Multilocus sequence analysis (MLSA) of Bradyrhizobium strains: revealing high diversity of tropical diazotrophic symbiotic bacteria (original) (raw)
Abstract
Symbiotic association of several genera of bacteria collectively called as rhizobia and plants belonging to the family Leguminosae (=Fabaceae) results in the process of biological nitrogen fixation, playing a key role in global N cycling, and also bringing relevant contributions to the agriculture. Bradyrhizobium is considered as the ancestral of all nitrogen-fixing rhizobial species, probably originated in the tropics. The genus encompasses a variety of diverse bacteria, but the diversity captured in the analysis of the 16S rRNA is often low. In this study, we analyzed twelve Bradyrhizobium strains selected from previous studies performed by our group for showing high genetic diversity in relation to the described species. In addition to the 16S rRNA, five housekeeping genes (recA, atpD, glnII, gyrB and rpoB) were analyzed in the MLSA (multilocus sequence analysis) approach. Analysis of each gene and of the concatenated housekeeping genes captured a considerably higher level of genetic diversity, with indication of putative new species. The results highlight the high genetic variability associated with Bradyrhizobium microsymbionts of a variety of legumes. In addition, the MLSA approach has proved to represent a rapid and reliable method to be employed in phylogenetic and taxonomic studies, speeding the identification of the still poorly known diversity of nitrogen-fixing rhizobia in the tropics.
Keywords: Biological nitrogen fixation, Bradyrhizobium, multilocus sequence analysis, phylogeny, taxonomy
INTRODUCTION
Species of the large family Leguminosae (Fabaceae in the USA) occupy a broad-range of terrestrial biomes, with several of them being capable of establishing symbioses with bacteria collectively called rhizobia, starting the process of fixing atmospheric nitrogen (N2). Diazotrophic bacteria were first isolated from root nodules and characterized in 1888 by Martinus Willem Beijerinck (2) as Bacillus radicicola, but then reclassified into the genus Rhizobium, with the taxonomy based on nodulation with certain host plants, establishing the "cross-inoculation group" concept (11). Taxonomical classification remained unaltered until the early 1980s, when numerical taxonomy considering morphological, physiological and genetic patterns led to the positioning of some strains into a new genus, Bradyrhizobium, with a unique defined species, B. japonicum (18). Bradyrhizobium includes slow growers that produce alkaline reaction in culture medium with mannitol as carbon source, while Rhizobium contains fast growing acid producers (18, 19).
A few years later, high diversity among Bradyrhizobium strains led to the description of a new species named Bradyrhizobium elkanii (23); however, the development of several molecular techniques helped to identify high genetic diversity among Bradyrhizobium strains isolated from a wide range of leguminous plants (e.g., 13, 28, 29, 46). Nowadays, there are nine defined Bradyrhizobium species (7), but certainly many more should be described in the next years.
Sequencing of the 16S rRNA has become the method of choice for tracing bacterial phylogenies (12, 50, 52), but in several genera including Bradyrhizobium variability in the 16S rRNA is often low and may not reflect the diversity detected by other morpho-physiological and genetic properties (e.g., 13, 28, 42, 46, 51). To detect higher diversity, other ribosomal genes or regions have been chosen, as the 23S rRNA and the 16S-23S rRNA intergenic transcribed spacer (ITS), since they evolve at a faster rate than the 16S rRNA, thus adding valuable information to the analysis (e.g., 29, 41, 42, 46, 51). However, proximity of ribosomal genes might not properly indicate the correct phylogeny in the case of horizontal gene transfer (43).
The multilocus sequence analysis (MLSA) method has been increasingly used in phylogeny and taxonomy studies. The method consists of the analysis of several conserved housekeeping genes dispersed in at least 100 kb of the genome (4, 6, 15, 25). Successful definitions of phylogenetic groups of rhizobia have also been achieved with the use of the MLSA (29, 31, 36).
Tropical rhizobia represent a key component for the sustainability of tropical soils, and the few results obtained so far clearly indicate that diversity is largely underestimated within the Bradyrhizobium genus (13, 28, 29). Therefore in this study the MLSA approach was applied to achieve a better phylogenetic resolution of twelve strains showing high genetic diversity of the 16S rRNA gene in relation to the described Bradyrhizobium species. The objective is to delineate strategies which may help to more promptly define diversity of tropical Bradyrhizobium.
MATERIALS AND METHODS
Strains
Twelve Bradyrhizobium strains were used in this study and are listed in Table 1. The strains were chosen from previous studies from our group (3, 13, 28, 29, 37), based on the high level of genetic diversity observed in comparison to the described species of Bradyrhizobium. The strains have been isolated from members of two subfamilies and five tribes of the family Leguminosae, and originated from four countries located in different continents: Australia, Brazil, Malaysia and Zimbabwe (Table 1). The strains were purified on yeast extract-mannitol agar (YMA) medium (45) containing Congo red (0.00125%) and stocks were prepared on YMA and kept at -80°C (under 30% of glycerol) for long-term storage and at 4°C as source cultures. The strains are deposited at the ''Culture Collection of Diazotrophic and PGPR Bacteria" of Embrapa Soja (http://www.bmrc.lncc.br) and at the Brazilian official culture collection "SEMIA" of rhizobial strains (IBP World Catalogue of Rhizobium Collections n°443 in the WFCC World Data Center on Microorganisms).
Table 1.
Information about the Bradyrhizobium strains from the Embrapa Soybean culture collection used in this study
| SEMIA number | Other designationsa | Origin of the strain | Country of origin | Host speciesb | Subfamilyb | Tribeb | Previous studiesc |
|---|---|---|---|---|---|---|---|
| 656 | SEMIA original, CNPSo 988 | FEPAGRO | Brazil | Neonotonia wightii (Wight & Arn.) Lackey | Papilionoideae | Phaseoleae | a, b, d |
| 662 | CB 188, CNPSo 990 | CSIRO | Australia | Vigna unguiculata (L.) Walp | Papilionoideae | Phaseoleae | a, b, d |
| 696 | CB 627, CNPSo 993 | CSIRO | Australia | Desmodium uncinatum (Jacq.) DC | Papilionoideae | Demodieae | a, b, d, e |
| 6002 | CB 756, TAL 309, RCR 3824, CNPSo 1092 | CSIRO | Zimbabwe | Vigna unguiculata (L.) Walp | Papilionoideae | Phaseoleae | a, b, d |
| 6028 | TAL 569, SPRL 472, MAR 472, CNPSo 1094 | NIFTAL | Zimbabwe | Desmodium uncinatum (Jacq.)DC | Papilionoideae | Demodieae | a, b, d |
| 6053 | TAL 827, UMKL 28, CNPSo 1095 | NIFTAL | Malaysia | Clitoria ternatea L. | Papilionoideae | Phaseoleae | a, b, d, e |
| 6144 | SMS 400, USDA 3187, MAR 11, CNPSo 1109 | IAC | Zimbabwe | Arachis hypogaea L. | Papilionoideae | Aeschynomeneae | a, b, d |
| 6145 | BR 2001, CNPSo 1110 | Embrapa Agrobiologia | Brazil | Crotalaria juncea L. | Papilionoideae | Crotalarieae | a, b, d |
| 6148 | SMS 303, CNPSo 1112 | IAC | Brazil | Neonotonia wightii (Wight & Arn.) Lackey | Papilionoideae | Phaseoleae | a, b, d |
| 6154 | BR 446, CNPSo 1117 | Embrapa Agrobiologia | Brazil | Stylosanthes spp. | Papilionoideae | Aeschynomeneae | a, c, e |
| 6160 | BR 5610, CNPSo 1123 | Embrapa Agrobiologia | Brazil | Albizia lebbeck (L.) Benth. | Mimosoideae | Ingeae | a, b, d |
| 6395 | BR 4301, CNPSo 1161 | Embrapa Agrobiologia | Brazil | Calliandra houstoniana (Mill.) Standl. | Mimosoideae | Ingeae | c |
DNA extraction and sequencing analysis of the 16S rRNA
The DNA samples were extracted from bacterial batch cultures grown in YM broth until late exponential phase (109 cells mL-1) as described before (20). The DNAs were submitted to the amplification with primers for the 16S rRNA, recA, atpD, glnII, gyrB and rpoB genes, as listed in Table 2. The PCR products were purified with the PureLinkTM PCR Purification Kit (Invitrogen), and the reactions for the sequencing analysis contained purified PCR products of each bacterium culture (80 ng per reaction), 3 μL of dye (DYEnamic ET terminator reagent premix for the MEGA BACE) and 3 pmol of each primer (Table 2). The conditions of PCR amplification were as described by Menna et al. (28). The sequencing was performed on a MEGA BACE 1000 (Amersham Biosciences) capillary sequencer according to the manufacturer's instructions.
Table 2.
Primers and DNA amplification conditions used in this study
| Primer | Sequence (5`- 3`)a | Target gene (position) † | PCR cycling | Reference |
|---|---|---|---|---|
| TSrecAf | CAACTGCMYTGCGTATCGTCGAAGG | recA (8-32) | 2 min 95°C, 35 X (45s 95°C, 30s 58°C, 1,5 min 72°C and 7 min 72°C. | Stepkowski et al. (2005) |
| TSrecAr | CGGATCTGGTTGATGAAGATCACCATG | recA (620-594) | ||
| TSatpDf | TCTGGTCCGYGGCCAGGAAG | atpD (189-208) | 2 min 95°C, 35 X (45s 95°C, 30s 58°C, 1,5 min 72°C and 7 min 72°C. | Stepkowski et al. (2005) |
| TSatpDr | CGACACTTCCGARCCSGCCTG | atpD (804-784) | ||
| TSglnIIf | AAGCTCGAGTACATCTGGCTCGACGG | glnII (13-38 | 2 min 95°C, 35 X (45s 95°C, 30s 58°C, 1,5 min 72°C and 7 min 72°C. | Stepkowski et al. (2005) |
| TSglnIIr | SGAGCCGTTCCAGTCGGTGTCG | glnII (681-660) | ||
| gyrB343F | TTCGACCAGAAYTCCTAYAAGG | gyrB (343-364) | 5 min 95°C, 5X (2 min 94°C, 2 min 58°C, 1 min 72°C) 28 X (30s 94°C, 1 min 58°C, 1 min 72°C and 5 min 72°C. | Martens et al. (2008) |
| gyrB1043R | AGCTTGTCCTTSGTCTGCG | gyrB (1061-1043) | ||
| rpoB83F | CCTSATCGAGGTTCACAGAAGGC | rpoB (1081-1061) | 5 min 95°C, 3X (2 min 94°C, 2 min 58°C, 1 min 72°C) 30 X (30s 94°C, 1 min 58°C, 1 min 72°C and 5 min 72°C. | Martens et al. (2008) |
| rpoB1061R | AGCGTGTTGCGGATATAGGCG | rpoB (83-103) | ||
| fD1 | AGAGTTTGATCCTGGCTCAG | 16S rRNA (9-29) | 2 min 95°C, 30 X (15s 94°C, 45s 93°C, 45s 55°C, 2 min 72°C and 5 min 72°C. | Weisburg et al. (1991) |
| rD1 | CTTAAGGAGGTGATCCAGCC | 16S rRNA (1474-1494) |
Cluster analyses
The 16S rRNA, recA, atpD, glnII, gyrB and rpoB sequences generated were analyzed with the programs Phred (8, 9), Phrap (http://www.phrap.org) and Consed (16). The consensus sequences obtained and confirmed in the 5´ and 3´ directions were submitted to the GenBank database and received the accession numbers listed in Table 3. Some genes have been previously sequenced by our group, but were re-sequenced in this study; as the identities were confirmed in 100% of the nucleotide bases, the original accession numbers were maintained (Table 3). Sequences for other reference/type strains were retrieved from the GenBank database and are also listed in Table 3. Caulobacter crescentus strain CB 15 (genome, AE005673), was used as an outgroup.
Table 3.
GenBank/EMBL/DDBJ accession numbers for the sequences of the Bradyrhizobium strains used in this study and of the reference/type strains
The MLSA analysis was performed considering only the complete aligned sequences (size among parenthesis) obtained for the Bradyrhizobium strains and for the type/reference strains retrieved from GenBank: 16S rDNA (1,347 bp), recA (293 bp), atpD (395 bp), glnII (442 bp), gyrB (434 bp) and rpoB (395 bp).
All sequences obtained in this study or retrieved from GenBank were analyzed individually and concatenated using the MEGA (Molecular Evolutionary Genetics Analysis) software version 4.0 with the default parameters, K2P distance model (21), and the Neighbor-Joining algorithm (38). Statistical support for tree nodes was evaluated by bootstrap (10) analyses with 1000 samplings (17).
RESULTS
Diversity in the 16S rRNA
The phylogenetic tree built with the 16S rRNA sequences split the Bradyrhizobium strains in two large groups, with final bootstrap supports for each group of 70 and 94%, respectively (Fig. 1). The first group (G-I) comprised eight SEMIA strains, all grouped with B. elkanii and also with B. pachyrhizi and B. jicamae. The second group (G-II) included SEMIAs 6395, 656, 6002 and 6144 showing closer relation with B. iriomotense, in addition to reference/type strains of B. betae, B. canariense, B. yuanmingense, B. liaoningense and B. japonicum. Fig. 1 also highlights that the clustering analysis based on the 16S rRNA gene did not define clear positions for the SEMIA strains.
Figure 1.
Phylogenetic relationships of Bradyrhizobium strains from this study and of reference/type rhizobial strains based on the 16S rRNA. Phylogeny was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). Phylogenetic analyses were conducted in MEGA4.
Diversity in the atpD, glnII, gyrB, recA and rpoB genes
The additional housekeeping genes selected to refine the phylogeny analysis in this study are highly conserved among bacteria of the order Rhizobiales, are dispersed in the genome of B. japonicum strain USDA 110 and encode important proteins. For each housekeeping gene, phylogenetic trees were constructed and resulted in distinct groups (Fig. 2).
Figure 2.
Phylogenetic relationships of Bradyrhizobium strains from this study and of reference/type rhizobial strains based on the (A) recA, (B) atpD, (C) glnII, (D) gyrB and (E) rpoB genes. Method and parameters of analysis were as described for Figure 1.
When compared to the 16S rRNA, higher variability was detected in the analysis of the recA gene, and strains also fit into two groups (Fig. 2A). All four strains previously positioned in G-II of the 16S rRNA were closer to B. iriomotense, but with the appearance of at least one subgroup including SEMIA 6002 and SEMIA 6144 strains. This first group of strains had a high bootstrap support, of 99%. Subgroups were even more evident with the strains previously positioned in G-I of the 16S rRNA, with the delineation of four subgroups based on the recA gene: one including strains SEMIAs 6160, 662 and B. pachyrhizi, the second with SEMIA 696 and B. elkanii and two new subgroups including exclusively SEMIA strains, one with SEMIAs 6148 and 6154 and the other with SEMIAs 6028, 6053 and 6145. These subgroups had bootstrap supports ranging from 81 to 99% (Fig. 2A).
Greater variability was also observed with atpD gene (Fig. 2B), when compared to the 16S rRNA. The tree built with the atpD gene resulted in the definition of three main groups (G-I, G-II and G-III), with a final bootstrap support of 81%. Although the DNA of strain SEMIA 696 amplified with the atpD primers, sequencing of the fragment failed, therefore the strain was not included in the analysis. In G-I, strains were split in two subgroups, the first clustering four strains with higher resemblance with B. elkanii, and the second with three strains grouping with B. pachyrhizi (Fig. 2B). The other four SEMIA strains from this study were positioned in G-III of the atpD tree, with SEMIA 6395 showing higher similarity with B. betae, while the other strains were grouped with B. liaoningense (Fig. 2B).
The tree built with the glnII gene also resulted in two groups. In G-I four SEMIA strains were clustered with B. pachyrhizi, SEMIA 696 was clustered with B. elkanii, followed by the inclusion of SEMIA 662 and B. jicamae (Fig. 2C). In G-II of the glnII, it is worth mentioning the clustering of strains SEMIAs 6002 and 6144 with a bootstrap support of 97% (Fig. 2C), also confirmed with high support in the previous trees of recA (Fig. 2A) and atpD (Fig. 2B), but not well defined in the 16S rRNA (Fig. 1).
In the trees built with the gyrB and rpoB genes the strains were also split in two groups, with the formation of a third group in the gyrB tree, which clustered only the type strains of B. betae and B. canariense. In general, a better definition of the strains positioned in G-I of the 16S rRNA (Fig. 1) was not achieved with the analysis of the gyrB (Fig. 2D) and rpoB (Fig. 2E) genes. The other four SEMIAs analyzed were positioned in G-II in both trees. In G-II of gyrB, the SEMIA 6395 clustered with B. iriomotense, while SEMIAs 656, 6002 and 6144 were closer to B. yuanmingense. In the rpoB tree, these four SEMIAs were clustered in two well defined subgroups (656-6395 and 6002-6144).
It is also interesting that, excepting for the atpD gene, B. pachyrhizi, B. jicamae and B. elkanii were positioned in the same group in the analysis of the 16S rRNA, recA, glnII, gyrB and rpoB (Fig. 1 and Figs. 2A to 2E). On the other hand, definition of the other Bradyrhizobium species considered in this study was not completely clear when considering those genes (Fig. 1 and Figs. 2A to 2E).
From the analysis of the 16S rRNA (Fig. 1) and of five housekeeping genes (Fig. 2), the division of Bradyrhizobium in two main groups was clear, with the appearance of a third group not completely well defined only in the trees built with the atpD and gyrB genes. Each SEMIA strain used in our study was always positioned in the same great group in all six trees.
Concatenated analysis of recA, atpD, glnII, gyrB and rpoB
All five sequences of the housekeeping genes were concatenated to gain a better understanding of the strains; SEMIA 696 was not included. A concatenated sequence with 1,959 bp was obtained and 2,028 sites were analysed, resulting in 1,496 conserved, 463 variable and 304 parsimony-informative sites (Table 4).
Table 4.
Sequence information obtained in this study. Twelve strains were analysed, together with nine type and reference strains, as described in Methods.
| Locus | Strains analysed (n) | Nucleotides (%) | Frequency T/C/A/G (%) | |||
|---|---|---|---|---|---|---|
| Conserved | Variable | Parsimony-informative | Total* | |||
| 16S rRNA | 21 | 1,266 (91,5) | 86 (6,2) | 52 (3,7) | 1,347/1,383 | 20.4/24.0/24.7/30.9 |
| atpD | 20 | 318 (77,6) | 77 (18,8) | 57 (13,9) | 395/410 | 15.7/33.3/19.6/31.3 |
| glnII | 21 | 334 (72,8) | 108 (23,5) | 77 (16,8) | 442/459 | 16.2/32.7/19.9/31.2 |
| gyrB | 21 | 338 (74,8) | 96 (21,2) | 56 (12,4) | 434/452 | 21.7/29.9/16.5/32.0 |
| recA | 21 | 217 (72,6) | 76 (25,4) | 52 (17,4) | 293/299 | 15.8/32.3/17.3/34.7 |
| rpoB | 21 | 287 (70,8) | 108 (26,7) | 63 (15,5) | 395/405 | 17.4/32.8/18.7/31.1 |
| Concatenated genes | 20 | 1,496 (73,7) | 463 (22,8) | 304(15) | 1,959/2,028 | 17.5/32.2/18.4/31.9 |
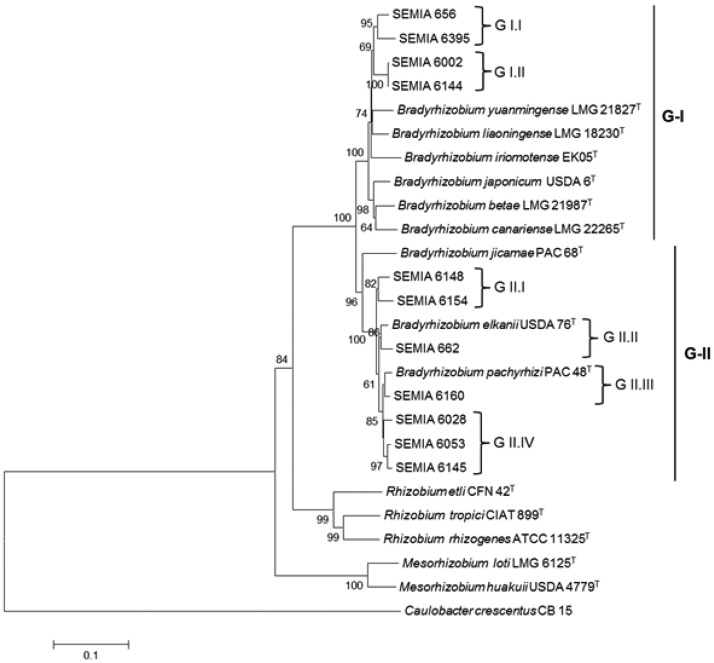
The tree built with the concatenated genes resulted in two great groups, with a bootstrap support of 100% (Fig. 3). G-I assembled SEMIAs 656, 6395, 6002 and 6144 together with type/reference strains of B. yuanmingense, B. liaoningense, B. iriomotense, B. japonicum, B. betae and B. canariense. Within G-I, subclusters that were not well defined in the 16S rRNA (Fig. 1) tree were now shown (Fig. 3), including the pairs of strains SEMIAs 656-6395 and SEMIAs 6002-6144, with bootstrap supports of 95 and 100%, respectively. The results highlight that these strains deserve further studies, as they may represent new species. The concatenated tree has also detected higher diversity of the strains occupying G-II, with the SEMIAs 6160, 6028, 6053 and 6145 showing similarity with the type strain of B. pachyrhizi. The pair of strains 6148-6154 could also represent a new species. Finally, it is important to mention that in all trees Bradyrhizobium was clearly apart from the other rhizobial genera.
Figure 3.
Evolutionary tree inferred using the Neighbor-Joining method for 22 strains based on concatenated genes (recA, atpD, glnII, gyrB, rpoB). The percentage of replicate trees in which the associated strains clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Codon positions included were 1st+2nd+3rd+noncoding. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). Phylogenetic analyses were conducted in MEGA4.
Another important observation is that clustering of strains by means of both 16S rRNA and housekeeping genes showed no relation with the host plant. Strains clustered in G-I of the MLSA were isolated from subfamilies Papilionoideae (SEMIAs 656, 6002 and 6144) and Mimosoideae (SEMIA 6395), and G-II also included strains from both subfamilies. Other examples refer to isolates from Vigna, clustered in both G-I (SEMIA 6002) and G-II (SEMIA 662), while isolates from tribe Ingeae were clustered in both G-I (SEMIA 6395) and G-II (SEMIA 6160) of the MLSA (Fig. 3).
DISCUSSION
Bradyrhizobium is an intriguing genus of bacteria encompassing a number of interesting features. First, the genus has been considered as the ancestor of all rhizobia (24, 32, 35, 49), and it has been isolated from a variety of legumes distributed worldwide. However, the great majority of the reports on diversity and genetics of diazotrophic symbiotic bacteria has been performed with fast-growing rhizobia, thus studies with Bradyrhizobium may reveal new insights into the evolution of rhizobia. The second important feature is that Bradyrhizobium has probably originated in the tropical region (24, 32), and indeed bradyrhizobia seem to represent the majority of the isolates from leguminous trees in Brazilian tropical forests (30). As it has been pointed out since the pioneer studies of ribosomal genes, apparently there are many more varieties of rhizobia in tropical and subtropical than in temperate regions (33, 46); therefore, studies on the genetic diversity of Bradyrhizobium may expose a high level of genetic diversity. Finally, the third important feature relies on the reports of high rates of nitrogen fixation related to bacteria belonging to the genus Bradyrhizobium (14), and excellent examples comprise the symbioses with cowpea (Vigna spp.) and soybean (Glycine max). Indeed, the SEMIA strains used in our study are highly effective in fixing nitrogen with their host plants.
Previous studies from our group employing analyses of the 16S rRNA, 23S rRNA, ITS and housekeeping genes have indicated an unexpected genetic diversity of Bradyrhizobium (13, 28, 29). In our study the use of five housekeeping genes in the MLSA approach highlighted a far higher diversity in comparison to the single analysis with the 16S rRNA, clearly indicating putative new species.
DNA-DNA hybridization is still required to define new species (12), but arguments against its obligatory use have been raised, including: high cost and intensive work (5, 44), existence of more accurate approaches (22), doubts about its adequacy (1). MLSA has then been proposed as a more accessible tool for assessing phylogeny and taxonomy of prokaryotes (4, 6, 15, 25). Also in this context, the use of at least four housekeeping genes for the phylogenetic analysis and taxonomic classification of bradyrhizobia has been proposed (29, 34, 39, 40, 48), and was confirmed as a successful approach in our study.
It is also interesting that host specificity was not related to genetic clustering, confirming previous reports from our group (3, 13, 28, 29, 37), and indicating that other genes must be searched aiming at getting a better understanding of the evolution of the symbioses, probably nodulation and nitrogen fixation genes.
Biological nitrogen fixation plays a key role in the environment and agriculture sustainability. The results from our study highlight the high genetic variability associated with Bradyrhizobium microsymbionts of a variety of legumes. Our results confirm that the MLSA approach can represent an important, effective, fast and low-cost strategy to reveal the still poorly known diversity of Bradyrhizobium and certainly other nitrogen-fixing rhizobial species (26, 29, 36, 40, 47). In our study MLSA clearly contributed to a better phylogeny definition, as well as to the identification of new subgroups indicative of new species.
Acknowledgments
The work was partially supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), CNPq-PNPD (558455/2008-5), CNPq/MCT/MAPA (577933/2008), CNPq-Universal (470162/2009) and CNPq-Repensa (562008/2010-1).
REFERENCES
- 1.Achtman M., Wagner M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 2.Beijerinck M.W. Cultur des Bacillus radicicola aus den Knöllchen. Bot. Ztg. 1888;46:740–750. [Google Scholar]
- 3.Binde D.R., Menna P., Bangel E.V., Barcellos F.G., Hungria M. rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl. Microbiol. Biotechnol. 2009;83:897–908. doi: 10.1007/s00253-009-1927-6. [DOI] [PubMed] [Google Scholar]
- 4.Brett P.J., Deshazer D., Woods D.E. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Evol. Microbiol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 5.Coenye T., Gevers D., Van de Peer Y., Vandamme P., Swings J. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 2005;29:147–167. doi: 10.1016/j.femsre.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Cooper J.E., Feil E.J. Multilocus sequence typing - what is resolved? Trends Microbiol. 2004;12:373–377. doi: 10.1016/j.tim.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.DSMZ – Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. 2010. (German Collection of Microorganisms and Cell Cultures) Available at: http://www.dsmz.de/index.htm. Accessed 11 Nov 2010.
- 8.Ewing B., Green P. Base-calling of automated sequencer traces using phred: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 9.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred: I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Fred E.B., Baldwin I.L., McCoy E. Root Nodule Bacteria of Leguminous Plant. Madison: University of Wisconsin Press; 1932. [Google Scholar]
- 12.Garrity G.M., Holt J.G. The roadmap to themanual. In: Boone D.R., Castenholz R.W., Garrity G.M., editors. Bergey's Manual of Systematic Bacteriology. 2. New York, USA: Springer; 2001. pp. 119–166. [Google Scholar]
- 13.Germano M.G., Menna P., Mostasso F.L, Hungria M. RFLP analysis of the rRNA operon of a Brazilian collection of bradyrhizobial strains from 33 legume species. Int. J. Syst. Evol. Microbiol. 2006;56:217–229. doi: 10.1099/ijs.0.02917-0. [DOI] [PubMed] [Google Scholar]
- 14.Giller K.E. Nitrogen fixation in tropical cropping systems. 2. Wallingford, UK: CABI Publishing; 2001. [Google Scholar]
- 15.Godoy D., Randle G., Simpson A.J., Aanensen D.M., Pitt T.L., Kinoshita R., Spratt B.G. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 17.Hedges S.B. The number of replications needed for accurate estimation of the bootstrap p-value in phylogenetic studies. Mol. Biol. Evol. 1992;9:366–369. doi: 10.1093/oxfordjournals.molbev.a040725. [DOI] [PubMed] [Google Scholar]
- 18.Jordan D.C. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 1982;32:136–139. [Google Scholar]
- 19.Jordan D.C. Family III: Rhizobiaceae Conn 1938. In: Krieg N.R., Holt J.G., editors. Bergey's Manual of Systematic Bacteriology. Baltimore: Williams & Wilkens Co.; 1984. pp. 234–235. [Google Scholar]
- 20.Kaschuk G., Hungria M., Andrade D.S., Campo R.J. Genetic diversity of rhizobia associated with common bean (Phaseolus vulgaris L.) grown under no-tillage and conventional systems in Southern Brazil. Appl. Soil Ecol. 2006;32:210–220. [Google Scholar]
- 21.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 22.Konstantinidis K.T., Tiedje J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuykendall L.D., Saxena B., Devine T.E., Udell S.E. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 1992;38:501–505. [Google Scholar]
- 24.Lloret L., Martínez-Romero E. Evolución y filogenia de Rhizobium. Rev. Latinoam. Microbiol. 2005;47:43–60. [PubMed] [Google Scholar]
- 25.Maiden M.C.J., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martens M., Delaere M., Coopman R., De Vos P., Gillis M., Willems A. Multilocus sequence analysis of Ensifer and related taxa. Int. J. Syst. Evol. Microbiol. 2007;57:489–503. doi: 10.1099/ijs.0.64344-0. [DOI] [PubMed] [Google Scholar]
- 27.Martens M., Dawyndt P., Coopman R., Gillis M., De Vos P., Willems A. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium) Int. J. Syst. Evol. Microbiol. 2008;58:200–214. doi: 10.1099/ijs.0.65392-0. [DOI] [PubMed] [Google Scholar]
- 28.Menna P., Hungria M., Barcellos F.G., Bangel E.V., Hess P.N., Martínez-Romero E. Molecular phylogeny based on the 16S rRNA gene of elite rhizobial strains used in Brazilian commercial inoculants. Syst. Appl. Microbiol. 2006;29:315–332. doi: 10.1016/j.syapm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Menna P., Barcellos F.G., Hungria M. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on Multilocus Sequence Analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int. J. Syst. Evol. Microbiol. 2009;59:2934–2950. doi: 10.1099/ijs.0.009779-0. [DOI] [PubMed] [Google Scholar]
- 30.Moreira F.M.S. Caracterização de estirpes de rizóbio isoladas de espécies florestais pertencentes a diversos grupos de divergência de Leguminosae introduzidas ou nativas da Amazônia e Mata Atlântica. Rio de Janeiro, Brasil: 1991. (Ph.D. Thesis. Universidade Federal Rural do Rio de Janeiro. UFRRJ) [Google Scholar]
- 31.Moulin L., Béna G., Boivin-Masson C., Stepkowski T. Phylogenetic analysis of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol. Phylogenet. Evol. 2004;30:720–732. doi: 10.1016/S1055-7903(03)00255-0. [DOI] [PubMed] [Google Scholar]
- 32.Norris D.O. Acid production by Rhizobium: a unifying concept. Plant Soil. 1965;22:143–166. [Google Scholar]
- 33.Oyaizu H., Naruhashi N., Gamou T. Molecular methods of analysing bacterial diversity: the case of rhizobia. Biodivers. Conserv. 1992;1:237–249. [Google Scholar]
- 34.Parker M.A. rRNA and dnaK relationships of Bradyrhizobium sp. nodule bacteria from four Papilionoid legume trees in Costa Rica. Syst. Appl. Microbiol. 2004;27:334–342. doi: 10.1078/0723-2020-00266. [DOI] [PubMed] [Google Scholar]
- 35.Provorov N.A., Vorob'ev N.I. Evolutionary genetics of nodule bacteria: molecular and populational aspects. Russ. J. Genet. 2000;36:1323–1335. [Google Scholar]
- 36.Ribeiro R.A., Barcellos F.G., Thompson F.L., Hungria M. Multilocus sequence analysis of Brazilian Rhizobium microsymbionts of common bean (Phaseolus vulgaris L.) reveals unexpected taxonomic diversity. Res. Microbiol. 2009;160:297–306. doi: 10.1016/j.resmic.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Roma Neto I.V., Ribeiro R.A., Hungria M. Genetic diversity of elite rhizobial strains of subtropical and tropical legumes based on the 16S rRNA and glnII genes. World J. Microbiol. Biotechnol. 2010;26:1291–1302. doi: 10.1007/s11274-009-0300-3. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Stepkowski T., Czaplínska M., Miedzinska K., Moulin L. The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha Proteobacteria. Syst. Appl. Microbiol. 2003;26:483–494. doi: 10.1078/072320203770865765. [DOI] [PubMed] [Google Scholar]
- 40.Stepkowski T., Moulin L., Krzyzanska A., McInnes A., Law I.J, Howieson J. European origin of Bradyrhizobium populations infecting lupins and serradella in soils of western Australia and South Africa. Appl. Environ. Microbiol. 2005;71:7041–7052. doi: 10.1128/AEM.71.11.7041-7052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tesfaye M., Holl F.B. Group-specific differentiation of Rhizobium from clover species by PCR amplification of 23S rDNA sequences. Can. J. Microbiol. 1998;44:1102–1105. [PubMed] [Google Scholar]
- 42.van Berkum P., Fuhrmann J.J. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 2000;50:2165–2172. doi: 10.1099/00207713-50-6-2165. [DOI] [PubMed] [Google Scholar]
- 43.van Berkum P., Terefework Z., Paulin L., Suomalainen S., Lindstrom K., Eardly B.D. Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 2003;185:2988–2998. doi: 10.1128/JB.185.10.2988-2998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandamme P., Pot B., Gillis M., De Vos P., Kersters K., Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent J.M. Manual for the practical study of root nodule bacteria. Oxford, UK: Blackwell Scientific; 1970. (IBP Handbook No. 15) [Google Scholar]
- 46.Vinuesa P., Rademaker J.L.W., de Bruijn F.J., Werner D. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the canary islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl. Environ. Microbiol. 1998;64:2096–2104. doi: 10.1128/aem.64.6.2096-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vinuesa P., León-Barrios M., Silva C., Willems A., Jarabo-Lorenzo A., Pérez-Galdona R., Werner D., Martínez-Romero E. Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int. J. Syst. Evol. Microbiol. 2005;55:569–575. doi: 10.1099/ijs.0.63292-0. [DOI] [PubMed] [Google Scholar]
- 48.Vinuesa P., Silva C., Werner D., Martínez-Romero E. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 2005;34:29–54. doi: 10.1016/j.ympev.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 49.Wang E.T., Martínez-Romero E. Phylogeny of root- and stem-nodule bacteria associated with legumes. In: Triplett E.W., editor. Prokaryotic Nitrogen Fixation: a model system for analysis of a biological process. Horizon Scientific: Wymondham; 2000. pp. 177–186. [Google Scholar]
- 50.Weisburg W.G., Barns S.M., Pelletier D.A, Lane D.J. 16S ribossomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems A., Doignon-Bourcier F., Goris J., Coopman R., de Lajudie P., De Vos P., Gillis M. DNA-DNA hybridization study of Bradyrhizobium strains. Int. J. Syst. Evol. Microbiol. 2001;51:1315–1322. doi: 10.1099/00207713-51-4-1315. [DOI] [PubMed] [Google Scholar]
- 52.Woese C.R. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]