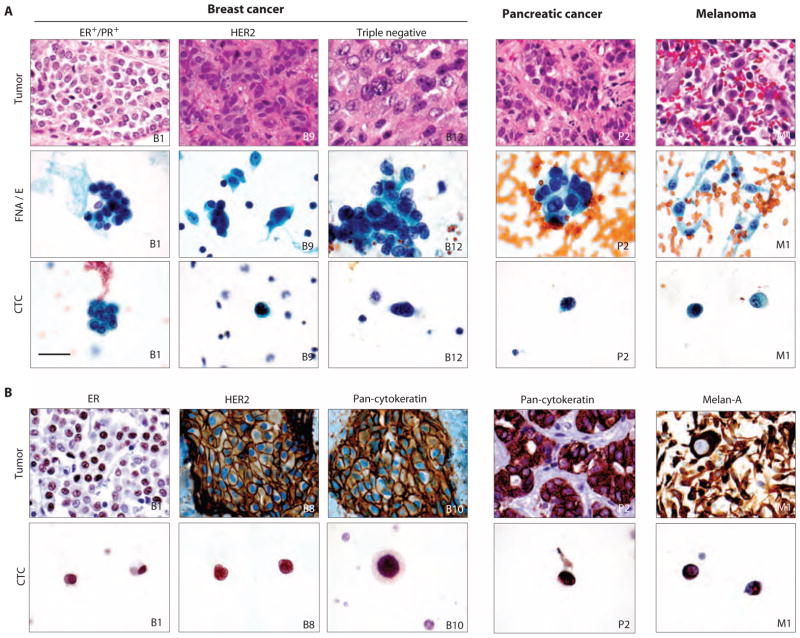
Classification of CTCs with cytopathology and ICC. (A) Specimens from H&E-stained primary and metastatic tumors (upper row) are compared with matched Pap-stained cytology samples from FNAs or pleural effusions (FNA/E) (middle row) and Pap-stained CTCs enriched from blood samples of the same patient using negCTC-iChip (lower row). Marked morphological similarity is seen between isolated CTCs and main tumors or cytology samples. (B) ICC profiles of primary and meta-static tumors (upper panel) matched to CTCs from the same patient (lower panel). All images: ×1000 original magnification. Scale bar is 30 μm and valid for all images.
We extended these morphological analyses to pancreatic cancer and melanoma with similar findings (Fig. 5A). For these, pancreatic adenocarcinoma showed CTCs of comparable size to the primary tumor by both histology and Pap cytology. Conversely, melanoma consisted of dyshesive tumor cells. The spindled cytoplasm in melanoma was also seen on the cytology preparation, but the CTCs appeared round. As a neural crest–derived malignancy, melanoma cells do not express EpCAM, and hence, their detection requires the negCTC-iChip isolation mode. Nevertheless, on the basis of established cellular and nuclear morphology criteria, our CTC analyses were considered to be of sufficient quality to enable a clinical diagnosis of suspicious for malignancy.
Pap-stained CTC slides were destained and then subjected to immunocytochemistry (ICC), which was first validated through cell lines (fig. S9). ICC of CTCs identified estrogen receptor (ER) protein in luminal breast cancer cells, keratin in triple-negative cells, and strong HER2 staining in cells from HER2+ breast cancers (Fig. 5B and fig. S10). Similarly, CTCs from patients with pancreatic cancer stained positive by ICC for CK, and CTCs from melanoma patients stained positive for the melanocytic marker Melan-A (Fig. 5B and fig. S10). The combination of Pap staining followed by ICC enabled enumeration of CTCs isolated by negCTC-iChip despite the presence of surrounding leukocytes.
Not all cytologically suspicious cells (for example, large cells with large, irregular nuclei as identified on the Pap-stained CTC slide) could be confirmed as tumor cells by ICC staining. Conversely, cells that were not scored as CTCs on initial cytological evaluation were subsequently identified as tumor cells by ICC, reflecting substantial heterogeneity in CTC size and morphology (fig. S11). Thus, by not relying exclusively on immunofluorescence-based scoring of CK+ cells, we were able to apply to CTCs the same rigorous morphological and immunohistochemical criteria used by clinical cytopathologists in the diagnosis of malignancy.
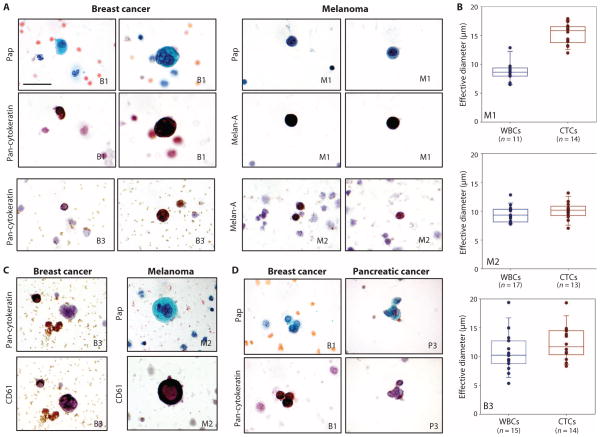
We observed large variation in CTC size among different cancer types. Although some CTCs were larger than leukocytes, there was considerable overlap between the two cell populations (Fig. 6). The variation in CTC size was not restricted to different cancer histologies. In one patient with ER+/PR+ breast cancer whose CTCs were isolated using the negCTC-iChip and analyzed using a combination of Pap stain and ICC, we identified CTCs ranging from 9 to 19 μm in diameter. Although most melanoma CTCs were large in size (>12 μm), one patient with metastatic melanoma had numerous CTCs less than 10 μm in diameter, detected using Pap and ICC for Melan-A (Fig. 6, A and B). In breast cancer and melanoma patients, some very large atypical cells (>30 μm) identified by Pap staining as having multilobed nuclei were at first assumed to be CTCs. However, ICC staining for the platelet marker CD61 confirmed their identity as megakaryocytes (Fig. 6C). Finally, application of the negCTC-iChip platform identified clusters of two to six CK+ CTCs in breast and pancreatic cancers, consistent with our previous detection of CTC clusters using the HbCTC-Chip (16) (Fig. 6D). The negative selection mode of the CTC-iChip thus provided a comprehensive and unbiased view of nonhematological cells in the bloodstream of cancer patients.
Fig. 6.
Variation of CTC sizes and morphologies. (A) CTCs from breast cancer and melanoma patients consecutively stained with Pap and either anti-CK (breast) or anti–Melan-A (melanoma) antibodies. (B) Quantitative analysis of the effective diameter (maximum feret diameter) for individual cells isolated in three cases. The top two panels are from different melanoma patients (M1 and M2). The bottom panel is from a breast cancer patient (B3). (C) Occasional very large cells with ample cytoplasm and multilobed nuclei were initially considered suspicious but were CK−. The same cells were subsequently restained for the platelet marker CD61, which supports their identification as circulating megakaryocytes. (D) CTCs were occasionally observed as clusters and confirmed by positive CK staining. All images: ×1000 original magnification. Scale bar, 30 μm.
Single-cell RNA expression in CTCs
Global CTC expression analyses may identify major pathways involved in metastasis (20), but the inherent heterogeneity of CTCs necessitates the identification of expression patterns and signaling pathways within individual cells. We therefore applied a series of single-cell micro-manipulation approaches to interrogate individual CTCs isolated from a patient with prostate cancer using the negCTC-iChip. Although micromanipulation approaches require expertise and can be time-consuming, the fact that the CTCs are unadulterated allows for more accurate RNA-based expression profiling than isolated fixed cells. EpCAM+ CTCs were distinguished from contaminating CD45+ leukocytes within the negCTC-iChip product by immunostaining (Fig. 7, A and B). CTCs identified as EpCAM+/CD45− were individually isolated and subjected to RNA analysis by multigene microfluidic quantitative RT-PCR (qRT-PCR), profiling for a panel of transcripts implicated in androgen receptor (AR) signaling, cellular proliferation, stem cell, epithelial and mesenchymal cell fates, and leukocyte-specific lineage (Fig. 7C). Single cells from the human prostate cancer cell line LNCaP were used to optimize assay conditions (fig. S12).
Fig. 7.
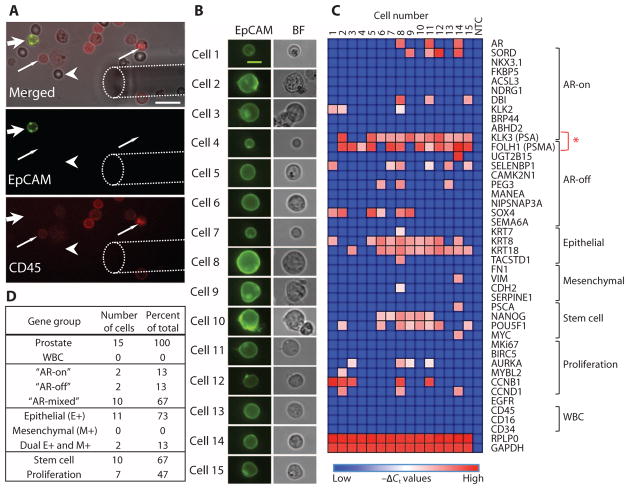
Heterogeneity of RNA expression between CTCs isolated from a prostate cancer patient. (A) Micromanipulation of single CTCs isolated from a blood specimen of a patient with prostate cancer using the negCTC-iChip and stained in solution with anti-EpCAM (green) and anti-CD45 (red) antibodies. Top panel shows a bright-field image merged. Wide arrow points to an EpCAM+/CD45− CTC. Thin arrow points to EpCAM−/CD45+ leukocytes. Arrowhead denotes an erythrocyte. Dashed line outlines the micromanipulator needle tip. Bottom two panels show distinct imaging channels. Scale bar, 20 μm. (B) EpCAM and bright-field images of 15 single prostate cancer CTCs from a single patient selected for transcriptional profiling. Scale bar, 10 μm. (C) Heat map of normalized gene expression (−Δ_C_t) of 43 genes in each of the single CTCs measured by microfluidic qRT-PCR. Columns list each individual prostate CTC, and rows show the panel of genes assayed, grouped thematically. The red asterisk highlights the gene expression patterns of PSA and PSMA, which provide a measure of AR signaling activity. NTC, no-template control. (D) Table listing the proportional distribution of various gene groups expressed in single CTCs isolated from the prostate cancer patient.
A marked heterogeneity was apparent among 15 CTCs isolated from a single patient with metastatic CRPC who had progressed through multiple lines of therapy, including androgen deprivation therapy with leuprolide, the chemotherapeutic drug docetaxel, and the second-line androgen biosynthesis inhibitor abiraterone acetate. Consistent with EpCAM+ immunostaining, 13 of the 15 CTCs were positive for epithelial gene expression, of which 2 CTCs were dual positive for epithelial as well as mesenchymal markers vimentin and N-cadherin (Fig. 7D). Thus, a subset of CTCs appears to have undergone partial EMT. CTC heterogeneity was also evident with expression of stem cell markers [Nanog, Oct-4 (POU5F1), and c-Myc] in 10 of the 15 CTCs, which overlapped primarily with epithelial markers within individual CTCs (Fig. 7C). Proliferation markers cyclin B, cyclin D, Aurora A kinase, and MYBL2 were detected in another subset of seven CTCs.
AR activity, previously defined in CTCs as the ratio of androgen-driven PSA to androgen-repressed PSMA expression (21), was heterogeneous among CTCs. The “AR on” phenotype (PSA expression only) was only seen in 2 of the 15 CTCs, whereas the “AR-off” state (PSMA only) was evident in 2 CTCs, and the “mixed AR” state (PSA+/PSMA+) in 10 CTCs (Fig. 7D). This distribution is concordant with single-cell immunofluorescence analysis of AR signaling status in CTCs from patients with CRPC (21).
DISCUSSION
The CTC-iChip described here has the ability to process large volumes of whole blood (8 ml/hour), with high throughput (107 cells/s) and at high efficiency, in positive selection (tumor antigen–dependent) and negative depletion (tumor antigen–independent) modes, thus enabling cytopathological and molecular characterization of both epithelial and nonepithelial cancers. Traditional magnetophoresis requires the attachment of either hundreds of beads per cell or very large beads to provide sufficient magnetic moment for cell isolation (11, 29). In contrast, by virtue of its ability to precisely position cells within the channel using inertial focusing, the fluidic design of the CTC-iChip allows for efficient fractionation of cells with only a few 1-μm beads, resulting in high yields and purity of CTC isolation.
We have tested initial “proof-of-principle” clinical applications of both the positive and negative selection modes of the CTC-iChip. The posCTC-iChip isolated CTCs at a purity of >0.1%, which is sufficient for molecular analyses, including detection of the EML4-ALK fusion transcript in NSCLC. Total CTC capture yield is critical to both genotyping and other applications, including enumeration for either prognostic or drug response measurements. The median number of CTCs detected by CK staining of posCTC-iChip product was 3.2 CTCs/ml, with 90% of clinical samples having CK+ cells above the threshold set using healthy donors. In a similar cohort using the CellSearch system, a median of 1.7 CTCs/ml was detected, with 57% of samples above the threshold (30). In our direct comparison between the posCTC-iChip and the CellSearch system, the microfluidic device was significantly more sensitive at low CTC numbers (<30 CTC/7.5 ml). These results suggest that a subpopulation of EpCAMlow cells was missed by the CellSearch bulk processing approach. Thus, whereas current commercially available approaches may be effective in patients with EpCAMhi CTCs, the CTC-iChip displayed increased sensitivity for patients with low numbers of circulating cancer cells, which may also have EpCAMlow expression.
Previously, we demonstrated the efficacy of two microfluidic systems to isolate CTCs from whole blood. CK+ CTCs were detected in 99% of patients with high purity (18 to 70%) in the first-generation micropost chip (15), and application of disease-specific markers for staining (PSA) and computer-assisted enumeration methods were later found to improve system reliability and specificity (19). Building on the improved heuristics and staining, CTCs were subsequently detected in 64% of prostate patients using the first-generation micropost chip (19), and in 93% of patients using the second-generation herringbone chip (16). Yet, these systems remain limited by low throughput (~1 to 2 ml of blood/hour), the inability to conduct single-cell or slide-based analyses, the requirement for three-dimensional image scanning platforms, and the availability of only a positive selection mode. The CTC-iChip system presented here thus encompasses major advances over our previous methods. Whole blood is now processed through a microscale system at speeds comparable to bulk systems (8 ml/hour) while preserving the high sensitivity afforded by microfluidic isolation techniques. Furthermore, rapid and gentle isolation of CTCs, as well as their collection in suspension, increases the integrity of these cells and their RNA quality, which are crucial for downstream analyses, such as cytopathology and single-cell expression profiling.
Moreover, the system can be run in either a positive selection or a negative depletion mode, thus broadening its potential application in the clinic and in basic research studies. The negCTC-iChip allows for depletion of normal blood cells, uncovering an unselected population of nonhematopoietic cells for analysis. The robustness of this platform was demonstrated by staining CTCs per clinical pathology protocols, which yielded high-quality diagnostic images. The negCTC-iChip allowed for isolation of CTCs from a nonepithelial cancer (melanoma) and from cancer that has undergone EMT and lost virtually all detectable EpCAM expression (triple-negative breast cancer). Hence, the negCTC-iChip will be broadly applicable to all cancers that demonstrate vascular invasion, a major limitation of current technologies.
However, several additional optimizations should be considered before the CTC-iChip technology can be deployed for large-scale clinical applications. These include further improvements in CTC purity to facilitate routine molecular analyses of CTCs and in total blood volume processed to enable early cancer detection. From a manufacturing standpoint, we envision the CTC-iChip being integrated into a single monolithic device made of plastic and incorporating all three components of the CTC-iChip within a single footprint. Integration of such an economical chip into a fully automated device would potentially enable broad dissemination of this technology.
The emerging field of CTC biology brings with it unprecedented insight into the mechanisms underlying the blood-borne metastasis of cancer, as well as powerful new clinical applications to help diagnose and manage disease. As the technology matures, these are likely to include the initial genotyping and molecular characterization of cancer, as well as repeated noninvasive sampling of tumors during treatment. Because targeted therapies increasingly shape the clinical paradigm of cancer therapeutics, such serial “real-time” monitoring of cancer for indicators of drug response and emerging resistance is likely to become a mainstay of clinical oncology. The integrated microfluidic technology platform presented here provides a major step in this direction by enabling processing of large blood volumes with high throughput and efficiency, isolating CTCs regardless of tumor surface epitopes, and providing an end product that is compatible with both standardized clinical diagnostics and advanced molecular analyses. Because rare cell detection technologies continue to improve in sensitivity, they may ultimately provide novel approaches for early detection of invasive cancer before the establishment of metastatic disease.
MATERIALS AND METHODS
Samples
MDA-MB-231, SKBR3, and MCF10A cell lines were obtained from the American Type Culture Collection. PC3-9 cells were obtained from Veridex, LLC, and LBX1-expressing MCF10A cells were derived from a stable cell line previously published by our laboratory (27). Device performance was evaluated by prelabeling the cell lines with a fluorescent marker and spiking them into whole blood at ~200 to 1000/ml of whole blood (Supplementary Materials and Methods).
Fresh whole blood was collected from healthy volunteers under an Institutional Review Board (IRB)–approved protocol or commercially sourced from Research Blood Components. Samples from metastatic breast, colorectal, pancreas, lung, melanoma, and prostate cancer patients were collected under a separate IRB-approved protocol.
Chip design and fabrication
Hydrodynamic sorting chips were designed at Massachusetts General Hospital (MGH) and fabricated by Silex with deep reactive ion etching on silicon wafers. The chip was sealed with anodically bonded glass cover to form the microfluidic chamber. A custom polycarbonate manifold was used to form the fluidic connections to the microchip (fig. S3). The inertial focusing and magnetophoresis chips were designed and fabricated at MGH with soft lithography and polydimethylsiloxane (fig. S4). The chip was placed within a custom stainless steel manifold that held four magnets in a quadrupole configuration to create a magnetic circuit enabling cell deflection (fig. S5) (Supplementary Materials and Methods).
Magnetic bead labeling of target cells in whole blood
Before processing the whole blood, samples were incubated with functionalized magnetic beads 1 μm in diameter (Dynal MyOne 656-01, Life Technologies) (fig. S2). For posCTC-iChip, beads were functionalized with a biotinylated anti-EpCAM antibody, and active magnetic mixing was applied to achieve good labeling of EpCAMlow cell lines. For negative depletion, anti-CD45 and anti-CD15 functionalized beads were used (Supplementary Materials and Methods).
Immunofluorescence staining of CTCs
For enumeration analysis, isolated cells were incubated with saponin, DAPI, and anti-CK [phycoerythrin (PE)] and anti-CD45 [allophycocyanin (APC)] antibodies, acquired from Veridex, still in suspension. Cells were plated on a poly-L-lysine–functionalized glass slide with a closed chamber (fig. S8), and glass slide was scanned with the BioView imaging system while the chamber was still intact. Cells that were CK+/DAPI+/CD45− were scored as CTCs. Samples evaluated for PSA/PSMA expression were stained with a primary/secondary approach. All antibodies are catalogued in table S3.
Comparison to CellSearch
For the CellSearch and posCTC-iChip comparison, two blood tubes were drawn: one CellSave tube for CellSearch run and one EDTA tube for posCTC-iChip run. Samples in CellSave tubes were processed within 3 days after the draw as optimized and recommended for CellSearch approach, and EDTA samples were processed with the posCTC-iChip within 4 hours of draw. CellSearch product was scanned in Magnest cartridges with CellTracks system. posCTC-iChip product was plated and scanned with the BioView system.
RT-PCR analysis
RNA isolation was done with RNeasy Micro Kit (Qiagen). After RNA isolation, reverse transcription of RNA to complementary DNA (cDNA) using oligo(dT) was performed with SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). For detection of EML4-ALK fusion cDNAs, partial nested PCR analysis was done with FideliTaq PCR Master Mix (Affymetrix). PCR amplification was performed in a thermocycler (Peltier Thermal Cycler, MJ Research). Gel electrophoresis was done with an aliquot of RT-PCR products. The amplified EML4-ALK products were sequenced, and results were analyzed with the ABI PRISM DNA sequence analysis software (Applied Biosystems) (Supplementary Materials and Methods).
Cytology and ICC
CTCs were enriched via negCTC-iChip from the whole blood of cancer patients and plated on a poly-L-lysine surface (fig. S8). Plating chamber was removed after cell adhesion to facilitate standard cytopathology processing. Pap stain was done with hematoxylin, eosin-azure, and orange G and initially reviewed for suspicious cells by a certified cytotechnologist (N. Hartford, MGH) and then formally reviewed by a staff cytopathologist (E.B.). Slides were then destained and exposed to ICC process (Supplementary Materials and Methods).
Single-cell micromanipulation and qRT-PCR
Blood samples from a patient with metastatic prostate cancer were processed through the negCTC-iChip, and unfixed CTCs and contaminating leukocytes were stained in solution with fluorophore-conjugated antibodies against EpCAM and CD45. Single CTCs were identified based on an EpCAM+/CD45− phenotype and transferred under direct microscopic visualization to individual PCR tubes with a TransferMan NK2 micromanipulator (Eppendorf AG). Single-cell cDNA was prepared and amplified for single-cell transcriptome analysis, followed by specific target preamplification (Fluidigm Corp.). Microfluidic qRT-PCR was performed with the BioMark Real-Time PCR system (Fluidigm Corp.). The normalized gene expression in each cell (−Δ_C_t) was calculated as the negative of the difference between the _C_t value for each gene and the _GAPDH C_t value for the cell. Heat maps of normalized gene expression (−Δ_C_t) were generated with the Heat Map image module of GenePattern, with global color normalization.
Supplementary Material
Supplement
Acknowledgments
We express our gratitude to all patients who participated in this study and healthy volunteers who contributed blood samples. We thank C. Koris and the MGH clinical research coordinators for help with clinical studies; B. Brannigan and H. Zhu for support of molecular analyses; E. Lim, J. Martel, J. Oakey, and F. Fachin for assistance in microfluidic device development; H. Cho for contributions to illustrations; L. Libby, O. Hurtado, and A. J. Aranyosi for coordination of the research labs; B. Hamza for expertise in device fabrication; D. Chianese and T. Bendele from Johnson & Johnson for supplying reagents and materials; and N. Bander from Weill Cornell Medical College for supplying anti-PSMA antibody, clone J591.
Funding: This work was partially supported by NIH P41 Biotechnology Resource Center (M.T.), NIH National Institute of Biomedical Imaging and Bioengineering (M.T. and D.A.H.), Stand Up to Cancer (D.A.H., M.T., and S.M.), Howard Hughes Medical Institute (D.A.H.), Prostate Cancer Foundation and Charles Evans Foundation (D.A.H. and M.T.), Department of Defense Prostate Cancer Research Program (D.T.M. and R.J.L.), Mazzone-DF/HCC (Dana-Farber/Harvard Cancer Center) (D.T.M.), Conquer Cancer Foundation (R.J.L.), Prostate Cancer Foundation (R.J.L.), and Johnson & Johnson (M.T. and S.M.).
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/5/179/179ra47/DC1
Materials and Methods
Fig. S1. CTC-iChip system details.
Fig. S2. Optimization of labeling in whole blood.
Fig. S3. Hydrodynamic size–based separation.
Fig. S4. Inertial focusing and magnetophoresis channels.
Fig. S5. Magnetic configuration.
Fig. S6. Beads per cell distribution in deflected and undeflected outputs.
Fig. S7. WBC contamination in posCTC-iChip.
Fig. S8. Cell plating chamber.
Fig. S9. ICC stain validation through cell lines.
Fig. S10. Additional images of ICC-stained cells.
Fig. S11. Comparison of cell identification through Pap and ICC.
Fig. S12. Single-cell qRT-PCR optimization using cell lines.
Table S1. Contaminating cells in the negCTC-iChip product are leukocytes.
Table S2. CellSearch versus posCTC-iChip comparison.
Table S3. Antibodies used throughout the study.
Author contributions: E.O., A.M.S., B.L.E., D.T.M., E.B., M.Y., S.L.S., N.M.K., T.A.B., J.R.W., K.S., P.S.S., J.P.S., R.J.L., D.T.T., X.L., A.T.S., A.B., L.V.S., D.N.L., S.M., R.K., D.A.H., and M.T. designed the research. E.O., A.M.S., and J.C.C. were responsible for overall system design and characterization. B.L.E., A.K., and M.Y. optimized and performed cytology and ICC staining on slides. D.T.M. and J.T. executed single-cell micromanipulation and RNA analysis. S.S. conducted molecular detection of NSCLC patients. P.C., B.M., J.T., S.S., and J.C.C. executed chip operation and blood processing. N.M.K. completed characterization of cell plating chamber. E.O., P.C., and B.M. completed immunofluorescence staining, scanning, and enumeration. E.O., A.M.S., B.L.E., D.T.M., E.B., M.Y., S.L.S., N.M.K., L.V.S., S.M., R.K., D.A.H., and M.T. analyzed the research. E.O., A.M.S., B.L.E., D.T.M., S.M., R.K., D.A.H., and M.T. wrote the paper.
Competing interests: MGH filed for patent protection for the CTC-iChip technology. The authors declare that they have no competing interests.
Data and materials availability: Bio-tinylated anti-EpCAM mouse monoclonal IgG1 (clone VU-1D9), PE-conjugated anti-CK8/18 mouse monoclonal IgG1 (clone C11), PE-conjugated anti-CK19 mouse monoclonal IgG2a (clone A53-B/A2), APC-conjugated anti-CD45 mouse monoclonal IgG1 (clone HI30), nuclear staining reagent DAPI, and cell membrane permeabilization reagent saponin were obtained under a Materials Transfer Agreement from Johnson & Johnson (contact: David Chianese). Anti-PSMA monoclonal mouse IgG1 antibody (clone J591) was obtained under Materials Transfer Agreement from Weill Medical College (contact: Neil Bander).
REFERENCES AND NOTES
- 1.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 2.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the cellsearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang JM, Casavant BP, Beebe DJ. Circulating tumor cells: Getting more from less. Sci Transl Med. 2012;4:141ps13. doi: 10.1126/scitranslmed.3004261. [DOI] [PubMed] [Google Scholar]
- 6.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. A rare-cell detector for cancer. Proc Natl Acad Sci USA. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, Ko AH, Korn WM, Schram E, Coward M, Yang X, Metzner T, Lamy R, Honnatti M, Yoshioka C, Kunken J, Petrova Y, Sok D, Nelson D, Kuhn P. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-Hall detector. Sci Transl Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 10.Deng G, Herrler M, Burgess D, Manna E, Krag D, Burke JF. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Res. 2008;10:R69. doi: 10.1186/bcr2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lara O, Tong X, Zborowski M, Chalmers JJ. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp Hematol. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu CL, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, Bander NH, Wu CL, Sequist LV, Smith MR, Ramaswamy S, Toner M, Maheswaran S, Haber DA. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 23.Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc Natl Acad Sci USA. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang R, Barber TA, Schmidt MA, Tompkins RG, Toner M, Bianchi DW, Kapur R, Flejter WL. A microfluidics approach for the isolation of nucleated red blood cells (NRBCs) from the peripheral blood of pregnant women. Prenat Diagn. 2008;28:892–899. doi: 10.1002/pd.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segré G, Silberberg A. Radial particle displacements in Poiseuille flow of suspensions. Nature. 1961;189:209–210. [Google Scholar]
- 26.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JWM, Gratama JW, Sleijfer S, Foekens JA. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu M, Smolen GA, Zhang J, Wittner B, Schott BJ, Brachtel E, Ramaswamy S, Maheswaran S, Haber DA. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Plouffe BD, Lewis LH, Murthy SK. Computational design optimization for microfluidic magnetophoresis. Biomicrofluidics. 2011;5:13413. doi: 10.1063/1.3553239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement