Protean PTEN: Form and Function (original) (raw)
Abstract
Germline mutations distributed across the PTEN tumor-suppressor gene have been found to result in a wide spectrum of phenotypic features. Originally shown to be a major susceptibility gene for both Cowden syndrome (CS), which is characterized by multiple hamartomas and an increased risk of breast, thyroid, and endometrial cancers, and Bannayan-Riley-Ruvalcaba syndrome, which is characterized by lipomatosis, macrocephaly, and speckled penis, the PTEN hamartoma tumor syndrome spectrum has broadened to include Proteus syndrome and Proteus-like syndromes. Exon 5, which encodes the core motif, is a hotspot for mutations likely due to the biology of the protein. PTEN is a major lipid 3-phosphatase, which signals down the PI3 kinase/AKT pro-apoptotic pathway. Furthermore, PTEN is a protein phosphatase, with the ability to dephosphorylate both serine and threonine residues. The protein-phosphatase activity has also been shown to regulate various cell-survival pathways, such as the mitogen-activated kinase (MAPK) pathway. Although it is well established that PTEN’s lipid-phosphatase activity, via the PI3K/AKT pathway, mediates growth suppression, there is accumulating evidence that the protein-phosphatase/MAPK pathway is equally important in the mediation of growth arrest and other crucial cellular functions.
Introduction
Prior to 1996, when the susceptibility gene for Cowden syndrome (CS [MIM 158350]) was mapped to 10q22-q23 (Nelen et al. 1996), the molecular bases of the inherited hamartoma-tumor syndromes were obscure. CS is an autosomal dominant disorder that is characterized by multiple hamartomas that affect derivatives of all three germ layers and by a risk of breast, thyroid, and endometrial neoplasias (Appendix A) (Eng 2000). Germline mutations in PTEN/MMAC1/TEP1 (MIM 601728), a tumor-suppressor gene located on 10q23, have since been found in 80% of probands with CS (Liaw et al. 1997; Marsh et al. 1998_b_). PTEN encodes a lipid dual-specificity phosphatase and is the major 3-phosphatase in the phosphoinositol-3-kinase (PI3K)/AKT pro-apoptotic pathway (Li and Sun 1997; Li et al. 1997; Steck et al. 1997; Maehama and Dixon 1998; Stambolic et al. 1998). This represents the first phosphatase gene that has been implicated in the etiology of an inherited cancer syndrome. Subsequently, the clinical spectrum of disorders that are associated with germline PTEN mutations has expanded to include seemingly disparate syndromes.
Identification of PTEN
PTEN was first identified in 1997 by three independent groups, each of which had slightly different strategies. Two groups used positional-cloning approaches to map this gene to 10q23 (Li et al. 1997; Steck et al. 1997); sequence analysis showed a large region of homology to chicken tensin, bovine auxilin, and a protein tyrosine-phosphatase domain, from which the name “PTEN” was coined (for phosphatase and tensin homolog, deleted on chromosome 10 [ten]). A third group (Li and Sun 1997) identified PTEN by searching for genes with its biochemical properties. Li and Sun searched for novel human protein tyrosine phosphatases by using two different methods (Li and Sun 1997). By searching GenBank for entries that contain phosphatase motifs and using a PCR-based approach to screen a human cDNA library, they identified TEP1 (transforming growth factor [TGF]–regulated and epithelial cell-enriched phosphatase), which was also shown to be identical to PTEN. TEP1 was shown to dephosphorylate an in vitro substrate for tyrosine phosphatases (phosphotyrosyl-RCML). Furthermore, Li and Sun demonstrated that when the essential cysteine in the tyrosine-phosphatase motif was mutated, a mutation that occurs in CS, the phosphatase activity was abolished (Li and Sun 1997).
CS
CS exhibits variable expressivity, which makes the diagnosis of CS and the accurate measurement of its incidence a challenge. Before the identification of the susceptibility gene, the incidence was thought to be 1/1,000,000 (Nelen et al. 1996). However, after identification of PTEN as the gene for CS (Liaw et al. 1997), a molecular-based study revealed the incidence to be ⩾1/200,000 (Nelen et al. 1997, 1999), although this too is likely to be an underestimate. Because CS is underdiagnosed, a true count of the proportion of isolated cases (defined as having no obvious family history) and familial cases (defined as having two or more related affected individuals) cannot be performed. From the literature and the experience of major CS centers in the world, the majority of CS cases appear to be isolated. As a broad estimate, ∼10%–50% of CS cases are familial (Marsh et al. 1999).
Because of the lack of uniform diagnostic criteria for CS prior to 1995, the International Cowden Consortium arrived at a set of consensus operational diagnostic criteria culled from published data and expert opinion (Nelen et al. 1996; Eng 1998). This consortium represented a group of centers, mainly in North America and Europe, that were interested in systematically studying this syndrome to localize the susceptibility gene. These diagnostic criteria have been revised recently, in the context of new data, and are reflected in the practice guidelines of the U.S.-based National Comprehensive Cancer Network (NCCN) Genetics/High Risk Panel (Appendix B) (NCCN 1999; Eng 2000).
More than 90% of individuals affected with CS are believed to manifest a phenotype by the age of 20 years (Nelen et al. 1996; Eng 2000). By the end of the 3d decade of life (i.e., at 29 years of age), 99% of affected individuals are believed to have developed at least the mucocutaneous signs of the syndrome, although any of the other clinical features could also be present (Appendixes A and B). The most commonly reported manifestations are mucocutaneous lesions, thyroid abnormalities, fibrocystic disease, and carcinoma of the breast, multiple, early-onset uterine leiomyoma, and macrocephaly (specifically, megalencephaly) (Appendix A) (Starink et al. 1986; Hanssen and Fryns 1995; Mallory 1995; Longy and Lacombe 1996; Eng 2000). Mucocutaneous trichilemmomas and papillomatous papules are considered diagnostic of the syndrome (Appendix B).
The two most well-documented component malignancies of CS are carcinomas of the breast and epithelial thyroid gland (Starink et al. 1986). In women with CS, lifetime risks of breast cancer are estimated to be 25%–50% (Starink et al. 1986; Hanssen and Fryns 1995; Longy and Lacombe 1996; Eng 1997) in contrast to the 11% in the general population. The mean age at diagnosis is likely 10 years earlier than that in the general population, at ∼38–48 years of age, with a range of 14–65 years of age (Starink et al. 1986; Longy and Lacombe 1996). Until genotype-phenotype analyses were performed with the discovery of the susceptibility gene, it was thought that male breast cancer was not a component of CS. However, male breast cancer does occur in _PTEN-_mutation–positive CS, but with unknown frequency (Marsh et al. 1998_b;_ Fackenthal et al. 2001). The lifetime risk of epithelial thyroid cancer can be as high as 10% in males and females with CS. It is unclear if the age at onset is truly earlier than that of the general population. Histologically, CS-associated thyroid cancer is predominantly follicular carcinoma, although papillary histology has also been rarely observed (Starink et al. 1986; Hanssen and Fryns 1995; Longy and Lacombe 1996; C. Eng, unpublished data). After identification of PTEN as the susceptibility gene, preliminary data suggested that endometrial carcinoma is a component cancer of CS (Marsh et al. 1998_a_; De Vivo et al. 2000; Eng 2000). Its frequency in mutation carriers is as yet unknown.
Germline PTEN Mutations Cause CS
Germline mutations in PTEN, have been found in CS (Liaw et al. 1997; Lynch et al. 1997; Nelen et al. 1997, 1999; Tsou et al. 1997; Marsh et al. 1998_b_). Although the original linkage study mapped CS to 10q22-q23 without genetic heterogeneity (Nelen et al. 1996), one subsequent study suggested that rare locus heterogeneity may exist (Tsou et al. 1997). It is unclear if BMPR1A, which is on 10q22 and encodes a bone morphogenic protein receptor belonging to the TGF-β–receptor superfamily, is a susceptibility gene for juvenile polyposis syndrome (Eng 2001; Howe et al. 2001), is also a rare susceptibility gene for CS (Zhou et al. 2001_b_). MINPP1, another gene that maps to 10q23 upstream from PTEN and that encodes a phosphatase, has been excluded as a locus for CS (Dahia et al. 2000).
When CS is strictly defined by the operational diagnostic criteria of the International Cowden Consortium, 80% have been found to harbor germline PTEN mutations (Marsh et al. 1998_b_). Approximately two-thirds of these mutations were found in exons 5, 7, and 8 (fig. 1 and table 1). Approximately 40% of all CS germline mutations are located in exon 5, although exon 5 represents only 20% of the coding sequence. Genotype-phenotype analyses revealed an association between the presence of germline mutations and malignant breast disease (Marsh et al. 1998_b_). In other words, more malignant breast disease occurred in the 80% of families with CS diagnosed by the International Cowden Consortium criteria and who were mutation positive, compared to the 20% of families who also met International Cowden Consortium criteria but were mutation negative. In addition, missense mutations and those within and 5′ to the phosphatase core motif appear to be associated with involvement of five or more organs, a surrogate phenotype for severity of disease (Marsh et al. 1998_b_). Another group examined families for germline PTEN mutations and found mutations in only 13 (∼50%) probands (Nelen et al. 1999). They could not find any clear genotype-phenotype associations, almost certainly due to their small sample size and consequent lack of statistical power. The differences in mutation frequency between the studies by Marsh et al. (1998_b_) and Nelen et al. (1999) include a decreased stringency of ascertainment by the International Cowden Consortium criteria and, perhaps, small sample size in the latter study.
Figure 1.
Germline PTEN mutations in CS, BRRS, PS, and Proteus-like syndromes
Table 1.
Missense Mutations in PTEN Tested for Lipid Phosphatase Activity[Note]
| Mutation | Domain | PhosphataseActivitya |
|---|---|---|
| S10N | Phosphatase | Full |
| Y16C | Phosphatase | Null |
| G20E | Phosphatase | Partialb |
| Y27S | Phosphatase | Null |
| L42R | Phosphatase | Full |
| H61R | Phosphatase | Null |
| Y68R | Phosphatase | Null |
| C71Y | Phosphatase | Null |
| H93Y | Phosphatase/active core | Null |
| C105F | Phosphatase | Null |
| D107Y | Phosphatase | Null |
| L112P | Phosphatase | Null |
| L112R | Phosphatase | Null |
| A121 | Phosphatase | Null |
| C124R | Phosphatase/active core | Null |
| G129R | Phosphatase/active core | Null |
| G129E | Phosphatase/active corec | Null |
| R130G | Phosphatase/active core | Null |
| R130L | Phosphatase/active core | Null |
| R130Q | Phosphatase/active core | Null |
| V133I | Phosphatase | Null |
| M134L | Phosphatase | Partialb |
| C136 | Phosphatase | Null |
| Y155 | Phosphatase | Null |
| G165R | Phosphatase/active core | Null |
| S170N | Phosphatase | Null |
| S170R | Phosphatase | Null |
| R173C | Phosphatase | Null |
| R173H | Phosphatase | Null |
| R173P | Phosphatase | Null |
| Y174N | Phosphatase | Null |
| S227F | C2 | Partialb |
| G251C | C2 | Null |
| K289E | C2 | Full |
| D331G | C2 | Partial |
| F341V | C2 | Null |
| K342N | C2 | Partialb |
| V343E | C2 | Null |
| L345Q | C2 | Null |
| F347L | C2 | Partial |
| V369 | C terminus | Full |
| T401 | C terminus | Full |
Clinical Spectrum of **PTEN-**Defined Syndromes: The Concept of the PTEN Hamartoma-Tumor Syndrome (PHTS)
Bannayan-Riley-Ruvalcaba Syndrome (BRRS)
Germline PTEN mutations in families with BRRS (MIM 153480), characterized by macrocephaly, lipomatosis, hemangiomatosis and speckled penis (Gorlin et al. 1992), have also been found (fig. 1) (Marsh et al. 1997_a_). Thus, at least a subset of patients with BRRS and CS may be considered allelic. In contrast to patients with CS, 60% of patients with BRRS were found to have germline PTEN mutations (Marsh et al. 1999). These mutations included one with a cytogenetically detectable deletion of 10q23, encompassing PTEN, and a translocation involving 10q23 (Arch et al. 1997; Ahmed et al. 1999; Marsh et al. 1999). The mutational spectra of BRRS and CS seem to overlap, lending formal proof that CS and BRRS are allelic (Marsh et al. 1999). Thus, it has been suggested that syndromes that are characterized by the presence of germline PTEN mutations may be grouped by molecular definition and referred to as the “PHTSs” (Marsh et al. 1999).
Genotype-phenotype association analyses revealed several correlations in BRRS (Marsh et al. 1999). The presence of germline PTEN mutations was found to be correlated with the presence of breast tumors, the presence of breast fibroadenomas, and lipomatosis. In other words, there was a higher frequency of breast tumors, fibroadenomas, and lipomas among the group of patients with BRRS, all of whom met published diagnostic criteria (Gorlin et al. 1992) and were found to be mutation positive, compared to the 40% of patients with BRRS, who also met diagnostic criteria but were mutation negative. In this analysis, there were nine families with clinical overlap of CS and BRRS; eight of these nine families were found to harbor germline PTEN mutations (Marsh et al. 1999). This high frequency of mutation positivity among families with overlap continues to hold true, since such families continue to be accrued and subjected to mutation analysis (Celebi et al. 1999; Wanner et al. 2001; X. P. Zhou and C. Eng, unpublished data).
One report suggested that germline PTEN mutations occur only in familial BRRS but not isolated (i.e., nonfamilial) BRRS (Carethers et al. 1998). However, this finding has not held true in multiple other studies (Arch et al. 1997; Longy et al. 1998; Zori et al. 1998; Marsh et al. 1999). In the largest single series of patients with isolated BRRS, 7 of 16 unrelated probands with isolated BRRS were found to have germline intragenic PTEN mutations (Marsh et al. 1999). If we also considered large rearrangements, 9 of 18 patients with isolated BRRS harbored a germline PTEN alteration. This frequency of mutation-positive cases is not significantly different from that obtained for familial BRRS.
Currently, it is unclear if the 40% of BRRS without germline PTEN mutations have large rearrangements affecting PTEN or if genes other than PTEN may also be responsible for BRRS. MINPP1 has also been excluded as a susceptibility gene for _PTEN-_mutation–negative BRRS (Dahia et al. 2000).
Proteus Syndrome (PS) and Proteus-like Syndromes
Although PS (MIM 176920) had always been considered part of the genetic differential diagnosis of CS, it was often discarded as being part of PHTS. The mosaic distribution of affected tissues in PS and its sporadic occurrence strongly suggested either somatic mutation or germline mosaic mutation as its etiology. Interestingly, during the ascertainment of probands with the minimal clinical features of hamartomas, lipomas and overgrowth, an individual with a Proteus-like syndrome was found to carry a germline PTEN R335X mutation and a second-hit germline mosaic R130X mutation in his affected tissues (Zhou et al. 2000). This individual was born of normal parents and had marked hemihypertrophy, macrocephaly, epidermoid nevi, lipomas, and progressively worsening arterioventricular malformations, features that are reminiscent of PS but do not meet the full published criteria for the clinical diagnosis of PS (Biesecker et al. 1999). This individual with a Proteus-like syndrome also did not meet the criteria for the diagnosis of either CS or classic BRRS.
Given these observations, a series of nine unrelated patients with isolated PS and five individuals with Proteus-like syndromes were analyzed for the presence of germline PTEN mutations (fig. 1) (Zhou et al. 2001_a_). Of the nine patients with classic PS who met the published clinical criteria (Biesecker et al. 1999), two were found to have germline PTEN mutations, W211R and C211X, neither of which have previously been found in CS or BRRS (Zhou et al. 2001_a_). Three of the five individuals with a Proteus-like syndrome were also found to have germline PTEN mutations. Interestingly, R335X, which has also been found in CS and BRRS, was found in the germline of two of these individuals. The third, M35T, is, thus far, unique. There is at least one other small series of individuals with PS that was examined for the presence of PTEN mutations (Barker et al. 2001). Among eight individuals with PS, no mutations were found. This negative result is almost certainly due to small sample size and/or may be secondary to the mutation-scanning method chosen, conformation-specific gel electrophoresis, which has been shown to be relatively insensitive even in the best of hands (Eng et al. 2001). Together, these data suggest that subsets of both PS and Proteus-like syndromes may be considered PHTS. It is also clear that other susceptibility genes will be found for PS and Proteus-like syndromes.
Although _PTEN-_mutation–positive CS, BRRS, PS, and Proteus-like syndromes are considered in the PHTS continuum, the phenotypes associated with the same mutation can vary quite remarkably (Appendix A). This is particularly pronounced for the hotspot mutations on CpG islands, R233X, R235X, and R335X.
Other Clinical Syndromes?
Germline PTEN mutations have been uncovered in a single case of isolated hydrocephaly with VATER association (Reardon et al. 2001) and an individual with megalencephaly with autistic features (Dasouki et al. 2001). VATER association comprises vertebral and anal malformations, tracheoesophageal atresia, and radial and renal malformations. VATER associated with hydrocephaly is a distinct entity from VATER association alone, and, unlike VATER, familial cases have been reported (Iafolla et al. 1991; Devriendt et al. 1995; reviewed in Reardon et al. 2001). In the latter case, it is unclear if the autistic features are part of PHTS or if it is only the megalencephaly that is germane. Both of these types of clinical presentations are extremely rare, and further investigation is required to determine if these clinical syndromes are also prominent members of PHTS.
Clinical Syndromes That Are Not PHTS
The molecular classification of PHTS is important in two ways. First, the broadening phenotypic spectrum of PHTS yields clues to fundamental insights into the structure-function relationship of PTEN. Second, the PHTS molecular classification of clinical syndromes is important from a clinical management point of view. Genotype-phenotype analyses have suggested that the presence of germline mutations, in CS or BRRS, is associated with cancer, at least breast cancer (Marsh et al. 1998_b,_ 1999). Thus, a conservative clinician would recommend cancer surveillance for all individuals with PHTS, as is advocated for classic CS, irrespective of their clinical diagnosis (Eng 2000).
There are several inherited hamartoma-tumor syndromes that do not belong to PHTS. Peutz-Jeghers syndrome (PJS [MIM 175200]) is an autosomal dominant inherited cancer syndrome characterized by gastrointestinal hamartomatous polyposis, peroral pigmentation, and a risk of gastrointestinal and breast cancers. Its susceptibility gene is LKB1/STK11, on 19p, encoding a nuclear serine threonine kinase (Hemminki et al. 1997, 1998; Jenne et al. 1998). PTEN has been excluded as a locus in PJS (D. J. Marsh and C. Eng, unpublished data).
Juvenile polyposis syndrome (JPS [MIM 174900]) is a clinical diagnosis of exclusion, and there has been some confusion whether JPS is a PHTS (Eng and Ji 1998). JPS is an autosomal dominant disorder characterized by gastrointestinal hamartomatous polyps (“juvenile polyps”) and a risk of gastrointestinal cancers. Initial confusion stemmed from a paper that described germline PTEN mutations in two individuals who had been reported to have JPS (Olschwang et al. 1998). It became clear, however, that the insufficiently detailed clinical descriptions that were provided for these patients strongly suggested that these individuals had CS or BRRS (Eng and Ji 1998). Similarly, the title of one report referred to germline mutations in individuals with JPS, but it was obvious, from the text, that all of these individuals had CS (Lynch et al. 1997). Even recently, a small study reported that PTEN may be a rare _JPS-_susceptibility gene (Huang et al. 2000), but, again, insufficient clinical detail was given in this report to determine if these individuals had features of either CS or BRRS. At least one series systematically examined this issue. The first was a single hospital-based series that looked for germline PTEN mutations in diagnoses of JPS (Kurose et al. 1999). In this series, one individual was found to harbor a germline PTEN mutation. When that individual was recalled for thorough examination, classic cutaneous features of CS were found (Kurose et al. 1999). PTEN was formally excluded as a _JPS-_susceptibility gene (Marsh et al. 1997_b_), and we now know that germline mutations in MADH4 on 18q and BMPR1A on 10q21-q22 account for 40%–60% of JPS (Howe et al. 1998_a,_ 1998_b,_ 2001; Zhou et al. 2001_b_).
Overall, the data to date suggest that JPS is not a PHTS. However, the discovery of a germline PTEN mutation in an individual considered to have JPS should raise the suspicion that the clinical diagnosis is incorrect and that such an individual be medically managed in the same manner as all patients with PHTS.
Murine Models
Animal models of human disease are helpful when they faithfully recapitulate the human disease and/or when they can be utilized for fundamental research that can elucidate the biology of the gene in question. Three different groups have generated _Pten-_knockout mice. All three groups targeted exon 5 in the targeting construct (Di Cristofano et al. 1998; Suzuki et al. 1998; Podsypanina et al. 1999), and two of these groups also deleted extra exons (i.e., exons 3–5 [Suzuki et al. 1998] and exons 4 and 5 [Di Cristofano et al. 1998]). The third group targeted a frameshift mutation within the phosphatase motif (Podsypanina et al. 1999). These groups all reported that homozygous knockout of Pten was embryonic lethal within a range of embryonic days 6.5–9.5. Beyond this, however, the groups observed varied phenotypes in the heterozygous mice. Few features of CS or BRRS have been described in these models, although each model contains features that are reminiscent of these disorders. For example, one model is characterized by abnormal development of the three germinal layers, as well as skin hyperkeratosis and papillary-like thyroid carcinoma in heterozygous mice (Di Cristofano et al. 1998). Superficially, one may imagine that these are features found in human CS; however, they are not (e.g., papillary thyroid carcinoma is rarely observed in _PTEN-_mutation–positive CS or BRRS). In their mouse model, Suzuki et al. (1998) found defects in the generation of mesodermal lineages, as well as hamartomatous colonic polyps, which are not particularly reminiscent of the human counterparts typical in CS or BRRS. They also observed thymic lymphomas, which are not components of either CS or BRRS. Interestingly, with long follow-up, these mice developed breast cancer and endometrial cancers, both of which are components of CS (Stambolic et al. 2000). With follow-up, pheochromocytomas also developed, although, to date, these neuroendocrine tumors have never been observed in CS or BRRS. Finally, Podsypanina et al. (1999) observed follicular and papillary thyroid tumors and atypical hyperplasia of the endometrium, features that, again, are reminiscent of CS/BRRS but are not similar when examined in histologic detail. The type of mutation or differences in the genetic background of the mice may contribute to the differences observed in each model. The lack of significant similarity to CS and BRRS raises questions to the usefulness of these models in terms of representing the clinical conditions. Nonetheless, these models should prove useful, particularly when crossbred with other knockout mice and/or for fundamental biochemical studies, in elucidating information about the pathways with which PTEN interacts in situ.
PTEN Structure: Does It Explain Phenotype?
Clinical and genetic analyses have revealed that PTEN mutations result in an ever-widening spectrum of phenotypic features. Does the structure of PTEN yield clues to explain this? Analysis of the crystal structure of PTEN has identified two major domains (fig. 2).
Figure 2.
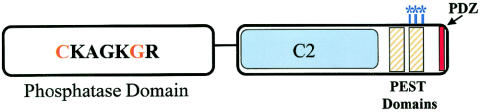
Protein domains of PTEN. The N-terminal phosphatase domain (amino acids 1–185) is shown with the catalytic core. The missense mutations which have been crucial for the elucidation of the cellular role of PTEN are highlighted in orange. Mutations at C124 render a lipid- and protein-phosphatase–inactive protein, whereas mutations at G129 result in a lipid-phosphatase–inactive yet protein-phosphatase–active PTEN. The C-terminal domain (amino acids 186–403) contains the lipid-binding C2 domain (amino acids 186–351); PEST domains (amino acids 350–375 and 379–396), which regulate protein stability; and the PDZ domain, which is important in protein-protein interactions. The CK2 phosphorylation sites (S380, T382, and T383), which are important for stability, are indicated by the blue asterisks (*).
The C-Terminal Domain
The C-terminal domain, in which ⩾43% of PTEN mutations occur, is composed of antiparallel β-sheets that are linked together by short α-helices (Lee et al. 1999). This domain contains many important subdomains that are common to other signal-transducing molecules. First, PTEN contains a C2 domain, which is associated with phospholipid-binding regions (Lee et al. 1999). C2 domains have been identified in many proteins involved in signal transduction and membrane localization (Rizo and Sudhof 1998). Indeed, the C2 domain in PTEN has been shown to have affinity for phospholipid membranes in vitro (Rizo and Sufhof 1998). Many germline PTEN mutations lie within the C2 domain, suggesting its functional significance (figs. 1 and 2 and table 1). However, germline mutations within the C2 domain have been found for the entire spectrum of PHTS phenotypes (figs. 1 and 2), without an obvious functional genotype-phenotype correlation.
Another feature of the C terminus is a PDZ-binding motif, which interacts strongly with the phosphatase domain (described below; fig. 2) by both hydrogen bonding and hydrophobic interactions (Lee et al. 1999). In addition, PDZ domains are significant regions for protein-protein interactions (Fanning and Anderson 1999; Kay et al. 2000), which play a vital role in cellular signal transduction. Removal of the PDZ domain reduces the ability of PTEN to inhibit one of its substrates, AKT (Wu X. et al. 2000), suggesting that PTEN interactions may be important in regulating PTEN activity. The C-terminal tail also contains PEST sequences, which are critical for PTEN stability (Georgescu et al. 1999). PEST sequences target proteins for short intracellular half-lives and protein degradation. Paradoxically, deletion of these regions leads to decreased protein expression versus the expected increase (Georgescu et al. 1999). Nonetheless, these studies point out that the PEST regions are necessary for PTEN stability.
The C-terminal tail also contains several phosphorylation sites located in the last 50 amino acids. Although this region or “tail” of PTEN is unnecessary for phosphatase activity and cell-growth suppression (Vazquez et al. 2000), it is critical for protein stability. Protein stability is dependent on the phosphorylation of S380, T382, and T383. Mutations of these sites reduced both the protein half-life and the PTEN levels (Vazquez et al. 2000). Torres and Pulido (2001) demonstrated that CK2, the protein kinase, phosphorylates PTEN in vivo on serine residues 370, 380, and 385 and on threonine residue 383. Furthermore, phosphorylation-defective mutants have decreased protein stability and provided evidence that dephosphorylated PTEN is degraded by proteasome-mediated mechanisms (Torres and Pulido 2001). Together, these data suggest that protein phosphorylation plays an important role in the regulation of PTEN by influencing protein levels. PTEN phosphorylation has also been shown to cause a conformational change that masks the PDZ domain, reducing PTEN’s ability to bind to PDZ-domain–containing proteins (Vazquez et al. 2000).
It is interesting to note that, to date, no germline or somatic mutations have been detected specifically in the PDZ, PEST, or phosphorylation sites, although there are many mutations, both germline and somatic, that would truncate the protein before these features. There are two obvious explanations for these observations. This may suggest that the presence of dominant negative mutations within each of these domains may be embryonic lethal while haploinsufficiency is viable, although it results in the PHTS phenotypes. It is difficult to imagine that the alternate hypothesis—that is, that they are not germane to carcinogenesis—could be true.
The N-Terminal Domain
The N-terminal domain contains the enzymatic side of PTEN (i.e., its phosphatase domain), which is composed of β-sheets surrounded by α-helices (Lee et al. 1999). The majority of PTEN mutations occur within this domain. Analysis of the crystal structure revealed that the PTEN phosphatase domain, although similar to those of other protein phosphatases, has a slightly larger active site. This enlarged active site allows for the accessibility of phospholipid substrates (Lee et al. 1999), thereby making PTEN a unique phosphatase. The active site is also a mutational hotspot, with ∼31% of germline and somatic mutations occurring in exon 5, which encodes the catalytic core. That 40% of germline mutations in CS lie in exon 5, which represents 20% of the coding sequence, undoubtedly reflects the biology of the resulting protein.
PTEN, a Dual-Specificity Phosphatase
Since the discovery of PTEN in 1997, abundant data have been presented that show that PTEN is a tumor suppressor in vitro and in vivo. PTEN can be regarded as a dual-specificity phosphatase on several levels. In particular, recombinant PTEN has been shown to dephosphorylate protein substrates in vitro on serine, threonine, and tyrosine residues (Myers et al. 1997). In this manner, PTEN is a dual-specificity protein phosphatase. One proposed PTEN substrate is focal adhesion kinase (FAK); PTEN interacts with FAK and decreases its tyrosine phosphorylation levels (Gu et al. 1998; Tamura et al. 1998). Dephosphorylation of FAK inhibits cell spreading, suggesting that the protein-phosphatase activity of PTEN may be important in the regulation of cellular interactions. This, however, may be cell line–specific, since others have been unable to reproduce this work.
In 1998, Maehama and Dixon reported another PTEN substrate. Previous work had shown that the PTEN mutant G129E, a mutation found in at least two families with CS, still had the ability to dephosphorylate peptide substrates in vitro (Maehama and Dixon 1998). This suggested that PTEN may also have a nonproteinaceous target in vivo. Maehama and Dixon observed that the overexpression of PTEN reduced the cellular levels of phosphoinositol 3,4,5-triphosphate (PIP3) in response to insulin. This occurred without a change in the activity of PI3K, the kinase that phosphorylates phosphoinositol-diphosphate. When the G129E mutant was expressed in cell lines, PIP3 levels increased. Together, these data suggested that PTEN was capable of dephosphorylating cellular phospholipids. Meahama and Dixon expanded their observations by showing that PTEN catalyzes, in vivo and in vitro, the removal of phosphate from the D3 position of the inositol ring. Therefore, PTEN is also a dual-specificity phosphatase in the sense that it dephosphorylates protein substrates in addition to lipid substrates.
PTEN Regulation of the PI3K Pathway
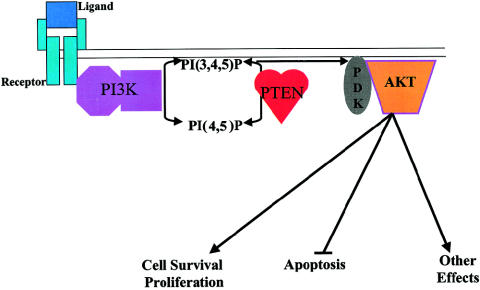
Phosphoinositide second messengers play an important role in signal transduction pathways that regulate cell growth, differentiation, apoptosis, metabolism, actin rearrangements, and membrane trafficking. They do this by directly activating enzymes or by directing proteins to various subcellular locations via lipid binding. Abundant data have shown that PIP3 is required for the activation of AKT/PKB, a serine/threonine protein kinase that plays a role in cell survival (Vazquez et al. 2000). PI3K activation, mainly by growth factors, results in the accumulation of PIP3 in cellular membranes (Kapeller and Cantley 1994) resulting in the translocation of AKT from cytoplasmic stores to cellular membranes. On membrane docking, AKT undergoes a conformational change and is phosphorylated by PDK1, thereby stimulating AKT and subsequent signaling via its downstream effectors (fig. 3). This stimulates pathways required for cell survival and proliferation (Downward 1998).
Figure 3.
PTEN as a regulator of the PI3K pathway. Ligand binding to membrane receptors results in the activation of PI3K and the subsequent increase in PIP3, which recruits PDK1 to the cellular membrane. PDK1 phosphorylates and activates AKT, which in turn regulates a variety of cellular processes. PTEN dephosphorylates PI3P, lowering its cellular levels and resulting in the down-regulation of AKT.
PTEN antagonizes the PI3K/AKT pathway by dephosphorylating PIP3, resulting in a decreased translocation of AKT to cellular membranes and subsequent down-regulation of AKT activation. Indeed, it has been shown that expression of PTEN in cells leads to decreased levels of phospho-AKT, and, therefore, to increased apoptosis (Davies et al. 1998; Myers et al. 1998). In addition, constitutively active, but not wild-type, AKT, can rescue cells from PTEN-mediated G1 arrest and apoptosis. Furthermore, PI3K inhibition by LY294002 could mimic the effects of PTEN (Li and Sun 1998). Together, these data demonstrate that PTEN exerts its effect upstream from PI3K.
The effect that PTEN has on the PI3K pathway is phosphatase dependent. C124 is the active-site cysteine of PTEN and has been found to be mutated in at least three probands with CS (Marsh et al. 1998_b;_ Bonneau and Longy 2000). The C124S mutation abolishes both the protein and lipid-phosphatase activities of PTEN (Myers et al. 1998). When the PTEN C124S mutant was introduced into cells, phospho-AKT levels remained constant, and cells did not undergo G1 arrest or apoptosis (Myers et al. 1998; Weng et al. 1999, 2001_b_).
Several groups have shown that PTEN coordinates G1 arrest through up-regulation of p27 and concomitant down-regulation of cyclin D1 (Cheney et al. 1999; Bruni et al. 2000; Medema et al. 2000; Persad et al. 2001; Weng et al. 2001_a_). The D cyclins are key regulators of progression through G1 of the cell cycle, whereas p27 is an inhibitor of cyclin-dependent kinases and acts as a negative regulator of the cell cycle. Another factor that influences G1 arrest may be the inhibition of retinoblastoma protein phosphorylation via the effects that PTEN has on the PI3K pathway (Paramio et al. 1999). Use of a G129E mutant has shed light on the mechanisms by which PTEN exerts some of these effects. Similar to C124, G129 is also one of the key residues in the active site of PTEN and has been found to be mutated in at least three probands with CS. Interestingly, this mutation renders the PTEN molecule _lipid-_phosphatase inactive but _protein-_phosphatase active. Weng et al. (2001_a_) used both the G129E and C124S mutants to determine that PTEN down-regulates cyclin D1 expression by its protein-phosphatase activity. In contrast, p27 levels are up-regulated downstream from the lipid-phosphatase activity (Weng et al. 2001_a_). In contrast, p27 levels, as well as p21 and p57 levels, are up-regulated downstream from the lipid-phosphatase activity. Increased transcription of these cyclin-dependent kinase inhibitors occurs in response to PTEN-mediated down-regulation of AKT. This is believed to be mediated by the Forkhead family of transcription factors, and it also contributes to G1 arrest. Together, these data indicate that PTEN’s lipid-phosphatase activity is critical to cell function. In light of this, it is interesting to note that 90% of PTEN missense mutations eliminate or reduce lipid-phosphatase activity (table 1), without affecting the ability of the protein to bind to the cellular membrane (Han et al. 2000). Since the PI3K pathway is involved in a variety of pathways, including those for hypoxia-induced protein regulation and obesity (reviewed in Katso et al. 2001; Minet et al. 2001), the phenotypic result of PTEN mutation could be compounded by the as-yet-unknown effects of multiple downstream and feedback pathways.
Most naturally occurring mutations are both lipid- and protein-phosphatase inactive (e.g., C124S; see table 1), although a minority are lipid-phosphatase inactive but protein-phosphatase active (e.g., G129E). It is interesting that no naturally occurring mutation that results in lipid-phosphatase–active but protein-phosphatase–inactive PTEN has been identified. This is most likely because a change in the active site that renders PTEN protein-phosphatase inactive would automatically render the protein lipid-phosphatase inactive. However, this does not necessarily mean that PTEN has no other function than to regulate the PI3K pathway, as has been suggested by emerging data (see next section).
Other Roles of PTEN
Although abundant data show that PTEN is a negative regulator of the PI3K/AKT pathway, it is also becoming clear that PTEN may be involved in other cellular functions. For example, FAK has been proposed as a protein substrate of PTEN (Tamura et al. 1998, 1999).
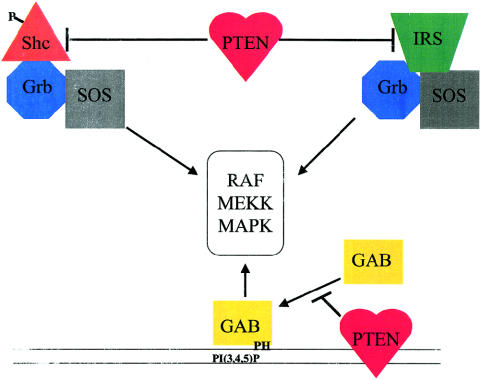
As is the PI3K pathway, the mitogen-activated kinase (MAPK) pathway is another critical pathway for proliferation and differentiation. PTEN also regulates this pathway. PTEN can dephosphorylate Shc, resulting in the inhibition of Grb2 and, ultimately, the down-regulation of MAPK. However, it has also been demonstrated that PTEN can inhibit the MAPK pathway in a Shc-independent manner (fig. 4). First, PTEN may regulate the MAPK pathway by modulating the movement of Gab1 to the plasma membrane. Gab1 contains a pleckstin-homology domain that interacts with PIP3-rich membranes (Ong et al. 2001). By dephosphorylating PIP3, PTEN would effectively prevent Gab1 from translocating to the membrane, thus decreasing the activation of MAPK (fig. 4) (Takahashi-Tezuka et al. 1998; Yart et al. 2001). Second, PTEN has been shown to inhibit insulin stimulation of the MAPK pathway. This inhibition results from the dephosphorylation of the insulin-receptor substrate–1 (IRS-1), which inhibits the formation of the IRS-1/Grb2/Sos complex, a complex that is required for MAPK activation. This indicates that PTEN can play a role in insulin signaling (Weng et al. 2001_c_).
Figure 4.
PTEN as a modulator of the MAPK pathway. PTEN can inhibit the activation of MAPK by several mechanisms. By dephosphorylating Shc and/or IRS-1, PTEN prevents the association of these proteins to the Sos:Grb complex, which is required for MAPK activation. Gab interacts with the membrane by binding to PI3P regions via the pleckstrin-homology domain. By decreasing the PI3P levels in the membrane, PTEN inhibits the translocation of Gab to the membrane and its subsequent activation of the MAPK pathways. The entire pathway for MAPK activation and protein-protein interactions has been omitted for clarity.
That PTEN has a central role in insulin signaling is supported by further evidence: (1) PTEN expression in adipocytes inhibits insulin-stimulated production of 2-deoxyglucose (Nakashima et al. 2000). (2) PTEN expression inhibits Glut4 translocation, which is a key event in insulin signaling (Nakashima et al. 2000); however, this line of evidence may be controversial, since PTEN does not have such an effect in adipose cells (Moser et al. 2001). (3) Insulin-receptor substrate–2 (IRS-2) has recently been shown to increase in the presence of PTEN (Simpson et al., in press). Together, this evidence suggests other roles for PTEN besides the regulation of the PI3K pathway.
An interesting observation—with regard to alternative roles for PTEN besides the regulation of PI3K or MAPK pathway—is that PTEN can be found in the nuclear compartment (Tamura et al. 1998; Gimm et al. 2000_b;_ Lachyankar et al. 2000; Perren et al. 2000; Whiteman et al., in press). PTEN lacks a clear nuclear-localization sequence, and the mechanism of its transport into the nuclear compartment is still being elucidated. Immunohistochemistry analysis demonstrated that PTEN expression in the nuclear compartment is higher in normal tissue than in counterpart neoplastic tissue found in the thyroid, endocrine pancreas, and primary cutaneous melanomas (Gimm et al. 2000_b;_ Perren et al. 2000; Whiteman et al., in press). The significance of this observation is still under investigation. Because PTEN can regulate transcription via the PI3K pathway, which is found predominantly at the plasma membrane (Datta et al. 1999; Dong et al. 1999), does the localization of PTEN to the nuclear compartment sequester PTEN and reduce PI3K-dependent transcription of pro-proliferative factors? Alternatively, nuclear PTEN could act on lipids in the nuclear membrane. Although the level of phosphoinositol in the nuclear membrane is low, it is present, along with PI3K (D'Santos et al. 1998; Marchisio et al. 1998; Martelli et al. 1999; Metjian et al. 1999). Thus, the trafficking of PTEN to the nuclear compartment may activate transcription of pro-apoptotic pathways, as it does at the plasma membrane. Many transcription factors are regulated by their phosphorylation, and a promising alternative hypothesis is that PTEN may regulate the activity of various transcription factors via direct dephosphorylation in the nucleus. Future investigations into the function of nuclear PTEN should prove to be enlightening.
The Present and the Future
What does the future hold for PTEN research? Undoubtedly, we will learn more about the mechanisms by which PTEN regulates the PI3K, MAPK, and insulin-signaling pathways, but it is naive to think that these are the only pathways that PTEN influences. Enough evidence is currently present to surmise that PTEN may play a role in the regulation of a variety of disease states and metabolic processes. In this era of genomics/proteomics, array analysis will certainly yield more clues to, as well as more puzzles about, the roles that PTEN plays. Indeed, the up-regulation of IRS-2 by PTEN was demonstrated by microarray expression analysis (Simpson et al., in press). Unoki and Nakamura have used global expression analysis to demonstrate that EGR2 and BPOZ are involved in PTEN signaling (Unoki and Nakamura 2001). EGR2 is part of a multigene family that contains C2H2-type zinc-finger proteins (Joseph et al. 1998), and germline mutations in this transcription factor gene have been implicated in Charcot-Marie-Tooth syndrome (MIM 118200) and congenital hypomyelinating neuropathy (MIM 605253) (Warner et al. 1998). The association between PTEN and EGR2 is interesting, because prominent PTEN expression has been demonstrated in the CNS and neural crest throughout human development (Gimm et al. 2000_a_) and because a subset of patients with CS have signs of neuropathy (C. Eng, unpublished data). Other expression-array studies have implicated PTEN in the regulation of members of the tumor necrosis factor (TNF)–receptor family, TNF-associated genes, and members of both the Mad and Notch signaling families (Hong et al. 2000; Matsushima-Nishiu et al. 2001). Additional array studies will undoubtedly discover more protean pathways, as well as “red herrings,” downstream from PTEN.
PTEN appears to be constitutively active. Thus, the regulation of PTEN levels via transcription, translation, and posttranslational means remains to be investigated in depth. We now know that phosphorylation of the C-terminal tail of PTEN by CK2 can negatively regulate the stability of PTEN (discussed in the “C-Terminal Domain” subsection of “PTEN Structure,” above). What we do not know is the identity of the phosphatase that dephosphorylates PTEN. Phosphorylation and dephosphorylation are the yin and yang of signal transduction; thus, the phosphatase must be there, yet it remains unidentified. Preliminary data have recently been published on the regulation of PTEN transcription. Activated PPARγ, p53, and EGR1 have been shown to up-regulate PTEN transcription (Patel et al. 2001; Stambolic et al. 2001; Virolle et al. 2001). Factors that may down-regulate transcription have yet to be identified.
Copious amounts of data have demonstrated that PTEN can influence signal transduction via its phosphatase activity. What remains to be determined is if PTEN can influence pathways by other mechanisms. As described above, PTEN contains many signaling domains (e.g., PEST and PDZ). It is interesting to postulate that these domains may also play a role in signal transduction. PTEN has been shown, via yeast two-hybrid studies, to associate with membrane-associated guanylate-kinase inverted (MAGI) proteins, which contain PDZ domains and are localized to tight junctions (Wu X. et al. 2000; Wu Y. et al. 2000). Additional studies suggest that the interaction between PTEN and MAGI may enhance the stability of PTEN (Wu X. et al. 2000). Although this interaction seems to regulate PTEN activity, an appealing proposal is that PTEN may interact with other signaling molecules to modulate specific pathways in a phosphatase-independent manner; however, this needs to be proved.
In this era of genetics, data generated in the laboratory should be translated to the practice of genomic medicine. Already, we are able to use _PTEN-_mutation analysis for predictive testing within _PTEN-_mutation–positive families, and we can use PTEN testing as a molecular-diagnostic tool to sort out a group of difficult-to-diagnose inherited hamartoma-tumor syndromes. Classification of these protean clinical syndromes by molecular identity is believed to be useful in the prediction of the development of neoplasias and the recommendation of surveillance. However, the mechanisms by which identical mutations result in phenotypes as diverse as CS, BRRS, and PS is, as yet, unknown. The challenge for the next decade is to determine the gene-gene interactions and gene-environment interactions that arise in different tissue-specific contexts to result in a specific phenotype.
The brave new world would be incomplete without an attempt to utilize molecular-based knowledge to create a novel treatment. Since overwhelming data demonstrate that the PI3K/AKT pathway lies downstream from PTEN and that mTOR (i.e., mammalian target of rapamycin) is downstream from—or, at least, acts in concert with—the PI3K/AKT pathway, rapamycin and its analogs have been hailed as the “magic bullet” that will cure all the ailments of a sick PTEN (Mills et al. 2001; Neshat et al. 2001). However, the PI3K/AKT and mTOR pathways merely lie downstream from PTEN’s lipid-phosphatase activity. Virtually all enzymatic activity–abrogating mutations affect the lipid- and protein-phosphatase activity. Thus, it would be predicted that drugs that target only downstream from the lipid-phosphatase activity would not be completely effective and might, in fact, cause harm. Nevertheless, this is an auspicious start. The challenge for the future is to have “designer drugs” that target in a manner that takes into account the genomic, epigenomic, and proteomic milieu. To achieve this, not only should efforts be made in genomic, epigenomic, and proteomic research, which is already happening, but these data must also be linked with comprehensive and meticulous clinical and cellular phenotypic data, which we call “phenomics.”
Acknowledgments
C.E. was partially supported by the National Institutes of Health (grant R01HD39058), the American Cancer Society (grant RPG98-211-01), the U.S. Army Breast Cancer Research Program (grant DAMD-00-1-0390), the Susan G. Komen Breast Cancer Research Foundation, and the V Foundation (the Jimmy V Golf Classic Award for Translational Cancer Research).
Appendix A: Common Manifestations of CS
- Mucocutaneous lesions (90%–100%):
- Trichilemmomas
- Acral keratoses
- Verucoid or papillomatous papules
- Thyroid abnormalities (50%–67%):
- Goiter
- Adenoma
- Cancer (3%–10%)
- Breast lesions:
- Fibroadenomas/fibrocystic disease (76% of affected females)
- Adenocarcinoma (25%–50% of affected females)
- Gastrointestinal lesions (40%): hamartomatous polyps
- Macrocephaly (38%)
- Genito-urinary abnormalities (44% of females): uterine leiomyoma (multiple, early onset)
Appendix B: International Cowden Consortium Operational Diagnostic Criteria: 2000 Version
Operational diagnostic criteria are reviewed and revised on a continuous basis, as new clinical and genetic information becomes available. The 1995 and 2000 versions have been accepted by the U.S.-based NCCN High Risk/Genetics Panel.
- Pathognomonic criteria (mucocutaneous lesions):
- Facial trichilemmomas
- Acral keratoses
- Papillomatous papules
- Mucosal lesions
- Major criteria:
- Breast carcinoma
- Thyroid carcinoma (nonmedullary), especially follicular thyroid carcinoma
- Macrocephaly (megalencephaly; e.g., ⩾97th percentile)
- Lhermitte-Duclos disease (LDD)
- Endometrial carcinoma
- Minor criteria:
- Other thyroid lesions (e.g., adenoma or multinodular goiter)
- Mental retardation (e.g., intelligence quotient ⩽75)
- Gastrointestinal hamartomas
- Fibrocystic disease of the breast
- Lipomas
- Fibromas
- Genito-urinary tumors (e.g., renal cell carcinoma or uterine fibroids) or malformation
- Operational diagnosis in an individual:
- 1.
Mucocutaneous lesions alone if there are (a) six or more facial papules, of which three or more must be trichilemmoma; (b) cutaneous facial papules and oral mucosal papillomatosis; (c) oral mucosal papillomatosis and acral keratoses; or (d) six or more palmar or plantar keratoses - 2.
Two major criteria, of which one must be macrocephaly or LDD - 3.
One major and three minor criteria - 4.
Four minor criteria
- 1.
- Operational diagnosis in a family in which one individual has had a diagnosis of CS:
- 1.
One or more of pathognomonic criteria - 2.
Any one major criterion with or without minor criteria - 3.
Two minor criteria
- 1.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Charcot-Marie-Tooth syndrome [MIM 118200], BRRS [MIM 153480], CS [MIM 158350], JPS [MIM 174900], PJS [MIM 175200], PS [MIM 176920], congenital hypomyelinating neuropathy [MIM 605253], and PTEN/MMAC1/TEP1 [MIM 601728])
References
- Ahmed SF, Marsh DJ, Weremowicz S, Morton CC, Williams DM, Eng C (1999) Balanced translocation of 10q and 13q, including the PTEN gene, in a boy with an HCG-secreting tumor and the Bannayan-Riley-Ruvalcaba syndrome. J Clin Endocrinol Metab 84:4665–4670 [DOI] [PubMed] [Google Scholar]
- Arch EM, Goodman BK, van Wesep RA, Liaw D, Clarke K, Parsons R, McKusick VA, Geraghty MT (1997) Deletion of PTEN in a patient with Bannayan-Riley-Ruvalcaba syndrome suggests allelism with Cowden disease. Am J Med Genet 71:489–493 [PubMed] [Google Scholar]
- Barker E, Martinez A, Wang R, Bevan S, Murday V, Shipley J, Houlston R, Harper J (2001) PTEN mutations are uncommon in Proteus syndrome. J Med Genet 38:480–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Happle R, Mulliken JB, Weksberg R, Graham JM, Viljoen DL, Cohen MM (1999) Proteus syndrome: diagnostic criteria, differential diagnosis and patient evaluation. Am J Med Genet 84:389–395 [DOI] [PubMed] [Google Scholar]
- Bonneau D, Longy M (2000) Mutations of the human PTEN gene. Hum Mutat 16:109–122 [DOI] [PubMed] [Google Scholar]
- Bruni P, Boccia A, Baldasarre G, Trapasso F, Santoro M, Chiappetta G, Fusco A, Viglietto G (2000) PTEN expression is reduced in a subset of sporadic thyroid carcinomas: evidence that PTEN-growth suppressing activity in thyroid cancer cells is mediated by p27kip1. Oncogene 19:3146–3155 [DOI] [PubMed] [Google Scholar]
- Carethers JM, Furnari FB, Zigman AF, Lavine JE, Jones MC, Graham GE, Teebi AS, SuHuang HJ, Ha HT, Chauhan DP, Chang CL, Cavanee WK, Boland CR (1998) Absence of PTEN/MMAC1 germline mutations in sporadic Bannayan-Riley-Ruvalcaba syndrome. Cancer Res 58:2724–2726 [PubMed] [Google Scholar]
- Celebi JT, Tsou HC, Chen FF, Zhang H, Ping XL, Lebwohl MG, Kezis J, Peacocke M (1999) Phenotypic findings of Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome in a family associated with a single germline mutation in PTEN. J Med Genet 36:360–364 [PMC free article] [PubMed] [Google Scholar]
- Cheney IW, Neuteboom STC, Vaillancourt M-T, Ramachandra M, Bookstein R (1999) Adenovirus-mediated gene entry of MMAC1/PTEN to glioblastoma inhibits S phase entry by the recruitment of p27Kip1 and cyclin E/CDK2 complexes. Cancer Res 59:2318–2323 [PubMed] [Google Scholar]
- Dahia PLM, Gimm P, Chi H, Marsh DJ, Reynolds PR, Eng C (2000) Absence of germline mutations in MINPP1, a phosphatase-encoding gene centromeric of PTEN, in patients with Cowden and Bannayan-Riley-Ruvalcaba syndrome without germline PTEN mutations. J Med Genet 37:715–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasouki MJ, Ishmael H, Eng C (2001) Macrocephaly, macrosomia and autistic behavior due to a de novo PTEN germline mutation. Am J Hum Genet Suppl 69:S280 [Google Scholar]
- Datta SR, Brunet A, Greenberg ME (1999) Cellular survival: a play in three Akts. Genes Dev 13:2905–2927 [DOI] [PubMed] [Google Scholar]
- Davies MA, Lu Y, Sano T, Fang X, Tang P, LaPuschin R, Koul D, Bookstein R, Stokoe D, Yung WKA, Mills GB, Steck PA (1998) Adenoviral transgene expression of MMAC/PTEN in human glioma cells inhibits Akt activation and induces anoikis. Cancer Res 58:5285–5290 [PubMed] [Google Scholar]
- De Vivo I, Gertig D, Nagase S, Hankinson SE, O'Brien R, Speizer FE, Parsons R, Hunter DJ (2000) Novel germline mutations in the PTEN tumour suppressor gene found in women with multiple cancers. J Med Genet 37:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, DeCock P, Fryns JP (1995) Hydrocephalus with features of VATER. J Genet Couns 6:69 [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP (1998) Pten is essential for embryonic development and tumour suppression. Nat Genet 19:348–355 [DOI] [PubMed] [Google Scholar]
- Dong Z, Huang C, Ma WY (1999) PI-3 kinase in signal transduction, cell transformation, and as a target for chemoprevention of cancer. Anticancer Res 19:3743–3747 [PubMed] [Google Scholar]
- Downward J (1998) Mechanisms and consequences of activation of protein kinase B/Akt. Curr Opin Genet Dev 8:49–549529605 [Google Scholar]
- D'Santos CS, Clarke JH, Divecha N (1998) Phospholipid signalling in the nucleus. Biochim Biophys Acta 1436:201–232 [DOI] [PubMed] [Google Scholar]
- Eng C (1997) Cowden syndrome. J Genet Couns 6:181–191 [DOI] [PubMed] [Google Scholar]
- ——— (1998) Genetics of Cowden syndrome: through the looking glass of oncology. Int J Oncol 12:701–710 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet 37:828–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2001) To be or not to BMP. Nat Genet 28:105–107 [DOI] [PubMed] [Google Scholar]
- Eng C, Brody LC, Wagner TMU, Devilee P, Vijg J, Szabo C, Tavtigian S, Nathanson KL, Ostrander E, Frank TS, Breast Cancer Information Core Consortium (2001) Interpretation of molecular epidemiologic research: blinded comparison of methods for detecting germline BRCA1 mutations. J Med Genet 38:824–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C, Ji H (1998) Molecular classification of the inherited hamartoma polyposis syndromes: clearing the muddied waters. Am J Hum Genet 62:1020–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackenthal J, Marsh DJ, Richardson AL, Cummings SC, Eng C, Robinson BG, Olopade OI (2001) Male breast cancer in Cowden syndrome patients with germline PTEN mutations. J Med Genet 38:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM (1999) Protein modules as organizers of membrane structure. Curr Opin Cell Biol 11:432–439 [DOI] [PubMed] [Google Scholar]
- Georgescu M-M, Kirsch KH, Akagi T, Shishido T, Hanafusa H (1999) The tumor-suppressor activity of PTEN is regulated by its carboxy-terminal region. Proc Natl Acad Sci USA 96:10182–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimm O, Attié-Bitach T, Lees JA, Vekemens M, Eng C (2000_a_) Expression of PTEN protein in human embryonic development. Hum Mol Genet 9:1633–1639 [DOI] [PubMed] [Google Scholar]
- Gimm O, Perren A, Weng LP, Marsh DJ, Yeh JJ, Ziebold U, Gil E, Hinze R, Delbridge L, Lees JA, Robinson BG, Komminoth P, Dralle H, Eng C (2000_b_) Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am J Pathol 156:1693–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Condon LM, Burke BA (1992) Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet 44:307–314 [DOI] [PubMed] [Google Scholar]
- Gu J, Tamura M, Yamada KM (1998) Tumor suppressor PTEN inhibits integrin- and growth factor-mediated mitogen-activated protein (MAP) kinase signaling pathway. J Cell Biol 143:1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S-Y, Kato H, Suzuki T, Shibata H, Ishii S, Shiba K, Matsuno S, Kanamaru R, Ishioka C (2000) Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res 60:3147–3151 [PubMed] [Google Scholar]
- Hanssen AMN, Fryns JP (1995) Cowden syndrome. J Med Genet 32:117–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Aminoff WM, Högland P, Järvinen H, Kristo P, Pelin K, Ridanpää M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA (1998) A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 391:184–187 [DOI] [PubMed] [Google Scholar]
- Hemminki A, Tomlinson I, Markie D, Järvinen H, Sistonen P, Björkqvist A-M, Knuutila S, Salovaara R, Bodmer W, Shibata D, de la Chapelle A, Aaltonen LA (1997) Localisation of a susceptibility locus for Peutz-Jeghers syndrome to 19p using comparative genomic hybridization and targeted linkage analysis. Nat Genet 15:87–90 [DOI] [PubMed] [Google Scholar]
- Hong TM, Yang PC, Peck K, Chen JJ, Yang YC, Chen YC, Wu CW (2000) Profiling the downstream genes of tumor suppressor PTEN in lung cancer cells by complementary DNA microarray. Am J Respir Cell Mol Biol 23:355–363 [DOI] [PubMed] [Google Scholar]
- Howe JR, Blair JA, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B (2001) Germline mutations of BMPR1A in juvenile polyposis. Nat Genet 28:184–187 [DOI] [PubMed] [Google Scholar]
- Howe JR, Ringold JC, Summers RW, Mitros FA, Nishimura DY, Stone EM (1998_a_) A gene for familial juvenile polyposis maps to chromosome 18q21.1. Am J Hum Genet 62:1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IPM, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA (1998_b_) Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280:1086–1088 [DOI] [PubMed] [Google Scholar]
- Huang SC, Chen CR, Lavine JE, Taylor SF, Newbury RO, Pham TT, Ricciardiello L, Carethers JM (2000) Genetic heterogeneity in familial juvenile polyposis. Cancer Res 60:6882–6885 [PubMed] [Google Scholar]
- Iafolla AK, McConkie-Rosell A, Chen YT (1991) VATER and hydrocephalus: distinct syndrome? Am J Med Genet 38:46–51 [DOI] [PubMed] [Google Scholar]
- Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Müller O, Back W, Zimmer M (1998) Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet 18:38–44 [DOI] [PubMed] [Google Scholar]
- Joseph JL, LeBeau MM, Jamieson GA, Acharya S, Shows TB, Rowley JD, Sukhatme VP (1998) Molecular cloning, sequencing, and mapping of EGR2, a human early growth response gene encoding a protein with “zinc-binding finger” structure. Proc Natl Acad Sci USA 85:7164–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller R, Cantley LC (1994) Phosphoinositol 3-kinease. Bioessays 16:565–576 [DOI] [PubMed] [Google Scholar]
- Katso R, Okkenbaug K, Ahmadi K, White S, Timms J, Waterfield MD (2001) Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol 17:615–675 [DOI] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14:231–241 [PubMed] [Google Scholar]
- Kurose K, Araki T, Matsunaka T, Takada Y, Emi M (1999) Variant manifestation of Cowden disease in Japan: hamartomatous polyposis of the digestive tract with mutation of the PTEN gene. Am J Hum Genet 64:308–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachyankar MB, Sultana N, Schonhoff CM, Mitra P, Poluha W, Lambert S, Quesenberry PJ, Litofsky NS, Recht LD, Nabi R, Miller SJ, Ohta S, Neel BG, Ross AH (2000) A role for nuclear PTEN in neuronal differentiation. J Neurosci 20:1404–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yang H, Georgescu M-M, Di Cristafano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99:323–334 [DOI] [PubMed] [Google Scholar]
- Li D-M, Sun H (1997) TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor B. Cancer Res 57:2124–2129 [PubMed] [Google Scholar]
- Li DM, Sun H (1998) PTEN/MMAC1/TEP1 suppresses the tumorigenecity and induces G1 cell cycle arrest in human glioblastoma cells. Proc Natl Acad Sci USA 95:15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittman M, Tycko B, Hibshoosh H, Wigler MH, Parsons R (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast and prostate cancer. Science 275:1943–1947 [DOI] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PLM, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16:64–67 [DOI] [PubMed] [Google Scholar]
- Longy M, Coulon V, Duboué DA, Larrègue M, Eng C, Amati P, Kraimps J-L, Bottani A, Lacombe D, Bonneau D (1998) Mutations of PTEN in patients with Bannayan-Riley-Ruvalcaba phenotype. J Med Genet 35:886–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longy M, Lacombe D (1996) Cowden disease: report of a family and review. Ann Genet 39:35–42 [PubMed] [Google Scholar]
- Lynch ED, Ostermeyer EA, Lee MK, Arena JF, Ji H, Dann J, Swisshelm K, Suchard D, MacLeod PM, Kvinnsland S, Gjertsen BT, Heimdal K, Lubs H, Møller P, King M-C (1997) Inherited mutations in PTEN that are associated with breast cancer, Cowden syndrome, and juvenile polyposis. Am J Hum Genet 61:1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger phosphoinositol 3,4,5-triphosphate. J Biol Chem 273:13375–13378 [DOI] [PubMed] [Google Scholar]
- Mallory SB (1995) Cowden syndrome (multiple hamartoma syndrome). Dermatol Clin 13:27–31 [PubMed] [Google Scholar]
- Marchisio M, Bertagnolo V, Colamussi ML, Capitani S, Neri LM (1998) Phosphotidylinositol 3-kinase in HL-60 nuclei is bound to the nuclear matrix and increases during granulocyte differentiation. Biochem Biophys Res Commun 253:346–351 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Caron S, Dahia PLM, Kum JB, Frayling IM, Tomlinson IPM, Hughes KS, Hodgson SV, Murday VA, Houlston R, Eng C (1998_a_) Germline PTEN mutations in Cowden syndrome-like families. J Med Genet 35:881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PLM, Zheng Z, Liaw D, et al (1998_b_) Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 7:507–515 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Dahia PLM, Zheng Z, Liaw D, Parsons R, Gorlin RJ, Eng C (1997_a_) Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet 16:333–334 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, et al (1999) PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461–1472 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Roth S, Lunetta K, Hemminki A, Dahia PLM, Sistonen P, Zheng Z, et al (1997_b_) Exclusion of PTEN and 10q22-24 as the susceptibility locus for juvenile polyposis syndrome (JPS). Cancer Res 57:5017–5021 [PubMed] [Google Scholar]
- Martelli AM, Capitani S, Neri LM (1999) The generation of lipid signaling molecules in the nucleus. Prog Lipid Res 38:273–308 [DOI] [PubMed] [Google Scholar]
- Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minagucki T, Kuramoto H, Nishida M, Satoh T, Tanaka T, Nakamura Y (2001) Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN. Cancer Res 61:3741–3749 [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Burgering BM (2000) AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782–787 [DOI] [PubMed] [Google Scholar]
- Metjian A, Roll RL, Ma AD, Abrams CS (1999) Agonists cause nuclear translocation of phosphotidylinositol 3-kinase γ: a Gβγ-dependent pathway that requires p110γ amino terminus. J Biol Chem 274:27943–27947 [DOI] [PubMed] [Google Scholar]
- Mills GB, Lu Y, Kohn EC (2001) Linking molecular therapeutics to molecular diagnostics: inhibition of the FRAP/RAFT/TOR component of the PI3K pathway preferentially blocks PTEN mutant cells in vitro and in vivo. Proc Natl Acad Sci USA 98:10031–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet E, Michel G, Mottet D, Raes M, Michiels C (2001) Transduction pathways involved in hypoxia-inducible factor-1 phosphorylation and activation. Free Radic Biol Med 31:847–855 [DOI] [PubMed] [Google Scholar]
- Mosser VA, Li Y, Quon MJ (2001) PTEN does not modulate GLUT4 translocation in rat adipose cells under physiological conditions. Biochem Biophys Res Commun 288:1011–1017 [DOI] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, van der Kaay J, Storalov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK (1998) The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc Natl Acad Sci USA 95:13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Stolarov J, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK (1997) PTEN, the tumor suppressor from human chromosome 10q23, is a dual specificity phosphatase. Proc Natl Acad Sci USA 94:9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Sharma PM, Imamura T, Bookstein R, Olefsky JM (2000) The tumor suppressor PTEN negatively regulates insulin signaling in 3T3-L1 adipocytes. J Biol Chem 275:12889–12895 [DOI] [PubMed] [Google Scholar]
- NCCN Genetics/High Risk Panel (1999) NCCN practice guidelines: genetics/familial high risk cancer. Oncology 13:161–186 [Google Scholar]
- Nelen MR, Kremer H, Konings IBM, Schoute F, van Essen AJ, Koch R, Woods CG, Fryns J-P, Hamel B, Hoefsloot LH, Peeters EAJ, Padberg GW (1999) Novel PTEN mutations in patients with Cowden disease: absence of clear genotype-phenotype correlations. Eur J Hum Genet 7:267–273 [DOI] [PubMed] [Google Scholar]
- Nelen MR, Padberg GW, Peeters EAJ, Lin AY, van den Helm B, Frants RR, Coulon V, Goldstein AM, van Reen MMM, Easton DF, Eeles RA, Hodgson S, Mulvihill JJ, Murday VA, Tucker MA, Mariman ECM, Starink TM, Ponder BAJ, Ropers HH, Kremer H, Longy M, Eng C (1996) Localization of the gene for Cowden disease to 10q22-23. Nat Genet 13:114–116 [DOI] [PubMed] [Google Scholar]
- Nelen MR, van Staveren CG, Peeters EAJ, Ben Hassel M, Gorlin RJ, Hamm H, Lindboe CF, Fryns J-P, Sijmons RH, Woods DG, Mariman ECM, Padberg GW, Kremer H (1997) Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet 6:1383–1387 [DOI] [PubMed] [Google Scholar]
- Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, Frost P, Gibbons JJ, Wu H, Sawyers CL (2001) Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci USA 98:10314–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschwang S, Serova-Sinilnikova OM, Lenoir GM, Thomas G (1998) PTEN germline mutations in juvenile polyposis coli. Nat Genet 18:12–14 [DOI] [PubMed] [Google Scholar]
- Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I (2001) Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc Natl Acad Sci USA 22:6074–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramio JM, Navarro M, Segrelles C, Gomez-Casero E, Jorcano JL (1999) PTEN tumour suppressor is linked to the cell cycle control through the retinoblastoma protein. Oncogene 18:7462–7468 [DOI] [PubMed] [Google Scholar]
- Patel L, Pass I, Coxon P, Downes CP, Smith SA, MacPhee CH (2001) Tumor suppressor and anti-inflammatory actions of PPARγ agonists are mediated via upregulation of PTEN. Curr Biol 11:764–768 [DOI] [PubMed] [Google Scholar]
- Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, Heitz PU, Eng C (2000) Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol 157:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persad S, Troussard AA, McPhee TR, Mulholland DJ, Dedhar S (2001) Tumor suppressor PTEN inhibits nuclear accumulation of β-catenin and T cell/lymphoid enhancer factor 1–mediated transcriptional activation. J Cell Biol 153:1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R (1999) Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA 96:1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Zhou XP, Eng C (2001) A novel germline mutation of the PTEN gene in a patient with macrocephaly, ventricular dilatation and features of VATER association. J Med Genet 38:820–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC (1998) C2 domains, structure and function of a universal C2+-binding domain. J Biol Chem 273:15879–15882 [DOI] [PubMed] [Google Scholar]
- Simpson L, Li J, Liaw D, Hennessey I, Oliner J, Christians F, Parsons R. PTEN expression causes a feedback upregulation of IRS-2 signaling. Mol Cell Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW (2001) Regulation of PTEN transcription by p53. Mol Cell 8:317–325 [DOI] [PubMed] [Google Scholar]
- Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Rulland J, Penninger JM, Siderovski DP, Mak TW (1998) Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95:1–20 [DOI] [PubMed] [Google Scholar]
- Stambolic V, Tsao MS, MacPherson D, Suzuki A, Chapman WB, Mak TW (2000) High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten +/− mice. Cancer Res 60:3605–3611 [PubMed] [Google Scholar]
- Starink TM, van der Veen JPW, Arwert F, de Waal LP, de Lange GG, Gille JJP, Eriksson AW (1986) The Cowden syndrome: a clinical and genetic study in 21 patients. Clin Genet 29:222–233 [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WKA, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DHF, Tavtigian SV (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 15:356–362 [DOI] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, Fukumoto M, Mak TW (1998) High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol 8:1169–1178 [DOI] [PubMed] [Google Scholar]
- Takahashi-Tezuka M, Yoshida Y, Fukada T, Ohtani T, Yamanaka Y, Nishida K, Nakajima K, Hibi M, Hirano T (1998) Gab1 acts as an adapter molecule linking the cytokine receptor gp130 to ERK mitogen-activated protein kinase. Mol Cell Biol 18:4109–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M, Gu J, Danen EHJ, Takino T, Miyamoto S, Yamada KM (1999) PTEN interactions with focal adhesion kinase and suppression of the extracellular matrix-dependent phosphotidyinositol 3-kinase/Akt cell survival pathway. J Biol Chem 274:20693–20703 [DOI] [PubMed] [Google Scholar]
- Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM (1998) Inhibition of cell migration, spreading and focal adhesions by tumor suppressor PTEN. Science 280:1614–1617 [DOI] [PubMed] [Google Scholar]
- Torres J, Pulido R (2001) The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem 276:993–998 [DOI] [PubMed] [Google Scholar]
- Tsou HC, Teng D, Ping XL, Broncolini V, Davis T, Hu R, Xie X-X, Gruener AC, Schrager CA, Christiano AM, Eng C, Steck P, Ott J, Tavtigian SV, Peacocke M (1997) Role of MMAC1 mutations in early onset breast cancer: causative in association with Cowden's syndrome and excluded in _BRCA1-_negative cases. Am J Hum Genet 61:1036–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M, Nakamura Y (2001) Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene 20:4457–4465 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy D, Nakamura N, Sellers WR (2000) Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol 20:5010–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I (2001) The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol 3:1124–1128 [DOI] [PubMed] [Google Scholar]
- Wanner M, Celebi JT, Peacocke M (2001) Identification of a PTEN mutation in a family with Cowden syndrome and Bannayan-Zonana syndrome. J Am Acad Dermatol 44:183–187 [DOI] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR (1998) Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet 18:382–384 [DOI] [PubMed] [Google Scholar]
- Weng LP, Brown JL, Eng C (2001_a_) PTEN coordinates G1 arrest by down regulating cyclin D1 via its protein phosphatase activity and up regulating p27 via its lipid phosphatase activity. Hum Mol Genet 10:599–604 [DOI] [PubMed] [Google Scholar]
- ——— (2001_b_) PTEN induces apoptosis and cell cycle arrest through phosphoinositol-3-kinase/Akt-dependent and independent pathways. Hum Mol Genet 10:237–242 [DOI] [PubMed] [Google Scholar]
- Weng LP, Smith WM, Brown JL, Eng C (2001_c_) PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet 10:605–616 [DOI] [PubMed] [Google Scholar]
- Weng L-P, Smith WM, Dahia PLM, Ziebold U, Gil E, Lees JA, Eng C (1999) PTEN suppresses breast cancer cell growth by phosphatase function-dependent G1 arrest followed by apoptosis. Cancer Res 59:5808–5814 [PubMed] [Google Scholar]
- Whiteman DC, Zhou XP, Cummings MC, Pavey S, Hayward NK, Eng C. PTEN expression and clinico-pathologic features in a population-based series of primary cutaneous melanoma. Int J Cancer (in press) [DOI] [PubMed] [Google Scholar]
- Wu X, Hepner K, Castello-Prabhu S, Do D, Kaye MB, Yuan XJ, Wood J, Ross C, Sawyers CL, Whang YE (2000) Evidence for regulation of PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAG1-2. Proc Natl Acad Sci USA 97:4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Dowbenko D, Spencer S, Laura R, Lee J, Gu Q, Lasky LA (2000) Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J Biol Chem 275:21477–21485 [DOI] [PubMed] [Google Scholar]
- Yart A, Laffargue M, Mayeux P, Chretien S, Peres C, Tonks N, Roche S, Payrastre B, Chap H, Raynal P (2001) A critical role for phosphoinositide 3-kinase upstream of Gab1 and SHP2 in the activation of ras and mitogen-activated protein kinases by epidermal growth factor. J Biol Chem 276:8856–8864 [DOI] [PubMed] [Google Scholar]
- Zhou XP, Hampel H, Thiele H, Gorlin RJ, Hennekam R, Parisi M, Winter RM, Eng C (2001_a_) Association of germline mutation in the PTEN tumour suppressor gene and a subset of Proteus sand Proteus-like syndromes. Lancet 358:210–211 [DOI] [PubMed] [Google Scholar]
- Zhou XP, Marsh DJ, Hampel H, Mulliken JB, Gimm O, Eng C (2000) Germline and germline mosaic mutations associated with a Proteus-like syndrome of hemihypertrophy, lower limb asymmetry, arterio-venous malformations and lipomatosis. Hum Mol Genet 9:765–768 [DOI] [PubMed] [Google Scholar]
- Zhou XP, Woodford-Richens K, Lehtonen R, Kurose K, Aldred M, Hampel H, Launonen V, et al (2001_b_) Germline mutations in BMPR1A/ALK3 cause a subset of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am J Hum Genet 69:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zori RT, Marsh DJ, Graham GE, Marliss EB, Eng C (1998) Germline PTEN mutation in a family with Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet 80:399–402 [PubMed] [Google Scholar]