Parental Origin and Timing of De Novo Robertsonian Translocation Formation (original) (raw)
Abstract
Robertsonian translocations (ROBs) are the most common chromosomal rearrangements in humans. ROBs are whole-arm rearrangements between the acrocentric chromosomes 13–15, 21, and 22. ROBs can be classified into two groups depending on their frequency of occurrence, common (rob(13q14q) and rob(14q21q)), and rare (all remaining possible nonhomologous combinations). Herein, we have studied 29 case subjects of common and rare de novo ROBs to determine their parental origins and timing of formation. We compared these case subjects to 35 published case subjects of common ROBs and found that most common ROBs apparently have the same breakpoints and arise mainly during oogenesis (50/54). These probably form through a common mechanism and have been termed “class 1.” Collectively, rare ROBs also occur mostly during oogenesis (7/10) but probably arise through a more “random” mechanism or a variety of mechanisms and have been termed “class 2.” Thus, we demonstrate that although both classes of ROBs occur predominantly during meiosis, the common, class 1 ROBs occur primarily during oogenesis and likely form through a mechanism distinct from that forming class 2 ROBs.
De novo chromosomal rearrangements provide excellent reagents for investigating the molecular mechanisms of their formation. The first step toward determining the mechanisms is to identify the “breakpoint.” For many rearrangements, it appears that there may be order to the apparent randomness of the breakpoints of different human constitutional rearrangements (Shaffer and Lupski 2000). Clearly, DNA architectural features in certain parts of the genome result in susceptibility to rearrangement (Stankiewicz and Lupski 2002).
The frequency of various chromosomal rearrangements in the general population varies from 1/625 to 1/5,000. Reciprocal translocations occur more frequently than ROBs. However, except for the t(11; 22), most reciprocal translocations are nonrecurring (or “private”) rearrangements. ROBs are found in ∼1 in 1,000 individuals (Hamerton et al. 1975). Therefore, ROBs are the most common, recurrent structural rearrangements in humans.
ROBs are whole-arm exchanges between the short arms of the acrocentric chromosomes (human chromosomes 13–15, 21, and 22). The p11 region of these chromosomes includes satellite DNAs I, II, III, IV, and β; the p12 region, referred to as “the stalks,” contains multiple copies of the genes coding for the 18S and 28S ribosomal RNA (nucleolar organizer region); and the p13 region terminates with β-satellite DNA and telomeric sequences (reviewed in Page et al. 1996; Bandyopadhyay et al. 2001_b_). Thus, the short-arm regions of the five pairs of human acrocentric chromosomes have extensive sequence homology, although some sequences are not common to all acrocentric chromosomes (Bandyopadhyay et al. 2001_b_).
Although all acrocentric chromosomes may participate in ROBs, the distribution of the different possible translocations in the population is nonrandom (Therman et al. 1989). Specifically, rob(13q14q) and rob(14q21q) are the most common, constituting ∼85% of all ROBs (Therman et al. 1989). The rate of de novo formation of ROBs is estimated to be ∼3.9 × 10−4 mutations per gamete per generation, a mutation rate one- to twofold higher than that estimated for autosomal dominant mutations (Vogel and Motulsky 1997). De novo formation of rob(13q14q) accounts for the largest proportion of the mutation rate of ROBs (∼1.5 × 10−4 mutations per gamete per generation [Jacobs 1981]).
On the basis of the nonrandom participation of certain acrocentric chromosomes in ROB formation, ROBs can be broadly classified into two groups: (1) common, which includes rob(13q14q) and rob(14q21q), and (2) rare, which includes all other possible nonhomologous ROBs. Studies of the common rob(13q14q) and rob(14q21q) found breakpoints in the same chromosomal region in the majority of cases (Han et al. 1994; Page et al. 1996); whereas the rare ROBs revealed highly variable breakpoint locations (Page et al. 1996). The breakpoints on the common ROBs cluster between repetitive classes of DNA. The breakpoint on 14p lies between two satellite III subfamilies, pTRS-47 and pTRS-63 (Earle et al. 1992; Kalitsis et al. 1993; Han et al. 1994; Page et al. 1996). For 13p and 21p, the breakpoints are between a satellite I sequence, pTRI-6, and the rDNA (Han et al. 1994; Page et al. 1996). Clustering of breakpoints may indicate a specific mechanism of translocation formation (Shaffer and Lupski 2000).
In ∼50% of cases of ROBs, the rearrangements occur de novo (Shaffer et al. 1992) and in ∼95% of the de novo cases, rob(13q14q) and rob(14q21q) originate during maternal meiosis (Page and Shaffer 1997). Determining the parental origin and the stage of translocation formation (meiotic or mitotic) is an important aspect of understanding the mechanisms for these common translocations. Studies to determine the parental origin and time of translocation formation have not been performed for rare ROBs. Here, we have studied both common and rare ROBs to determine the parental origin and stage of translocation formation, which will ultimately help us understand the mechanisms of formation of this common type of chromosomal rearrangements.
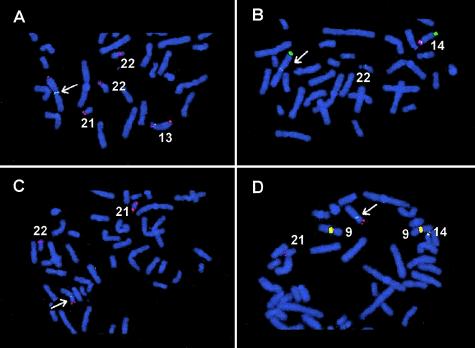
Twenty-nine de novo ROBs were ascertained through cytogenetics laboratories throughout the United States, including 19 common ROBs and 10 rare ones (table 1). All samples were collected after informed consent, using a Baylor College of Medicine Institutional Review Board approved protocol. We studied 15 de novo instances of rob(13q14q) and 4 de novo instances of rob(14q21q) using FISH and DNA probes that map to the short arms of the acrocentric chromosomes (Bandyopadhyay et al. 2001_b_). All ROBs showed breakpoints within the same regions, as published elsewhere (Han et al. 1994; Page et al. 1996) (fig. 1). Satellite III subfamily pTRS-47 is retained on all chromosomes 14 in the de novo ROBs, but the satellite III subfamily pTRS-63 was lost during the translocation event. The satellite I subfamily pTRI-6 was retained on the chromosomes 13 and 21 of the de novo ROBs, whereas the rDNA sequences were lost. These results confirm the homogeneous population of the common ROBs used in our studies (Han et al. 1994; Page et al. 1996; present study).
Table 1.
De Novo Robertsonian Translocation (ROB) Case Subjects Investigated in the Current Study
| ROB | No. of Case Subjects (_n_=29) |
|---|---|
| rob(13q14q) | 15 |
| rob(14q15q) | 4 |
| rob(14q21q) | 4 |
| rob(14q22q) | 1 |
| rob(15q21q) | 3 |
| rob(15q22q) | 2 |
Figure 1.
Partial metaphase spreads showing FISH with specific probes for satellite DNA subfamilies. The Robertsonian translocations are indicated by arrows. The same de novo rob(13q14q) is shown in panels A and B. A, FISH with pTRI-6 (green signals), rDNA (red signals), and a 13q subtelomeric probe (Knight et al. 2000; red signals) shows that the de novo translocation breakpoint is between pTRI-6 (retained) and the rDNA (lost) on chromosome 13. B, FISH with pTRS-47 (green signals), pTRS-63 (red signals), and a 14q subtelomeric probe (Knight et al. 2000; green signals) shows that the translocation breakpoint is between pTRS-47 (retained) and pTRS-63 (lost) on chromosome 14. For panels C and D, the same de novo rob(14q21q) was used. C, FISH with pTRI-6 (green signals), rDNA (red signals), and a 21q subtelomeric probe (Knight et al. 2000; red signals) shows that the translocation breakpoint was between pTRI-6 (retained) and rDNA (lost) on chromosome 21. D, FISH with pTRS-47 (green signals), pTRS-63 (red signals), and a 21q subtelomeric probe (red signals) shows that the translocation breakpoint was between pTRS-47 (retained) and pTRS-63 (lost) on chromosome 14. Note that probes pTRS-47 and pTRS-63 cross hybridize to the pericentromeric region of chromosome 9 (yellow signals).
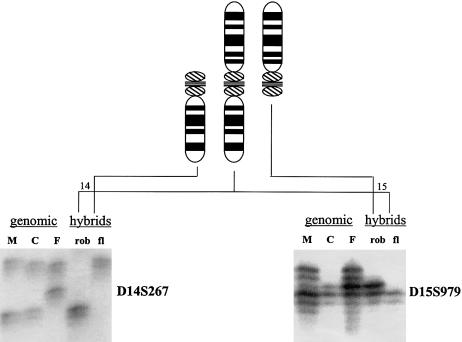
Somatic cell hybrids were constructed to separate the translocations from their homologues for determining the parental origin of the de novo ROBs (Page and Shaffer 1997). The parental origins of the de novo translocations were identified using two fully informative polymorphic microsatellite markers for each rearranged chromosome in the hybrid DNA and genomic DNA derived from the de novo ROB carrier as compared to the genomic DNA from their chromosomally normal parents (fig. 2). ROBs can form postzygotically (Robinson et al. 1994; Page and Shaffer 1997) or during meiosis (Page and Shaffer 1997). The timing of the translocation formation can be inferred by comparing the alleles present on the two rearranged chromosomes to the normal parental chromosomes. Under the assumption of a postzygotic model with a random chance for the acrocentric chromosomes to form nonhomologous ROBs, 50% of the rearrangements are predicted to contain a chromosome from each parent, 25% would contain both maternally inherited chromosomes, and 25% would contain both paternally inherited chromosomes. A χ2 goodness-of-fit test was used to evaluate the timing of formation and any parental origin biases. A probability of P<.05 allowed for rejection of a mitotic model in favor of a meiotic model of translocation formation.
Figure 2.
Parental origin determination by PCR analysis using polymorphic markers on somatic cell hybrids. Shown are two markers used to determine the parental origins of the chromosomes involved in this case of a de novo rob(14q15q). The top of the figure shows the idiograms for chromosomes 14 (left), 15 (right), and the rob(14q15q) (middle). The left of the figure shows the results for D14S267 on DNA from the mother (M), child (C) with the de novo rob(14q15q), father (F), somatic cell hybrid carrying the rob(14q15q) (rob), and somatic cell hybrid carrying the free-lying chromosome 14 (fl). The child inherited the upper allele from the father and the lower allele from the mother. The chromosome 14 involved in the rob(14q15q) was derived from the mother, since it shares this lower allele. The free-lying chromosome 14 was inherited from the father. The right of the figure shows the results for D15S979. The child inherited the upper allele from the father and the lower allele from the mother. The chromosome 15 involved in the rob(14q15q) was derived from the father, as it carries the father’s allele. The free-lying chromosome 15 was inherited from the mother. Thus, this rob(14q15q) was derived postzygotically from the maternally derived 14 and the paternally derived 15.
Of 15 instances of rob(13q14q), 12 involved two maternal chromosomes, 1 involved two paternal chromosomes, and 2 consisted of one paternal chromosome and one maternal chromosome, indicating a postzygotic origin (table 2). For all four instances of rob(14q21q), both chromosomes were maternal in origin (table 2). For the rare translocations, two instances of rob(15q22q), three instances of rob(15q21q), and one instance of rob(14q22q) were maternal in origin (table 3). Of the four instances of rob(14q15q) studied, one was completely maternal, two were paternal, and one was postzygotic in origin, containing one maternal and one paternal chromosome (table 3; fig. 2).
Table 2.
Results on Common Robertsonian Translocations (ROBs) Investigated in the Present Study and Those Reported Elsewhere[Note]
| ROB | No. ofCase Subjects | Mat/Mat | Pat/Pat | Mat/Pat |
|---|---|---|---|---|
| rob(13q14q) | 23 (15) | 19 (12) | 2 (1) | 2 (2) |
| rob(14q21q) | 31 (4) | 31 (4) | … | … |
| Total | 54 (19) | 50 (16) | 2 (1) | 2 (2) |
Table 3.
Results on Rare ROBs Investigated in the Present Study
| ROB | No. ofCase Subjects | Mat/Mat | Pat/Pat | Mat/Pat |
|---|---|---|---|---|
| rob(14q15q) | 4 | 1 | 2 | 1 |
| rob(14q22q) | 1 | 1 | … | … |
| rob(15q21q) | 3 | 3 | … | … |
| rob(15q22q) | 2 | 2 | … | … |
| Total | 10 | 7 | 2 | 1 |
Analyses were performed on the present study population (tables 2 and 3) and then performed combining all cases analyzed in our laboratory (Page and Shaffer 1997; S. A. Berend, S. L. Page, W. Atkins, C. McCaskill, N. E. Lamb, S. L. Sherman, and L. G. Shaffer, unpublished data) (table 4) to identify the time of translocation formation. Analysis of the common ROBs in this study showed that the majority comprised two chromosomes from the same parent (table 2). A mitotic model of postzygotic ROB formation could be rejected in favor of a meiotic model for this study (χ22=35.5, P<.0001) and over all studies (χ22=131.6, P<.0001). The results also show that in the majority of the cases, the translocation is maternal in origin (χ21=13.2, P<.0001 for this study and χ21=44.3, P<.0001 for the combined studies of all common ROBs).
Table 4.
Published De Novo Robertsonian Translocation Case Subjects
| No. of Case Subjects (_n_=35) | ROB | References |
|---|---|---|
| 8 | rob(13q14q) | Page and Shaffer 1997 |
| 3 | rob(14q21q) | Page and Shaffer 1997; S. A. Berend, S. L. Page, W. Atkins, C. McCaskill, N. E. Lamb, S. L. Sherman, and L. G. Shaffer, unpublished data |
| 4 | rob(14q21q) | Page and Shaffer 1997 |
| 20 | rob(14q21q) | S. A. Berend, S. L. Page, W. Atkins, C. McCaskill, N. E. Lamb, S. L. Sherman, and L. G. Shaffer, unpublished data |
Analysis of the rare ROBs showed that the majority comprised two chromosomes from the same parent (table 3). Although the number of cases is relatively small for the rare ROBs as compared to the common ROBs, a meiotic model of formation was still favored over a mitotic model of translocation formation (χ22=11.4, P<.005). Of the 10 rare ROBs studied, 7 were maternal in origin, 2 were paternal in origin, and 1 was postzygotic. Among those rare ROBs derived from a single parent, there was no significant difference between parental origin (χ21=2.8, 0.10>_P_>.05). Although there were apparently more maternally derived rare ROBs, the number of case subjects was too small to reach statistical significance. Among all ROBs comprising chromosomes from a single parent in all studies, 57 were maternal in origin, and 4 were paternal in origin (χ21=46.0, P<.0001) (tables 2 and 3). Thus, significantly more resulted from maternally derived chromosomes.
The mechanisms of ROB formation are not yet identified. However, the vast majority of common ROBs have breakpoints in a consistent location, whereas the rare translocations can have highly variable breakpoint locations (Page et al. 1996; Sullivan et al. 1996). Herein, we have studied an additional 19 common ROBs and have determined the locations of the breakpoints and the parental origins for each translocation. Our intention was twofold. First, this study represents a greater number of common ROBs and confirms the fact that the majority of these translocations have consistent breakpoints that likely indicate a common mechanism of formation. Second, these studies confirm that this is a homogeneous group of common ROBs that can be used for determining the mechanism of translocation formation.
Combined with previous studies, we have shown that, for the common ROBs, the breakpoints in the majority of translocations (53/54) are within a consistent region and arise mainly during oogenesis (Page and Shaffer 1997; present study). One reported case of rob(13q14q) was shown elsewhere to be paternally derived and had an unusual breakpoint (Page and Shaffer 1997). Of three additional cases of rob(13q14q) reported here, two were postzygotically formed between a maternal and a paternal chromosome, and one was completely paternally derived, but all had breakpoints within the consistent breakpoint region. These cases likely reflect the dynamic nature of the acrocentric short-arm chromatin and the tendency for these regions to participate in exchanges. Thus, with the large series reported here of cases of rob(13q14q) (_n_=23), it is not surprising to find occasional “random” translocation events. Probably more surprising is the lack of any “random” events in rob(14q21q). In 31 cases of de novo rob(14q21q) studied by our laboratory, all are maternally derived and all have consistent breakpoint locations when a limited number of FISH probes are used. However, in a few cases at a higher resolution, using several satellite III subfamilies, we have shown that the breakpoints may vary within these regions (Bandyopadhyay et al. 2001_a_). Efforts are ongoing to sequence the translocation breakpoints. High-resolution analysis on additional case subjects will likely show that, within this highly repetitive DNA region between the satellite subfamilies and rDNA sequences, there will be differences in the placement of breakpoints. However, we speculate that the underlying mechanism (proposed as homologous recombination) will be the same in the majority of common ROBs.
Previous studies of rare ROBs have shown highly variable breakpoints (Page et al. 1996). This indicates that these translocations may form through different, more “random” mechanisms than the common ROBs. Among these translocations, rob(14q15q) was found to have the most variable breakpoint locations (Page et al. 1996). Four different exchanges were found among five different case subjects examined. In this study, we have determined the parental origin for four de novo case subjects and found that the origins were highly variable: one was maternally derived, two were paternally derived, and one was postzygotic in origin (table 3; fig. 2).
Though the number of cases is small for the rare translocations, the present data indicate that all ROBs can be broadly classified into two groups: class 1 are those ROBs that probably occur through a distinct, reproducible mechanism, and class 2 are those that appear, mechanistically, more random for translocation formation. Class 1 ROBs were found to occur during oogenesis and apparently have the same breakpoints within their type of ROB. These would include all cases of rob(14q21q) and the majority of cases of rob(13q14q). This class may also include some rare ROBs that involve chromosome 14 (e.g., rob(14q22q)) the breakpoints of which on chromosome 14 fall between pTRS-47 and pTRS-63. However, study of larger groups of each type of rare ROBs may uncover heterogeneity, as found for rob(14q15q). Class 2 ROBs are predicted to have varied breakpoints. These cases can also form during meiosis or mitosis and have variable parental origins. These findings indicate a more random process in translocation formation within this class. Class 2 rearrangements may also include some common ROBs that have different breakpoint locations or postzygotic formation.
The observations that (1) ROBs occur nonrandomly in the population, (2) a high mutation rate exists for de novo formation of common ROBs, (3) ∼93% of ROBs occur maternally, and (4) their breakpoints are in consistent locations suggest a specific mechanism of formation for class 1 ROBs. The mechanism proposed is recombination between homologous sequences that are shared on the short arms of the acrocentric chromosomes. This would lead to specific, recurrent breakpoints (e.g., those found in the class 1 rearrangements rob(13q14q) and rob(14q21q)) and may possibly account for the frequent mutation events seen in these translocations.
Although the mechanism of formation is unknown, these translocations may be due to a double-strand break (DSB) initiated by the genomic architectural features and/or DNA sequence in the short arm regions. Meiotic pairing between nonhomologous chromosomes and recombination between homologous sequences shared between these chromosomes may lead to the formation of the rearranged products. Because most class 1 ROBs probably occur during maternal meiosis, we restrict our comments to oogenesis. It may be that, during oogenesis, certain factors bring specific acrocentric chromosomes into close proximity, which facilitates the formation of these translocations. One such factor may be formation of the nucleoli, during which the acrocentric short arms associate, increasing the chances of pairing and recombination between paralogous segments. In the human female fetus, a few hundred germ cells are present by 3 wk after conception. In the human ovary, the transformation of oogonia to oocytes by entrance into meiotic prophase I is initiated in the 3rd mo of gestation. The oocytes proceed through initial phases of prophase I and then undergo meiotic arrest (dictyotene) until hormones stimulate the resumption of meiosis I at ovulation many years later. The DNA of the oocyte is synthesized prior to the first meiotic prophase in the fetus (Byskov 1982; Cohen and Pollard 2001). The “life cycle” of the female germ cell implies that these cells carry “aged” DNA by the time they finally ovulate and are fertilized. Thus, the genetic content of the oocyte was mostly decided when the mother was a fetus in the maternal grandmother.
Several studies have suggested that a block in premeiotic replication may prevent proper meiotic recombination (Borde et al. 2000; Davis et al. 2001; Murakami and Nurse 2001). DNA DSBs can arise from a stalled or damaged replication fork (Constantinou et al. 2001). A repeat-rich region, such as the satellite III regions of the acrocentric short arms, may produce unusual DNA structures, which can arrest the replication fork (Akgun et al. 1997). This could lead to the formation of DSBs. The aberrant repair of DSBs can lead to the formation of chromosomal rearrangements, including translocations (Richardson et al. 1998).
For the class 2 ROBs, both the breakpoints and timing of translocation formation revealed a high degree of variability. The most striking example of this variability was seen among cases of rob(14q15q), which was also the largest group of a particular type of rare ROB studied. One possible mechanism of formation is homologous recombination, which may lead to the formation of these ROBs as implicated in the common ROBs. The sequences involved may be located in multiple places on the participating chromosomes leading to an apparent randomness of the translocation breakpoints, but through a specific homologous sequence. The variable breakpoint could result from breakage and exchange in repetitive DNA, such as satellite III DNA sequences, that are common to all acrocentric short arms and the pericentromeric regions of these chromosomes (Bandyopadhyay et al. 2001_b_).
An alternative, not-yet-identified mechanism may involve a “short motif” for formation of rare ROBs. In studies that involved translocations between the X chromosome and an autosome, it was suggested that very short segments of homologous sequence (4–6 bp) may play a role in forming translocations (Bodrug et al. 1987, 1991; Giacalone and Francke 1992). Because the acrocentric short arms consist mainly of repetitive sequences, such a sequence may be responsible for the formation of these translocations. Several subfamilies of the satellite III DNA have been cloned and shown to contain a consensus 5′-GGAAT-3′ monomer (Bandyopadhyay et al. 2001_b_). The GGAAT motif was found at the breakpoints in a t(X;4) (Giacalone and Francke 1992). This model could explain the randomness of the breakpoint observed in the class 2 translocations. Finally, hypervariable minisatellite DNA is a hotspot for homologous recombination in humans (Wahls et al. 1990). The characterization of the DNA that constitutes the short arm of the acrocentric chromosomes may lead to identification of such repetitive sequences, specific motifs, or minisatellites in these regions that could be investigated as sites of breakpoints in class 2 ROBs. Mechanisms such as nonhomologous end joining and illegitimate recombination are other pathways of repairing spontaneous DNA DSBs (Roth and Wilson 1988; Jackson and Jeggo 1995). DSBs stimulate illegitimate recombination more than homologous recombination (Sargent et al. 1997), which suggests that this mechanism may play a role in the formation of the rare ROBs.
In summary, the common class 1 ROBs form through a distinct and reproducible mechanism in oogenesis. Unique features of oogenesis, such as the dictyotene arrest, may provide the environment for these translocations to form. Given that the evidence indicates a specific process, it is likely that recombination through a homologous sequence on these nonhomologous chromosomes is the mechanism through which these common translocations arise. In contrast, the rare ROBs that comprise the class 2 rearrangements may form through a variety of mechanisms, resulting in the apparent variability in breakpoint locations, parental origins, and timing of formation.
Acknowledgments
We thank M. M. Madrigal for constructing many of the somatic cell hybrids used in this investigation. This research was supported in part by grants from the National Institutes of Health (to L.G.S.) and Wilhelm Sander-Stiftung (99.105.1) and Boehringer Ingelheim Funds (to A.H.).
References
- Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M (1997) Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17:5559–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay R, Berend SA, Page SL, Choo KHA, Shaffer LG (2001_a_) Satellite III sequences on 14p and their relevance to Robertsonian translocation formation. Chromosome Res 9:235–242 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay R, McQuillan C, Page SL, Choo KHA, Shaffer LG (2001_b_) Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chromosome Res 9:223–233 [DOI] [PubMed] [Google Scholar]
- Bodrug SE, Holden JJ, Ray PN, Worton RG (1991) Molecular analysis of X-autosome translocations in females with Duchenne muscular dystrophy. EMBO J 10:3931–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodrug SE, Ray PN, Gonzalez IL, Schmickel RD, Sylvester JE, Worton RG (1987) Molecular analysis of a constitutional X-autosome translocation in a female with muscular dystrophy. Science 237:1620–1624 [DOI] [PubMed] [Google Scholar]
- Borde V, Goldman AS, Lichten M (2000) Direct coupling between meiotic DNA replication and recombination initiation. Science 290:806–809 [DOI] [PubMed] [Google Scholar]
- Byskov AG (1982) Primordial germ cells and regulation of meiosis. In: Austin CR, Short RV (eds) Reproduction of mammals. VI: germ cells and fertilization. Cambridge University Press, Cambridge, pp 1–16 [Google Scholar]
- Cohen PE, Pollard JW (2001) Regulation of meiotic recombination and prophase I progression in mammals. BioEssays 23:996–1009 [DOI] [PubMed] [Google Scholar]
- Constantinou A, Davies AA, West SC (2001) Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104:259–268 [DOI] [PubMed] [Google Scholar]
- Davis L, Barbera M, McDonnell A, McIntyre K, Sternglanz R, Jin Q, Loidl J, Engebrecht J (2001) The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics 157:1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle E, Shaffer LG, Kalitsis P, McQuillan C, Dale S, Choo KHA (1992) Identification of DNA sequences flanking the breakpoint of human t(14q21q) Robertsonian translocations. Am J Hum Genet 50:717–724 [PMC free article] [PubMed] [Google Scholar]
- Giacalone JP, Francke U (1992) Common sequence motifs at the rearrangement sites of a constitutional X/autosome translocation and associated deletion. Am J Hum Genet 50:725–741 [PMC free article] [PubMed] [Google Scholar]
- Hamerton JL, Canning N, Ray M, Smith J (1975) A cytogenetic survey of 14,069 newborn infants: incidence of chromosomal abnormalities. Clin Genet 8:223–243 [DOI] [PubMed] [Google Scholar]
- Han J-Y, Choo KHA, Shaffer LG (1994) Molecular cytogenetic characterization of 17 rob(13q14q) Robertsonian translocations by FISH, narrowing the region containing the breakpoints. Am J Hum Genet 55:960–967 [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Jeggo PA (1995) DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci 20:412–415 [DOI] [PubMed] [Google Scholar]
- Jacobs PA (1981) Mutation rates of structural chromosome rearrangements in man. Am J Hum Genet 33:44–54 [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Earle E, Vissel B, Shaffer LG, Choo KHA (1993) A chromosome 13 specific human satellite I DNA subfamily with minor presence on chromosome 21: further studies on Robertsonian translocations. Genomics 16:104–112 [DOI] [PubMed] [Google Scholar]
- Knight SJL, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, Cardy DL, Nguyen H, Hudson TJ, Riethman HC, Ledbetter DH, Flint J (2000) An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 67:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Nurse P (2001) Regulation of premeiotic S phase and recombination related double-strand DNA breaks during meiosis in fission yeast. Nat Genet 28:290–293 [DOI] [PubMed] [Google Scholar]
- Page SL, Shaffer LG (1997) Nonhomologous Robertsonian translocations form predominantly during female meiosis. Nat Genet 15:231–232 [DOI] [PubMed] [Google Scholar]
- Page SL, Shin J-C, Han J-Y, Choo KHA, Shaffer LG (1996) Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum Mol Genet 5:1279–1288 [DOI] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, Jasin M (1998) Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev 12:3831–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, Bernasconi F, Basaran S, Yuksel-Apak M, Neri G, Serville F, Balicek P, Haluza R, Farah LMS, Luleci G, Schinzel AA (1994) A somatic origin of homologous Robertsonian translocations and isochromosomes. Am J Hum Genet 54:290–302 [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Wilson JH (1988) Illegitimate recombination in mammalian cells. In: Kucherlapati R, Smith GR (eds) Genetic recombination. American Society for Microbiology, Washington, DC, pp 621–653 [Google Scholar]
- Sargent RG, Brenneman MA, Wilson JH (1997) Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol 17:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Jackson-Cook CK, Stasiowski BA, Spence JE, Brown JA (1992) Parental origin determination in thirty de novo Robertsonian translocations. Am J Med Genet 43:957–963 [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR (2000) Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 34:297–329 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Jenkins LS, Karson EM, Leana-Cox J, Schwartz S (1996) Evidence for structural heterogeneity from molecular cytogenetic analysis of dicentric Robertsonian translocations. Am J Hum Genet 59:167–175 [PMC free article] [PubMed] [Google Scholar]
- Therman E, Susman B, Denniston, C (1989) The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations. Ann Hum Genet 53:49–65 [DOI] [PubMed] [Google Scholar]
- Vogel F, Motulsky AG (1997) Human genetics problems and approaches. Springer-Verlag, Berlin, pp 396–397 [Google Scholar]
- Wahls WP, Wallace LJ, Moore PD (1990) Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell 60:95–103 [DOI] [PubMed] [Google Scholar]