Recurrent Mutation of the Gene Encoding sequestosome 1 (SQSTM1/p62) in Paget Disease of Bone (original) (raw)
Abstract
Paget disease of bone (PDB) is a common disorder characterized by focal and disorganized increases of bone turnover. Genetic factors are important in the pathogenesis of PDB. We and others recently mapped the third locus associated with the disorder, PDB3, at 5q35-qter. In the present study, by use of 24 French Canadian families and 112 unrelated subjects with PDB, the PDB3 locus was confined to ∼300 kb. Within this interval, two disease-related haplotype signatures were observed in 11 families and 18 unrelated patients. This region encoded the ubiquitin-binding protein sequestosome 1 (SQSTM1/p62), which is a candidate gene for PDB because of its association with the NF-κB pathway. Screening SQSTM1/p62 for mutations led to the identification of a recurrent nonconservative change (P392L) flanking the ubiquitin-associated domain (UBA) (position 394–440) of the protein that was not present in 291 control individuals. Our data demonstrate that two independent mutational events at the same position in SQSTM1/p62 caused PDB in a high proportion of French Canadian patients.
Paget disease of bone (PDB [MIM 167250]) is a chronic disease of the skeleton that affects up to 2%–3% of the population aged >40 years (Siris and Canfield 1990; Klein and Norman 1995). The disorder is characterized by focal areas of increased and disorganized bone turnover, leading to bone pain, deformity, pathological fracture, neurological complications, and an increased risk of osteosarcoma (Hamdy 1995; Kanis 1998).
Genetic factors play an important role in the pathogenesis of PDB. The disease often segregates as an autosomal dominant trait manifesting genetic heterogeneity and incomplete penetrance (Haslam et al. 1998; Nance et al. 2000; Good et al. 2001; Hocking et al. 2001; Laurin et al. 2001; Leach et al. 2001). The disorder was initially linked to susceptibility loci at chromosomes 6p21.3 (PDB1) and 18q21-22 (PDB2) (Fotino et al. 1977; Tilyard et al. 1982; Cody et al. 1997; Haslam et al. 1998). PDB2 also contains the gene responsible for familial expansile osteolysis (FEO [MIM174810]) (Hughes et al. 1994), a rare bone dysplasia with some similarities to PDB. However, the involvement of PDB1 and PDB2 may be restricted to only a few families, since linkage analyses excluded these two loci in the vast majority of the pagetic kindreds subsequently studied (Breanndan Moore and Hoffman 1988; Hocking et al. 2000; Nance et al. 2000; Good et al. 2001; Laurin et al. 2001).
The primary cellular defect of PDB seems to reside in the osteoclasts. Within the pagetic lesion, osteoclasts are large, multinucleated, and overactive and contain paramyxovirus-like inclusions (Rebel et al. 1974; Mills and Singer 1976; Howatson and Fornasier 1982). Osteoclast formation and activation involve the receptor activator of NF-κB (RANK) pathway, a tumor-necrosis factor (TNF) family member (Hsu et al. 1999). Activating mutations of the TNFRSF11A gene encoding this receptor have been found to be responsible for FEO and rare cases of early-onset familial PDB (Hughes et al. 2000). Targeted disruption of several components of this signaling pathway in the mouse, including RANK, RANK ligand, TRAF6, and NF-κB1/NF-κB2, affects osteoclast differentiation and/or activity leading to osteopetrosis (reviewed by McLean and Olsen [2001]).
Exploiting a genomewide-scan approach and large French Canadian families, we recently mapped two new PDB-causing genes, at 5q35-qter (PDB3) and 5q31 (PDB4) (Laurin et al. 2001). Using a different set of families, Hocking et al. also described a candidate locus at chromosome 5q35 (Hocking et al. 2001). We report here the confinement of the PDB3 locus to ∼300 kb and the identification of a recurrent mutation in SQSTM1/p62, a gene also involved in the NF-κB signaling pathway.
Blood samples were obtained from 479 family members, 112 sporadic patients, and 205 individuals from the general population (108 men and 97 women, mean age 62 years, residing within a 50-km radius of Quebec City, contacted at random, and almost all French Canadian). All affected individuals displayed the typical phenotype for adult-onset PDB, with characteristic radiographic and scintigraphic abnormalities. A detailed clinical description of the families and clinical procedures has been presented elsewhere (Laurin et al. 2001). A full set of pedigree drawings for families included in the present study is also available at the CHUL Research Center Web site.
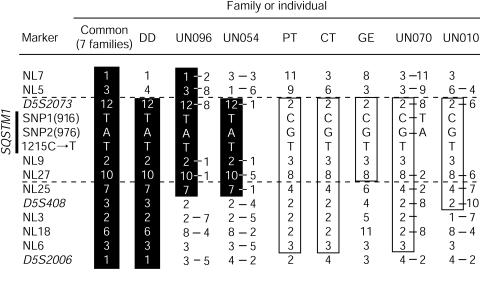
We previously reported that one founder disease haplotype cosegregated with Paget disease in eight families showing linkage at PDB3. One ancestral recombination event in one of these families restricted the disease interval to within 6 cM. To further refine this interval, we developed eight new microsatellite markers, using the Human Genome Working Draft sequence, and performed saturation mapping. We searched for (CA)n and (TG)n repeats on sequences of BAC clones included within the contig covering the 6-cM interval. Flanking primers were designed by use of Primer 3 software (Primer 3 Software Distribution Web site). The more-informative markers were retained. The sequences of these primers are available at the CHUL Research Center Web site. Purified PCR products were pooled and genotyped on a 377 ABI sequencer. Data were analyzed with GENOTYPER 2.5 (ABI). Haplotypes were constructed by use of SIMWALK 2.8 (Sobel and Lange 1996). This analysis strengthened the inference of a common founder haplotype in the eight families originally linked at PDB3. This haplotype was named PDB3H1 (fig. 1, alleles within black vertical bars). These new markers allowed us, by means of the DD family, to position the disease interval between NL5 and the telomere (fig. 1).
Figure 1.
Haplotype analysis in French Canadian families with PDB (common, DD, PT, CT, and GE families) and allele combinations of informative sporadic cases (UN096, UN054, UN070, and UN010). Markers are indicated on the left, with the 1215C→T mutation and two informative SNPs at position 916 and 976 in the coding region of SQSTM1/p62. The marker order is based on haplotype analysis of 24 families (379 individuals), allowing the lower level of recombination. Shared haplotypes are represented by vertical black bars (PDB3H1) or are boxed (PDB3H2). Because haplotypes of sporadic cases could not be determined, both alleles for heterozygotic markers are depicted.
We then exploited a haplotype signature strategy to find ancestral haplotypes shared by affected individuals. The French Canadian population, which frequently shows founder effects, is particularly well suited for such a procedure (Scriver 2001). Genotyping of 112 unrelated patients revealed that 8 of them harbored part of the PDB3H1 haplotype for several contiguous markers. A second signature was also detected in 10 patients with sporadic PDB, and its counterpart haplotype was observed in 10 affected members of three additional kindreds. This haplotype was named “PDB3H2” (fig. 1, boxed alleles). In one of these three families, three of eight patients did not share any of the disease-related haplotypes, suggesting the presence either of a second disease gene, located at a different PDB locus, or of nongenetic (environmental) phenocopies (see pedigree CT at the CHUL Research Center Web site). As is depicted in figure 1, four patients with sporadic PDB who carried short portions of either of the two haplotype signatures and the pedigree GE haplotype allowed us to confine the PDB3 locus within an interval of ∼300 kb, between markers NL5 and NL25.
We searched the Unigene and the LocusLink databases, from the National Center for Biotechnology Information (NCBI) server, for genes and ESTs mapping within the PDB3 locus. Particular interest was put on those genes mapping within the reduced interval (fig. 2A, B, and C). Two of these were sequestosome 1 (SQSTM1/p62), which mediates intracellular signaling through the IL-1/TNF pathways towards NF-κB (Sanz et al. 1999, 2000) and MAPK9, a potential component of the RANK signaling pathway. On the basis of the association between these pathways and osteoclast differentiation/activation, we thoroughly screened these two genes for mutations. We did not find any sequence variation within the MAPK9 gene.
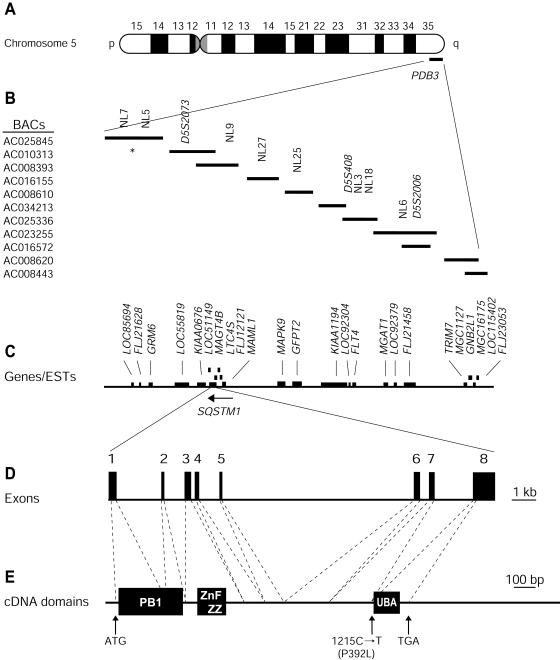
Figure 2.
Physical map of the PDB3 locus at 5q35-qter and genomic organization of SQSTM1/p62. We retrieved a contig of candidate BACs, overlapping the PDB interval from the Human Genome Project Working Draft (Golden Path). We localized Généthon microsatellite markers (Dib et al. 1996), present within this interval, using an ePCR approach (Schuler 1998). Genotyping additional markers using the same approach confirmed order of the BAC clones. A, Chromosome 5. B, BAC contig of the PDB3 linkage region and localization of microsatellite markers. Eight new markers were developed for this study; NL5 and NL25 defined the 300-kb PDB critical interval. The asterisk (*) indicates the NL7–NL5 subcontig being in a different order relative to the Golden Path (August 2001 release). C, Genes present within the PDB3 linkage region. D, Genomic structure of SQSTM1/p62. Exons are represented as boxes and intronic sequences as thin bars. E, cDNA and mutation localization. The segments encoding the Phox and Bem1p domain (PB1), zinc finger ZZ, and UBA domains (Schultz et al. 1998; SMART Web site) are shown by black boxes.
The 2,870-nt SQSTM1/p62 transcript is contained within eight exons distributed over a 16-kb genomic segment (fig. 2_D_). Exon 1 encodes the methionine start codon at position 41, whereas exon 8 encompasses the stop codon at position 1363 and the entire 3′ UTR (fig. 2_D_ and 2_E_). We initially tested five affected members of the DD and TH families, who carried the PDB3H1 haplotype, as well as two unaffected individuals >54 years old that we investigated. One of these five symptomatic individuals, TH-143, was homozygous for PDB3H1 (Laurin et al. 2001). This homozygote was a member of a severely affected nucleus, and his phenotype was similar to that of his heterozygotic sibs. We designed primer pairs for amplification of the eight coding exons of SQSTM1/p62 and at least 30 nt of each of the neighboring intronic sequences. Amplification products were purified and sequenced on a 3700 ABI sequencer using the Big Dye Deoxy Terminator Cycle Sequencing kit (ABI). Both strands were analyzed using STADEN package 1.1 (Staden 1996).
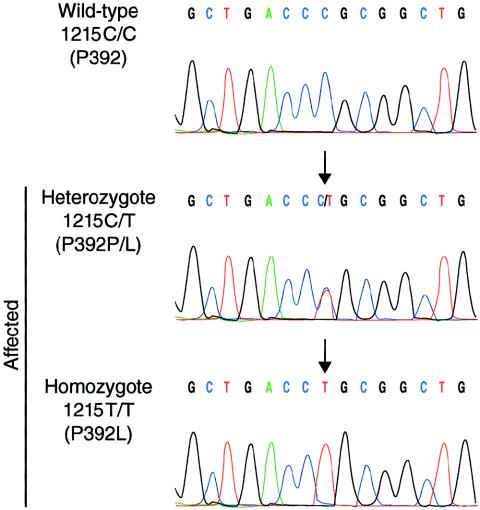
This screening revealed a C→T transversion at position 1215 in SQSTM1/p62 exon 8 in all five affected individuals (fig. 3). This change caused the substitution of proline 392 to a leucine (P392L). Sequencing of exon 8 in the eight families sharing the PDB3H1 disease haplotype demonstrated that the P392L variant was present in all affected individuals carrying the disease haplotype and therefore cosegregated with PDB. Genomic sequence analysis of exon 8 in 86 nonaffected spouses and 205 individuals from the general population revealed no sequence variations at this position, indicating that this sequence change was unlikely to be a single-nucleotide polymorphism.
Figure 3.
SQSTM1/p62 mutation P392L. Sequence electrophoregram of a wild-type unaffected subject (top panel) and heterozygous or homozygous patients (middle and lower panels). Nucleotide and predicted amino acid changes are indicated on the left. The arrows point to the mutation. The nucleotide sequences of primers annealing to exon 8–flanking intronic DNA used for amplification and sequencing of exon 8 were as follows: forward, 5′-CACTGTGGCCTGTGAGGAC-3′, and reverse, 5′-CAGTGAGCCTTGGGTCTCG-3′. A full description of the primers used for amplification and sequencing is available at the CHUL Research Center Web site.
To further test this association between SQSTM1/p62 and PDB, we screened for mutations among 112 sporadic cases and the remaining 16 families. The same 1215C→T substitution was detected in 10 affected individuals from three additional families and in 18 patients with sporadic PDB, all of them harboring one of the two haplotypic signatures described above. The SQSTM1/p62 P392L mutation was thus found in 18 (16%) of the 112 sporadic cases and in 11 (46%) of the 24 families tested. We did not find additional disease-causing mutations. We did, however, identify five SNPs: 380C→T (A117V), in exon 3; 862G→C (Q274E), in exon 6; 916C→T (E292E), in exon 6; 976G→A (R312R), in exon 6; and 994C→A (S318S), in exon 6. Frequencies of these five SNPs were similar between affected and unaffected subjects. None of these SNPs had alleles associated with the mutation. However, as is depicted in fig. 1, two of these SNPs, 916C→T and 928G→A, showed distinct alleles cosegregating with the PDB3H1 and PDB3H2 disease haplotypes. These two SNPs were therefore used as proximal markers to further dissociate the two disease haplotypes. For instance, the PDB3H1 haplotype harbored a T and an A at positions 916 and 976, respectively, whereas the PDB3H2 signature displayed a C and a G at these two positions.
The 1215C→T transversion in exon 8 resulted in the substitution of a cyclic amino acid proline residue at position 392 for a leucine. This residue was conserved in the mouse and rat homologues, Osi (oxydative stress induced) (GenBank accession number NM_011018) and Zip (PKC-zeta-interacting protein) (GenBank accession number Y08355), respectively. The P392 residue being located within the C-terminal end of the protein flanking the ubiquitin associated domain (UBA) (Hofmann and Bucher 1996; Schultz et al. 1998; Wilkinson et al. 2001) at position 394–440 could thus be of particular importance for the conformation and/or function of this region.
Cosegregation of the P392L variant with the phenotype, its absence in 86 spouses and 205 individuals from the general population, the nonconservative nature of the amino acid substitution, and the conservation of the UBA domain, including the P392 residue, among other species (mouse and rat) provided support for the idea that this amino acid change was causing PDB. Except for homozygote TH-143 (fig. 3), pagetic individuals carrying the P392L variant were all heterozygotes, confirming the dominant mode of action of this mutation in a high proportion of our French Canadian patients. As 13 of the 24 families investigated did not harbor any mutation in the SQSTM1/p62 coding sequence, it is likely that other disease gene(s) are also associated with PDB, in agreement with the well-established genetic heterogeneity of the disorder (Haslam et al. 1998; Hocking et al. 2001; Laurin et al. 2001; Good et al. 2002). Indeed, we recently mapped 2 of these 13 families to a fourth locus for the disorder, PDB4, at 5q31 (Laurin et al. 2001), and there is evidence for three other loci associated with the disorder: PDB5 at 2q36, PDB6 at 10p13, and a locus at 18q23 (Hocking et al. 2001; Good et al. 2002).
Haplotype analysis by means of intragenic SNPs was useful in determination of the origin of the mutation and the genetic relatedness of the patients carrying it. Indeed, we found that the P392L mutation, associated with two distinct haplotypes, probably originated from two independent events. Since two different intragenic SNPs haplotypes were identified, our data do not support a very ancient common founder for this mutation. This mutation, occurring at a hypermutable CpG dinucleotide sequence (Green et al. 1990; Koeberl et al. 1990), might have arisen by deamination of a methylated cytosine. For other disorders, several authors already suggested that such mutational hotspots may explain recurrent mutations (Glaser et al. 1999; Rizzo et al. 1999; Aksentijevich et al. 2001; Lund et al. 2001). Additional mutational events may also be found in SQSTM1/p62, since one other study showed linkage between pagetic families of different ethnic origins and markers at PDB3 (Hocking et al. 2001).
It has been proposed that SQSTM1/p62 selectively interacts with TRAF6 and is thus an important intermediary in interleukin-1 (IL-1) and TNFα signaling toward NF-κB activation (Sanz et al. 1999, 2000). TRAF6 interacts with RANK, a member of the TNF receptor family. Its binding is essential for RANK-induced NF-κB activation (Darnay et al. 1999), and TRAF6-deficient mice exhibit severe osteopetrosis and are defective in osteoclast formation because of impaired signaling induced by RANK (Naito et al. 1999). The functional importance of SQSTM1/p62 in NF-κB activation has been further highlighted by the observation that its depletion severely abrogated NF-κB activation through the TNFα or IL-1 pathways (Sanz et al. 1999, 2000). The precise role of SQSTM1/p62 remains to be elucidated, but our data suggest that SQSTM1/p62 may be involved in signaling pathways that control osteoclast activity, differentiation or survival. Further studies in both transgenic animals and cell-culture systems are required to understand the cell biology of this disease and the effect of the SQSTM1/p62 P392L mutation.
The present study emphasized the importance of heredity in the pathogenesis of adult-onset PDB. Indeed, the SQSTM1/p62 P392L mutation was causing, respectively, 16% and 46% of sporadic and familial cases of the disorder in the population tested. As two distinct disease-related haplotypes, encompassing two intragenic SNPs, shared the same SQSTM1/p62 P392L mutation generated by two independent events, our data suggest that this recurrent variation may be frequently associated with PDB in other white populations.
Acknowledgments
The authors would like to thank all the families and patients who participated in this study. The authors also thank Claire Brousseau and Marc Gendreau, for their outstanding support in clinical data collection and analysis, and Evelyne Lejeune and her team of research nurses, for their excellent logistical support. Acknowledgments also go to Annie Duchesne and Marc-André Rodrique, for their expert technical assistance in the sequencing and genotyping laboratory. N.L. was a recipient of a studentship from the Fonds pour la Formation des Chercheurs et l’Aide à la Recherche (FCAR) and the Fonds de la Recherche en Santé du Québec (FRSQ). V.R. was an FRSQ National Investigator. Funding to initiate this work was provided by a John G. Haddad Jr. Award granted by the Paget Foundation for Paget’s Disease of Bone and Related Disorders. Subsequent funding was provided by Canadian Institutes for Health Research grant MOP-3804 and Canadian Foundation for Innovation grant 548.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CHUL Research Center Web site, http://www.crchul.ulaval.ca/public/articles/Laurin2002a.htm (for pedigree drawings of families included in this study and for sequences of primers)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC clone RP11-281O15 [accession number AC025845], CTB-77M18 [accession number AC010313], CTC-241N9, [accession number AC008393], CTD-2376K3, [accession number AC016155], RP11-101O23 [accession number AC008610], CTC-573N18 [accession number AC 034213], RP11-17A5 [accession number AC025255], CTB-22L19 [accession number AC016572], CTB-14A14 [accession number AC008620], and CTC-338M12 [accession number AC008443], for human SQSTM1/p62 cDNA [accession number NM_003900], and mouse homologue Osi [accession number NM_011018], rat homologue ZIP [accession number Y08355])
- Généthon, http://www.genethon.fr (for the reference genetic map)
- Human Genome Project Working Draft at UCSC, http://genome.ucsc.edu/ (August 2001 release)
- NCBI Human Genome Resources, http://www.ncbi.nlm.nih.gov/genome/guide/human/ (for annotated sequences and identification of candidate genes)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PDB [MIM 167250], PDB1 [MIM 167250], PDB2 [MIM 602080], PDB3 [MIM 606262], PDB4 [MIM 606263], FEO [MIM 174810], and TNFRSF11A [MIM 603499])
- Primer 3 Software Distribution Web site, http://www-genome.wi.mit.edu/genome_software/other/primer3.html (for Primer 3 software)
- SMART, http://smart.embl-heidelberg.de/ (for prediction of PB1, ZZ, and UBA domains)
References
- Aksentijevich I, Galon J, Soares M, Mansfield E, Hull K, Oh HH, Goldbach-Mansky R, Dean J, Athreya B, Reginato AJ, Henrickson M, Pons-Estel B, O'Shea JJ, Kastner DL (2001) The tumor-necrosis-factor receptor-associated periodic syndrome: new mutations in TNFRSF1A, ancestral origins, genotype-phenotype studies, and evidence for further genetic heterogeneity of periodic fevers. Am J Hum Genet 69:301–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breanndan Moore S, Hoffman DL (1988) Absence of HLA linkage in a family with osteitis deformans (Paget's disease of bone). Tissue Antigens 31:69–70 [DOI] [PubMed] [Google Scholar]
- Cody JD, Singer FR, Roodman GD, Otterund B, Lewis TB, Leppert M, Leach RJ (1997) Genetic linkage of Paget disease of the bone to chromosome 18q. Am J Hum Genet 61:1117–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnay BG, Ni J, Moore PA, Aggarwal BB (1999) Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-kappaB-inducing kinase: identification of a novel TRAF6 interaction motif. J Biol Chem 274:7724–7731 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Fotino M, Haymovits A, Falk CT (1977) Evidence for linkage between HLA and Paget's disease. Transplant Proc 9:1867–1868 [PubMed] [Google Scholar]
- Glaser B, Furth J, Stanley CA, Baker L, Thornton PS, Landau H, Permutt MA (1999) Intragenic single nucleotide polymorphism haplotype analysis of SUR1 mutations in familial hyperinsulinism. Hum Mutat 14:23–29 [DOI] [PubMed] [Google Scholar]
- Good D, Busfield F, Duffy D, Lovelock PK, Kesting JB, Cameron DP, Shaw JT (2001) Familial Paget's disease of bone: nonlinkage to the PDB1 and PDB2 loci on chromosomes 6p and 18q in a large pedigree. J Bone Miner Res 16:33–38 [DOI] [PubMed] [Google Scholar]
- Good DA, Busfield F, Fletcher BH, Duffy DL, Kesting JB, Andersen J, Shaw JTE (2002) Linkage of Paget disease of bone to a novel region on human chromosome 18q23. Am J Hum Genet 70:517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PM, Montandon AJ, Bentley DR, Ljung R, Nilsson IM, Giannelli F (1990) The incidence and distribution of CpG–TpG transitions in the coagulation factor IX gene: a fresh look at CpG mutational hotspots. Nucleic Acids Res 18:3227–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy RC (1995) Clinical features and pharmacologic treatment of Paget's disease. Endocrinol Metab Clin North Am 24:421–436 [PubMed] [Google Scholar]
- Haslam SI, Van Hul W, Morales-Piga A, Balemans W, San-Millan JL, Nakatsuka K, Willems P, Haites NE, Ralston SH (1998) Paget's disease of bone: evidence for a susceptibility locus on chromosome 18q and for genetic heterogeneity. J Bone Miner Res 13:911–917 [DOI] [PubMed] [Google Scholar]
- Hocking L, Slee F, Haslam SI, Cundy T, Nicholson G, van Hul W, Ralston SH (2000) Familial Paget's disease of bone: patterns of inheritance and frequency of linkage to chromosome 18q. Bone 26:577–580 [DOI] [PubMed] [Google Scholar]
- Hocking LJ, Herbert CA, Nicholls RK, Williams F, Bennett ST, Cundy T, Nicholson GC, Wuyts W, Van Hul W, Ralston SH (2001) Genomewide search in familial Paget disease of bone shows evidence of genetic heterogeneity with candidate loci on chromosomes 2q36, 10p13, and 5q35. Am J Hum Genet 69:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci 21:172–3 [PubMed] [Google Scholar]
- Howatson AF, Fornasier VL (1982) Microfilaments associated with Paget's disease of bone: comparison with nucleocapsids of measles virus and respiratory syncytial virus. Intervirology 18:150–159 [DOI] [PubMed] [Google Scholar]
- Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ (1999) Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci USA 96:3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Ralston SH, Marken J, Bell C, MacPherson H, Wallace RG, Van Hul W, Whyte MP, Nakatsuka K, Hovy L, Anderson DM (2000) Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat Genet 24:45–48 [DOI] [PubMed] [Google Scholar]
- Hughes AE, Shearman AM, Weber JL, Barr RJ, Wallace RG, Osterberg PH, Nevin NC, Mollan RA (1994) Genetic linkage of familial expansile osteolysis to chromosome 18q. Hum Mol Genet 3:359–361 [DOI] [PubMed] [Google Scholar]
- Kanis JA (1998) Pathophysiology and treatment of Paget's disease of bone, 2d ed. Martin Dunitz, London [Google Scholar]
- Klein RM, Norman A (1995) Diagnostic procedures for Paget's disease: radiologic, pathologic, and laboratory testing. Endocrinol Metab Clin North Am 24:437–450 [PubMed] [Google Scholar]
- Koeberl DD, Bottema CD, Ketterling RP, Bridge PJ, Lillicrap DP, Sommer SS (1990) Mutations causing hemophilia B: direct estimate of the underlying rates of spontaneous germ-line transitions, transversions, and deletions in a human gene. Am J Hum Genet 47:202–217 [PMC free article] [PubMed] [Google Scholar]
- Laurin N, Brown JP, Lemainque A, Duchesne A, Huot D, Lacourciere Y, Drapeau G, Verreault J, Raymond V, Morissette J (2001) Paget disease of bone: mapping of two loci at 5q35-qter and 5q31. Am J Hum Genet 69:528–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach RJ, Singer FR, Roodman GD (2001) The genetics of Paget's disease of the bone. J Clin Endocrinol Metab 86:24–28 [DOI] [PubMed] [Google Scholar]
- Lund A, Udd B, Juvonen V, Andersen PM, Cederquist K, Davis M, Gellera C, Kolmel C, Ronnevi LO, Sperfeld AD, Sorensen SA, Tranebjaerg L, Van Maldergem L, Watanabe M, Weber M, Yeung L, Savontaus ML (2001) Multiple founder effects in spinal and bulbar muscular atrophy (SBMA, Kennedy disease) around the world. Eur J Hum Genet 9:431–436 [DOI] [PubMed] [Google Scholar]
- McLean W, Olsen BR (2001) Mouse models of abnormal skeletal development and homeostasis. Trends Genet 17:S38–S43 [DOI] [PubMed] [Google Scholar]
- Mills BG, Singer FR (1976) Nuclear inclusions in Paget's disease of bone. Science 194:201–202 [DOI] [PubMed] [Google Scholar]
- Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, Nakao K, Nakamura K, Katsuki M, Yamamoto T, Inoue J (1999) Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells 4:353–362 [DOI] [PubMed] [Google Scholar]
- Nance MA, Nuttall FQ, Econs MJ, Lyles KW, Viles KD, Vance JM, Pericak-Vance MA, Speer MC (2000) Heterogeneity in Paget disease of the bone. Am J Med Genet 92:303–307 [DOI] [PubMed] [Google Scholar]
- Rebel A, Malkani K, Basle M, Bregeon C, Patezour A, Filmon R (1974) Ultrastructural characteristics of osteoclasts in Paget's disease. Rev Rhum Mal Osteoartic 41:767–771 [PubMed] [Google Scholar]
- Rizzo WB, Carney G, Lin Z (1999) The molecular basis of Sjögren-Larsson syndrome: mutation analysis of the fatty aldehyde dehydrogenase gene. Am J Hum Genet 65:1547–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J (2000) The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J 19:1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J (1999) The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J 18:3044–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD (1998) Electronic PCR: bridging the gap between genome mapping and genome sequencing. Trends Biotechnol 16:456–459 [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver CR (2001) Human genetics: lessons from Quebec populations. Annu Rev Genomics Hum Genet 2:69–101 [DOI] [PubMed] [Google Scholar]
- Siris ES, Canfield RE (1990) Paget's disease of bone. In: Becker KL (ed) Principles and practice of endocrinology and metabolism. JB Lippincott, Philadelphia, pp 504–512 [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5:233–241 [DOI] [PubMed] [Google Scholar]
- Tilyard MW, Gardner RJ, Milligan L, Cleary TA, Stewart RD (1982) A probable linkage between familial Paget's disease and the HLA loci. Aust N Z J Med 12:498–500 [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol 3:939–943 [DOI] [PubMed] [Google Scholar]