Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 8.
Summary
The spindle checkpoint is the prime cell cycle control mechanism that ensures sister chromatids are bi-oriented before anaphase takes place. Aurora B kinase, the catalytic subunit of the chromosome passenger complex, both destabilises kinetochore attachments that do not generate tension and simultaneously maintains the spindle checkpoint signal. However, it is unclear how the checkpoint is silenced following chromosome bi-orientation. We demonstrate that association of type 1 phosphatase (PP1Dis2) to both the N-terminus of Spc7 and the non-motor domains of the Klp5-Klp6 (Kinesin-8) complex are necessary to counteract Aurora B kinase to efficiently silence the spindle checkpoint. The role of Klp5 and Klp6 in checkpoint silencing is specific to this class of kinesin and independent of their motor activities. These data demonstrate that at least two distinct pools of PP1, one kinetochore associated and the other motor associated, are needed to silence the spindle checkpoint.
Introduction
The accurate and equal segregation of genetic material to daughter cells is ensured by the attachment of sister chromatids to dynamic spindle microtubules from opposite spindle poles, a process known as chromosome bi-orientation. Microtubules associate to a specialised multi-protein structure built on the centromeric DNA of each chromosome, known as the kinetochore. The establishment of chromosome bi-orientation requires that only tension bearing kinetochoremicrotubule connections are stabilised, whereas those that do not generate tension are destabilised. A recent model explains how this is achieved (Lampson and Cheeseman, 2010). The interaction of kinetochores with microtubules is governed by dynamic changes in the phosphorylation of key proteins at the microtubule-kinetochore interface. Phosphorylation of these proteins is largely catalysed by Aurora B kinase, a component of the chromosome passenger complex, which resides at the inner centromere region during early mitosis (Ruchaud et al., 2007). Microtubule connections that do not generate tension fail to separate Aurora B kinase from proteins at the outer kinetochore. This promotes phosphorylation of several components of the KMN complex, including Ndc80, KNL-1 and Dsn1, which inhibits microtubule binding to the kinetochore (Andrews et al., 2004; Welburn et al., 2010). When tension is applied across the sister chromatids (centromere stretch) the outer kinetochore is pulled away from the inner centromere, thereby separating Aurora B kinase from its substrates. The application of tension also promotes interaction of type 1 phosphatase, PP1, with the N-terminus of KNL-1 which reverses Aurora kinase-dependent phosphorylation of proteins at the outer kinetochore to stabilise tension bearing microtubule-kinetochore attachments (Liu et al., 2010).
The kinetochore not only provides the mechanical linkage between microtubules and chromosomes but also acts as a signal transducer to link the movement of chromosomes to cell cycle progression (Cheeseman and Desai, 2008). In particular, the kinetochore provides the platform for the action of a surveillance system, known as the spindle assembly checkpoint (SAC), which ensures sister chromatids do not separate until all chromosomes are correctly bi-oriented (Musacchio and Salmon, 2007). Components of the checkpoint include the Mad1, Mad2, BubR1(Mad3) and Bub3 proteins and the Bub1, Mps1(Mph1) and Aurora B kinases. The checkpoint is activated when individual kinetochores are not bound to spindle microtubules or not under tension. Upon activation at the kinetochore Mad2 and Mad3 bind to and inhibit Cdc20, an essential activator of an E3 ubiquitin ligase known as the anaphase-promoting complex/cyclosome (APC/C). When the checkpoint is satisfied the spindle checkpoint signal is silenced and the Cdc20-APC/C is activated. This triggers the polyubiquitination of securin and cyclin, which allows the dissolution of sister chromatid cohesion and mitotic progression. However, the molecular mechanism(s) by which the spindle checkpoint is switched off following chromosome bi-orientation are not well understood. Two recent studies indicate that intrakinetochore stretch, rather than centromere (inter-kinetochore) stretch, is necessary and sufficient to satisfy the spindle checkpoint (Maresca and Salmon, 2009; Uchida et al., 2009).
A number of spindle checkpoint silencing mechanisms have been proposed in mammalian cells. These include dynein-dependent stripping of checkpoint proteins, particularly Mad2, from the kinetochore. Dynein is recruited to the kinetochore by Spindly (Gassmann et al., 2010). Others have suggested that p31comet, a conformation specific inhibitor of Mad2, helps to silence the spindle checkpoint, but it is unclear how this pathway is controlled (Xia et al., 2004; Yang et al., 2007). Notably, these silencing mechanisms are unlikely to be evolutionarily conserved as fungi lack homologues of Spindly and p31comet, and dynein is not essential for spindle checkpoint inactivation in yeast (Courtheoux et al., 2007). By contrast, two recent reports indicate that, in budding and fission yeast, PP1 opposes Aurora B kinase in maintaining the spindle checkpoint (Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009). In this paper we examine whether association of PP1 to Spc7, the fission yeast homologue of KNL-1, controls spindle checkpoint signaling and whether other PP1 targeting factors are required.
Results
Association of PP1Dis2 to fission yeast kinetochores is tension dependent
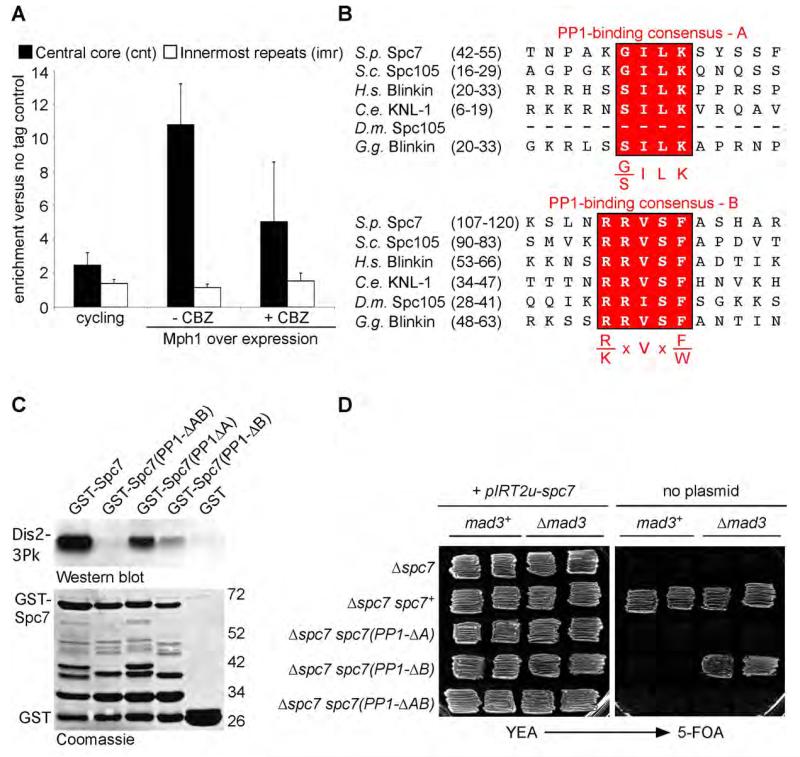
Fission yeast contains two PP1 enzymes, Dis2 and Sds21, which share an essential overlapping function in mitosis (Ohkura et al., 1989). PP1Dis2 binds the central core region of centromeres, whereas PP1Sds21 only does so in the absence of PP1Dis2 (Alvarez-Tabares et al., 2007). To examine whether interaction of PP1Dis2 with the fission yeast kinetochore is cell cycle regulated, we performed chromatin immunoprecipitation analysis in both log phase and cell cycle arrested nmt41-mph1 cells. Under repressive conditions cells cycle normally but when the nmt41 promoter is de-repressed cells arrest in mitosis, due to ectopic activation of the spindle checkpoint, but with short spindles and separated centromeres, indicating kinetochores are under tension (Karen May and KGH, unpublished data). We observed substantial enrichment of PP1Dis2 at centromeres 1 and 2 in _mph1_-overexpressing cells relative to log phase cultures, although some Dis2 was detected at kinetochores throughout the cell cycle (Figure 1A; Supplemental Figure 1A). To examine whether association of PP1Dis2 to the kinetochore in M phase is regulated by microtubule kinetochore attachment or tension, carbendazim (CBZ), a microtubule depolymerising agent, was added to M phase arrested cells. This caused a decrease, but not abolition, of the binding of PP1Dis2 to centromeres 1 and 2 (Figure 1A). These data suggest that, like in C. elegans and human cells, association of PP1 to fission yeast kinetochores is regulated by the application of tension across sister chromatids by spindle microtubules (Liu et al., 2010).
Figure 1. Association of Dis2 to Spc7 at kinetochores is tension regulated and essential for viability.
(A) Tension regulated binding of Dis2 to kinetochores in mitosis. Log phase dis2+ and dis2-gfp cultures (cycling) or the same cells arrested in mitosis by overexpression of mph1 either before (− CBZ) or after treatment with carbendazim (+ CBZ) for 10 minutes were fixed. Enrichment of Dis2 at the central core region (black bar) or innermost repeats (white bar) was analysed by ChIP. Error bars correspond to standard deviation. See also Supplemental Figure 1A.
(B) Protein alignments of the N-terminus of fission yeast Spc7 and its homologues from budding yeast, human, worm, fruit-fly and chicken. Two PP1-binding consensus sites are highlighted in red.
(C) Two PP1-binding sites of Spc7 contribute to the interaction of Spc7 and Dis2 in vitro. GST-Spc7 fusion protein or mutant proteins missing either one of both PP1-binding sites were mixed with cells extracts of dis2-3Pk cells. Interaction following affinity precipitation with GSH-agarose was analysed by western blot. Input proteins are shown by coomassie stain.
(D) Both PP1-binding sites in Spc7 are required for cell viability. Δspc7 cells expressing either nothing or a wild type copy of spc7 or mutant alleles lacking either one or both PP1-binding sites were kept alive by a ura4 containing episomal plasmid expressing wild type spc7. The phenotype of mutants was revealed by replica plating onto media containing 5-FOA.
Association of PP1Dis2 to Spc7 is required for efficient spindle checkpoint silencing
Regulated targeting of PP1 to the kinetochore protein KNL-1 opposes the activity of Aurora B kinase (Liu et al., 2010). Despite repeated efforts under different conditions, we have failed to identify PP1Dis2 in TAP-tag purifications of the Ndc80/Mis12/Spc7 (NMS) complex. Similarly Spc7, the fission yeast homologue of KNL-1, has not been identified in immunoprecipitates with PP1Dis2 (Liu et al., 2005; VV and KGH, unpublished data). Nevertheless, the N-terminus of Spc7 contains two conserved consensus PP1-binding sites, KGILK (referred to as motif A) and RRVSF (referred to as motif B) (Hendrickx et al., 2009; Wakula et al., 2003); Figure 1B). To assess whether fission yeast Spc7 can interact with PP1Dis2 in vitro, we purified a GST-Spc7(1-283) fusion protein from bacteria and incubated it with lysates from dis2-3Pk cells. PP1Dis2 bound strongly to the N-terminus of Spc7 following affinity purification and western blot (Figure 1C). To assess the role of the putative PP1-binding sites, fusion proteins were produced in which amino acids 46-50 (GST-Spc7(PP1-ΔA)) or amino acids 111-115 (GST-Spc7(ΔPP1-B)) or both (GST-Spc7(PP1-ΔAB)) were removed, and their ability to affinity precipitate PP1Dis2 from cell lysates assessed. Strikingly, PP1Dis2 bound to both Spc7(PP1-ΔA) and Spc7(PP1-ΔB) fusion proteins, but not to the Spc7(PP1-ΔAB) fusion protein, indicating that both A and B motifs bind PP1Dis2 in vitro (Figure 1C). Similar results were obtained by two-hybrid analysis (Supplemental Figure 1B and 1C). We note that binding of PP1Dis2 to Spc7(PP1-ΔA) and Spc7(PP1-ΔB) single mutants is considerably weaker than to wild type GST-Spc7, suggesting that the two motifs may bind co-operatively to PP1Dis2 (Figure 1C).
To assess the role of PP1Dis2 binding to Spc7 in vivo, we introduced a copy of either wild type spc7+, spc7(PP1-ΔA), spc7(PP1-ΔB) or spc7(PP1-ΔAB) mutants into a spc7::natMX6 haploid strain expressing wild type spc7+ on an episomal plasmid containing the ura4+ gene (pIRT2u-spc7). These strains were replica plated to media containing 5-fluoroorotic acid (5-FOA) to select against the pIRT2u-spc7 plasmid. This revealed that both PP1-binding motif A and motif B are essential for cell proliferation (Figure 1D). The experiment was repeated in cells deleted for mad3 or mad2, components of the mitotic checkpoint complex (MCC). Remarkably, spc7(PP1-ΔB) Δmad3 and spc7(PP1-ΔB) Δmad2 cells readily lost the pIRT2u-spc7 plasmid, suggesting that the prime defect in spc7(PP1-ΔB) cells is arrest due to hyperactivation of the spindle checkpoint (Figure 1D, Supplemental Figure 2A). By contrast, neither spc7(PP1-ΔA) nor spc7(PP1-ΔAB) mutants were rescued by loss of mad2 or mad3, indicating that the A motif has an important role(s) in establishing stable microtubule-kinetochore attachments (Figure 1D, Supplemental Figure 2A), although we cannot formally assign this to a failure to bind PP1Dis2.
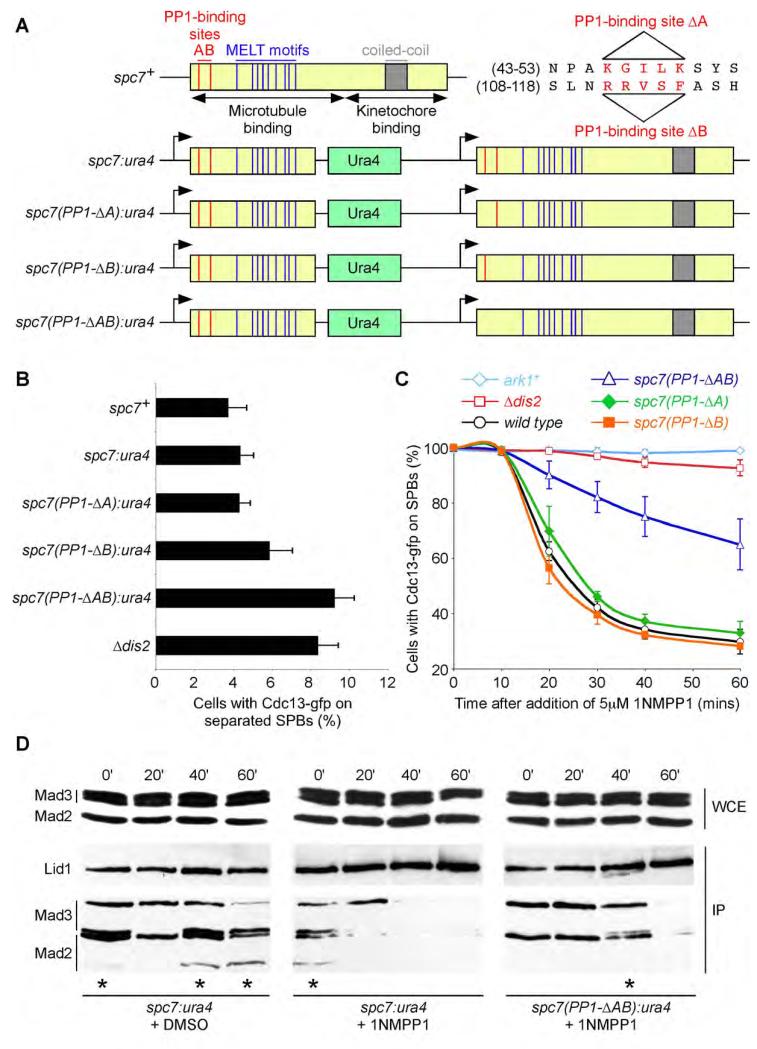
To directly assess the role of the Spc7 bound pool of PP1 in checkpoint silencing we employed a recently described assay which relies on monitoring Cdc13 (cyclin B) degradation by fluorescence microscopy in single nda3-KM311 (β-tubulin) cells, which are arrested in mitosis at the restrictive temperature due to a lack of spindle microtubules (Vanoosthuyse and Hardwick, 2009). In this assay, the spindle assembly checkpoint is inactivated by addition of 1NMPP1 to mitotically arrested nda3-KM311 ark1-as3 cdc13-GFP cells. Addition of 1NMPP1 selectively inhibits analogue-sensitive Ark1 kinase (Hauf et al., 2007) and, in doing so, promotes silencing of the spindle checkpoint signal. In this situation PP1Dis2 is essential for dissociation of Mad2 and Mad3 from the APC/C and for activation of the APC/C complex, which triggers cyclin B destruction (Vanoosthuyse and Hardwick, 2009). However, since both PP1-binding motifs in Spc7 are essential for viability, we needed to construct a conditional mutant to assess the role of PP1-binding mutants in this assay. Notably, we find that spc7(PP1-ΔA), spc7(PP1-ΔB) and spc7(PP1-ΔAB) alleles can rescue the temperature sensitivity of an spc7-23 mutant (Kerres et al., 2007) at high temperature (Supplemental Figure 2B). Since the Spc7-23 protein has a mutation which prevents it binding to kinetochores at high temperature, this hetero-allelic complementation suggests that a non-kinetochore bound population of Spc7, which can bind PP1Dis2, can compensate for kinetochore bound Spc7, which cannot bind PP1Dis2. Indeed, we find that expression of just the N-terminal 666 amino acids of spc7 (spc7(1-666)) rescues the lethality of a spc7(PP1-ΔAB) mutant, but not if the N-terminal fragment also lacks the ability to interact with PP1Dis2 (Supplemental Figure 3A). This suggests that the Spc7(1-666) protein recruits sufficient PP1 to the microtubule-kinetochore interface to compensate for the lack of PP1 associated to kinetochore-bound Spc7. This allowed us to construct a series of mutants (Figure 2A, Supplemental Figure 3B). Although all four mutants are viable, the truncated spc7(1-666) allele does not fully rescue spc7(PP1-ΔAB) function as cells spend longer in pro-metaphase than control cells, as judged by kinetochore and spindle pole position and the presence of Cdc13 (Cyclin B) on spindles and spindle poles (Figures 2B, Supplemental Figure 4A). This analysis also indicates the B motif (RRVSF) has a more prominent role than the A motif (KGILK), but that both contribute to the timing of anaphase onset (Figure 2B, Supplemental Figure 4A). Nevertheless, we were able to assess the role of spc7-PP1 binding mutants in checkpoint silencing using these constructions since the spc7(1-666) allele does not alter the efficiency of checkpoint silencing and the Spc7(1-666) protein is not bound to kinetochores in the absence of microtubules (Supplemental Figures 4B and 4C). Importantly, following forced inactivation of the checkpoint by chemical inhibition of Ark1 kinase in nda3-KM311 ark1-as3 cdc13-GFP cells, we find that the rate of Cdc13 destruction is unaltered in either spc7(PP1-ΔA) or spc7(PP1-ΔB) mutants compared to wild type, suggesting that a very limited amount of PP1Dis2 on kinetochores is sufficient to silence the checkpoint, but is dramatically slowed in the spc7(PP1-ΔAB) mutant, although not as effectively as in the absence of PP1Dis2 (Figure 2C). Under the same conditions, the rate of dissociation of the Mad2 and Mad3 checkpoint proteins from the APC/C is also slower in the spc7(PP1-ΔAB) mutant compared to wild type, but again is not blocked (Figure 2D). Together these data suggest that association of PP1 to Spc7 is important, but not absolutely necessary, for spindle checkpoint silencing, possibly due to the existence of another spindle checkpoint silencing pathway.
Figure 2. Association of PP1 to Spc7 is required for efficient checkpoint silencing.
(A) Schematic of the conditional spc7:ura4 allele or variants lacking either PP1-binding site A or B or both are shown. PP1-binding sites are shown in red, MELT motifs (Cheeseman et al., 2004) in blue and the conserved kinetochore-binding domain in grey.
(B) Log phase cultures of wild type, Δdis2 or spc7 alleles shown in (A) expressing cdc13-gfp were fixed and the percentage of cells with Cdc13 (Cyclin B) on separated spindle poles and spindles assessed. All error bars correspond to standard deviation.
(C) Association of PP1 to Spc7 is partly required for checkpoint silencing. nda3-KM311 ark1-as3 cdc13-gfp cells expressing the spc7 alleles in (A) were arrested in mitosis and the percentage of cells with Cdc13 on spindle poles assessed after addition of 1NMPP1. Cells lacking Dis2 or containing an analogue-insensitive allele of ark1 are shown as controls.
(D) Association of PP1 to Spc7 is partly required for dissociation of the Mad2 and Mad3 checkpoint proteins from the anaphase promoting complex. nda3-KM311 lid1-TAP mad2-gfp mad3-gfp ark1-as3 cells either expressing spc7:ura4 or spc7(PP1-ΔAB):ura4 alleles were arrested in mitosis and association of Mad2 and Mad3 to Lid1 was assessed by immunoprecipitation and western blot following addition of either DMSO or 1NMPP1. Stars indicate artefactual cleavage of Mad2-gfp and Mad3-gfp during immunoprecipitation.
The Klp5, Klp6 kinesin complex is required for efficient spindle checkpoint silencing
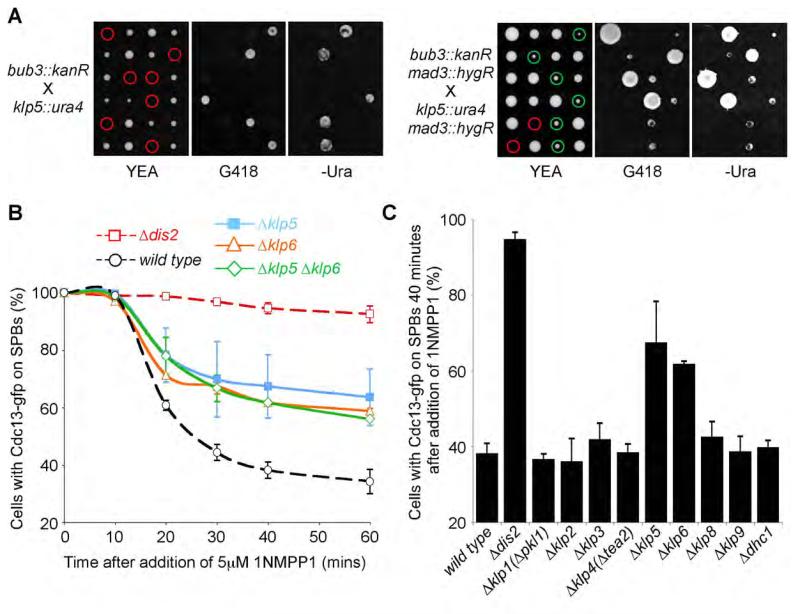
Our observation that an Spc7-bound pool of PP1 is only partially required for PP1-dependent spindle checkpoint silencing persuaded us to search for other potential PP1Dis2 targeting proteins involved in this process. In fission yeast, Bub3 is not a core component of the MCC, nor is it required for the establishment of the spindle checkpoint, but is instead required for efficient spindle checkpoint silencing (Tange and Niwa, 2008; Vanoosthuyse et al., 2009; Windecker et al., 2009). We sought additional components of the spindle silencing machinery by isolating mutants that were synthetic lethal with Δbub3. We reasoned that if the lethality of a double mutant between Δbub3 and another silencing factor were due solely to hyperactivation of the spindle checkpoint, one would expect the synthetic lethality to be bypassed by inactivation of mad3, a core component of the MCC. We identified two mutants, Δklp5 and Δklp6 (kinesin-8), which satisfied these criteria. Tetrad analyses reveal that Δbub3 Δklp5 double mutants arrest as inviable micro-colonies of cells, whereas Δbub3 Δklp5 Δmad3 triple mutants are viable but slow growing (Figure 3A). Similar results were obtained by random spore analyses (Supplemental Figure 5A). Klp5 and Klp6 are members of the kinesin-8 class of plus-end directed microtubule motor proteins that act at the microtubule-kinetochore interface to direct chromosome congression in mitosis (Garcia et al., 2002; Tischer et al., 2009; West et al., 2002). Δklp5 and Δklp6 mutants have indistinguishable effects on spindle microtubule dynamics, the timing of mitosis and accuracy of chromosome segregation, strongly suggesting Klp5 and Klp6 act primarily, if not exclusively, as a heterodimer during mitosis (Garcia et al., 2002; West et al., 2002). Indeed localisation of Klp5 and Klp6 both to kinetochores and the mitotic spindle is dependent on the presence of both proteins (Unsworth et al., 2008). Strikingly, deletion of either klp5 or klp6 alone or deletion of both klp5 and klp6 partly inhibited spindle checkpoint silencing in nda3-KM311 ark1-as3 cdc13-GFP on addition of 1NMPP1 (Figure 3B). This effect is specific to the Klp5 and Klp6 kinesins, as deletion of other non-essential kinesins, or the dynein motor (Dhc1), had no effect on spindle checkpoint silencing (Figure 3C). Notably, although loss of Klp5 only has a marginal effect on the dissociation of Mad2 and Mad3 from APC/C, re-introduction of wild type copies of klp5 into Δklp5 cells or klp6 into Δklp6 cells or both genes into Δklp5 Δklp6 cells rescued the silencing defect, suggesting that efficient checkpoint silencing requires the Klp5-Klp6 kinesin heterodimer (Figures 4C-E, Supplemental Figure 5B).
Figure 3. Klp5 and Klp6 kinesins are required for efficient checkpoint silencing.
(A) Abrogation of the spindle assembly checkpoint rescues the synthetic lethality of a Δklp5 Δbub3 strain. Tetrad analysis of a cross between Δklp5 and Δbub3 cells (left panels) or Δklp5 Δmad3 and Δbub3 Δmad3 cells (right panels) is shown. Genotype was scored by replica plating to selective medium. Red and green circles highlight inviable and viable meiotic products respectively. See also Supplemental Figure 5A.
(B) Cells lacking Klp5 or Klp6 or both are partially deficient in checkpoint silencing. nda3-KM311 ark1-as3 cdc13-gfp cells or the same cells deleted for Klp5 (Δklp5), Klp6 (Δklp6) or both (Δklp5 Δklp6) were arrested in mitosis and the percentage of cells with spindle pole associated Cdc13 (Cyclin B) assessed after the addition of 1NMPP1. Dashed lines indicate data from control cultures reproduced from a previous figure. All error bars correspond to standard deviation.
(C) The role of Klp5 and Klp6 in checkpoint silencing is not shared by other fission yeast motor proteins. nda3-KM311 ark1-as3 cdc13-gfp cells bearing deletions in all non-essential kinesins (Klps) or dynein heavy chain (Dhc1) were arrested in mitosis and the percentage of cells with spindle pole associated Cdc13 (Cyclin B) assessed at time zero and 40 minutes after the addition of 1NMPP1.
Figure 4. The motor activities of Klp5 and Klp6 are not required for checkpoint silencing.
(A) Serial dilutions reveal that motor-inactive (SwII) klp5 and klp6 mutants are unable to rescue the resistance of Δklp5 cells (top panels) and Δklp6 cells (bottom panels) to thiabendazole (TBZ).
(B) Representative images of either metaphase or anaphase B cells from the strains in (A) showing that kinesin-8 heterodimers comprising one fluorescently-tagged protein (green) and one motor-dead (SwII) protein localise to the entire anaphase B spindle. Chromatin is stained with DAPI (blue). Bar, 2μm.
(C-E) The motor-inactive klp5(SwII) and klp6(SwII) mutants are competent for checkpoint silencing. (C) nda3-KM311 ark1-as3 cdc13-gfp Δklp5 cells or (D) nda3-KM311 ark1-as3 cdc13-gfp Δklp6 cells or (E) nda3-KM311 ark1-as3 cdc13-gfp Δklp5 Δklp6 cells expressing either wild type klp5 or klp6 either individually or in combination or expressing motor inactive klp5(SwII) or klp6(SwII) mutants either individually (C and D) or in combination (E) were arrested in mitosis and the percentage of cells with spindle pole associated Cdc13 (Cyclin B) assessed after the addition of 1NMPP1. Dashed lines indicate data from control cultures reproduced from a previous figure. Error bars correspond to standard deviation.
The motor activity of Klp5 and Klp6 is not required for checkpoint silencing
The involvement of Klp5 and Klp6 kinesins in microtubule-independent checkpoint silencing is unexpected as these proteins are microtubule-dependent molecular motors. To assess the role of Klp5 and Klp6 motor domains in spindle checkpoint silencing, we mutated residues G297 and E299 in Klp5 and residues G296 and E298 in Klp6 in the switch II region (Browning et al., 2003) of each kinesin motor protein to alanine (hereafter called klp5(SwII) and klp6(SwII)). Expression of a klp5(SwII) mutant restored kinetochore localisation of Klp6 during metaphase, but failed to rescue the resistance of Δklp5 cells to thiabendazole (TBZ), a microtubule depolymerising agent, and caused Klp6 to spread along the entire length of the anaphase B spindle in Δklp5 klp6-gfp cells (Figures 4A and 4B). Expression of a klp6(SwII) mutant in Δklp6 klp5-gfp cells yielded similar results (Figures 4A and 4B). These data suggest that the motility of the Klp5-Klp6 heterodimer is dependent on the ATPase activity of both motor heads and is not required for its association to kinetochores. However the silencing defect of Δklp5 and Δklp6 cells is rescued by re-introduction of klp5(SwII) and klp6(SwII) mutants, respectively (Figures 4C and 4D). Moreover, re-introduction of both klp5(SwII) and klp6(SwII) mutants into Δklp5 Δklp6 cells fully restored spindle checkpoint silencing (Figure 4E), indicating that the checkpoint silencing function of the Klp5-Klp6 heterodimer is independent of its motor activity.
Association of PP1Dis2 to Klp5-Klp6 is required for efficient spindle checkpoint silencing
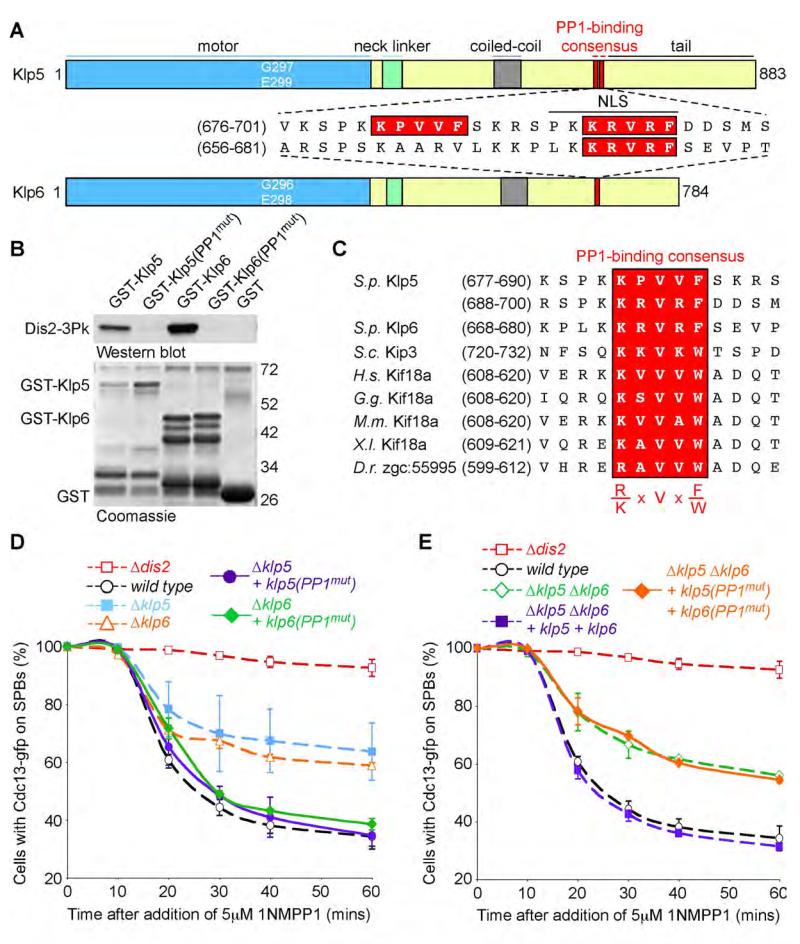
Sequence analyses reveal that Klp5 and Klp6 contain several potential binding motifs for PP1. Klp5 contains two closely spaced motifs (residues 681-685 and 692-696) whereas Klp6 contains one motif (residues 672-676) (Figure 5A). All three motifs are located in the regions of Klp5 and Klp6 that have been implicated in nuclear localisation (Unsworth et al., 2008). To examine whether Klp5 and Klp6 interact with PP1Dis2 in vitro, we constructed GST-Klp5, GST-Klp5(V683A, F685A, V694A, F696A) (hereafter GST-Klp5(PP1mut)), GST-Klp6 and GST-Klp6(V674A, F676A) (hereafter GST-Klp6(PP1mut)) fusion proteins containing only the tail domains of Klp5 (residues 631-883) and Klp6 (residues 623-784). These were incubated in cell lysates from dis2-N3Pk cells and association of PP1Dis2 to Klp5 and Klp6 assessed after affinity precipitation and western blot. This revealed that wild type GST-Klp5 and GST-Klp6 proteins, but not GST-Klp5(PP1mut) or GST-Klp6(PP1mut) mutant proteins, strongly interact with PP1Dis2 in vitro (Figure 5B). We have been unable to detect association of PP1Dis2 to Klp5 or Klp6 in vivo as the Klp5 and Klp6 proteins are acutely sensitive to proteolysis in native extracts. Strikingly, budding yeast KIP3 and human, chicken, mouse, frog and zebrafish KIF18A proteins, but not Drosophila Klp67A or human KIF18B proteins, contain an [RK]X(0-1)VX[FW] motif in their non-catalytic domains. Moreover, human KIF18A has been found to interact with the catalytic subunit of PP1α by two-hybrid analysis (Colland et al., 2004), suggesting that PP1 may associate to some, but not all, members of the kinesin-8 family (Figure 5C).
Figure 5. Association of PP1 to Klp5-Klp6 is required for spindle checkpoint silencing.
(A) Schematic of S. pombe Klp5 and Klp6 kinesins showing motor domains (blue), neck linker domains (green), coiled-coil domains (grey) and PP1-binding consensus sites (red). Nuclear localisation sequences are underlined.
(B) Klp5 and Klp6 bind Dis2 in vitro. GST-Klp5 and GST-Klp6 fusion proteins or proteins mutated in both PP1-binding sites in Klp5 or the single site in Klp6 were mixed with cell extracts of dis2-3Pk cells. Interaction following affinity precipitation with GSH-agarose was analysed by western blot. Input proteins are shown by coomassie stain.
(C) Protein alignments showing the homology of the PP1-binding consensus (highlighted in red) in kinesin-8 family members from fission and budding yeast, human, chicken, mouse, frog and zebrafish.
(D) Checkpoint silencing assay in nda3-KM311 ark1-as3 cdc13-gfp cells shows that expression of klp5(PP1mut) rescues the silencing deficiency of a Δklp5 mutant and that expression of klp6(PP1mut) does likewise in a Δklp6 mutant. All error bars correspond to standard deviation.
(E) Co-expression of both klp5(PP1mut) and klp6(PP1mut) mutants does not rescue the silencing defect of a Δklp6 Δklp6 double mutant.
Next we directly tested whether association of PP1Dis2 to Klp5-Klp6 influences spindle checkpoint silencing. Notably, re-introduction of full length klp5(PP1mut) gene restored checkpoint silencing in Δklp5 cells as did re-introduction of full length klp6(PP1mut) in Δklp6 cells, suggesting that binding of PP1Dis2 to either Klp5 or Klp6 is sufficient to satisfy the role of Klp5-Klp6 complex in spindle checkpoint silencing (Figure 5D). Neither Klp5(PP1mut) nor Klp6(PP1mut) alter the nuclear localisation of their heterodimer partner (Supplemental Figure 6). Importantly, however, expression of both klp5(PP1mut) and klp6(PP1mut) mutants failed to rescue the spindle checkpoint silencing defect in Δklp5 Δklp6 cells (Figure 4E). These results indicate that association of PP1 to kinesin-8 motors plays an important role in spindle checkpoint silencing.
Association of PP1Dis2 to Klp5-Klp6 is required for chromosome bi-orientation and timely anaphase onset
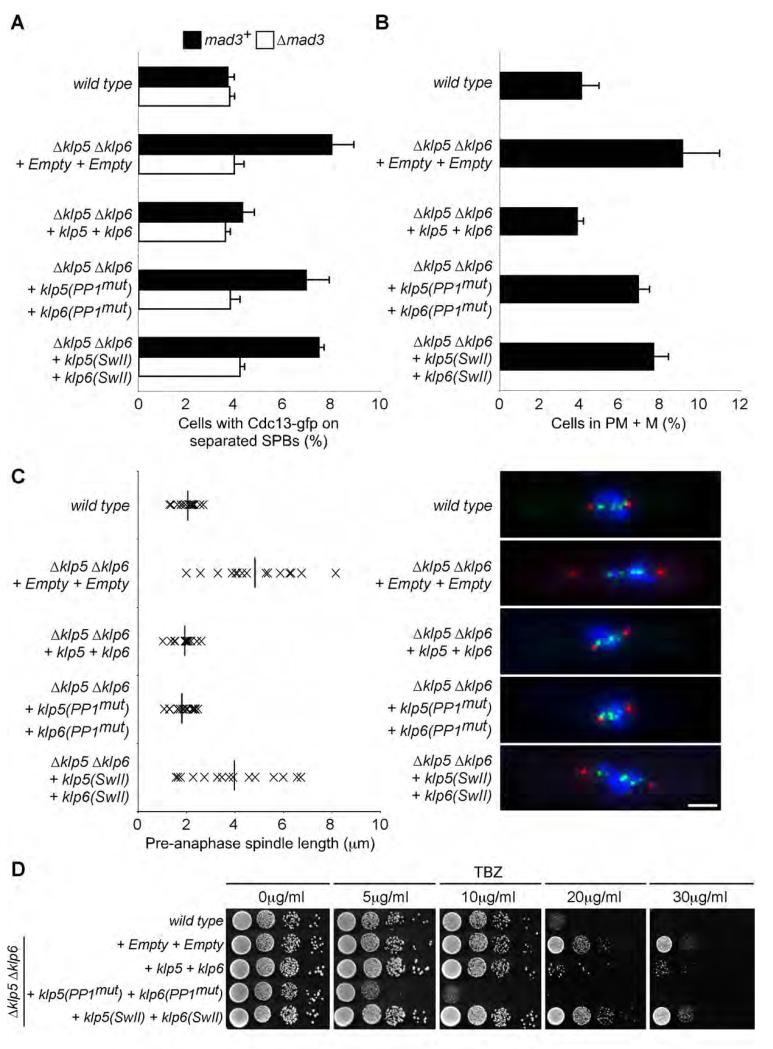
Cells lacking either Klp5 or Klp6 are resistant to thiabendazole, a microtubule depolymerising agent, display a prolonged delay in pro-metaphase and metaphase with highly elongated spindles and are essential for viability in the absence of either Bub3 or the Dam1 complex (Figure 3A, Garcia et al., 2002; Sanchez-Perez et al., 2005; West et al., 2002). It is presently unclear why mitotic spindles are elongated in the absence of Klp5 and Klp6, since neither protein displays microtubule depolymerase activity in vitro (Grissom et al., 2009). To assess the roles of both the motor activity of the Klp5-Klp6 complex and its ability to associate with PP1Dis2 on mitotic progression, we monitored kinetochore and spindle pole position and Cdc13 (cyclin B) levels in log phase cultures of klp5(SwII) klp6(SwII) and klp5(PP1mut) klp6(PP1mut) mutants. We find that both mutants display a checkpoint-mediated delay over the timing of anaphase onset, although neither effect is as pronounced as that observed in the complete absence of Klp5 and Klp6 (Figures 6A and 6B). Strikingly, however, although spindles are highly elongated at anaphase onset in klp5(SwII) klp6(SwII) mutants, no such defect is observed in klp5(PP1mut) klp6(PP1mut) mutants, suggesting that the motor activity of the Klp5-Klp6 complex is responsible for this phenotype (Figure 6C). Moreover, klp5(SwII) klp6(SwII) mutants, but not klp5(PP1mut) klp6(PP1mut) mutants, are inviable in the absence of either Dam1 or Bub3, indicating that the motor activity of the Klp5-Klp6 complex is important for its mitotic function (Table 1). Indeed re-introduction of wild type klp5 and klp6 genes, but not re-introduction of klp5(SwII) and klp6(SwII) mutants, suppressed the resistance of Δklp5 Δklp6 mutants to thiabendazole, indicating that this phenotype is also associated with the motor activity of the Klp5-Klp6 complex (Figure 6D). In stark contrast, however, we find that klp5(PP1mut) klp6(PP1mut) mutants are hypersensitive to thiabendazole (more sensitive than wild type cells), and that klp5(PP1mut) klp6(PP1mut) cells display a profound defect in chromosome bi-orientation (worse than Δklp5 Δklp6 mutants) (Figure 6D, Table 1). These data indicate that both the motor activity of Klp5-Klp6 and its association to PP1 are required for normal mitotic progression and, secondly, that the Klp5-Klp6 bound pool of PP1 controls both silencing of the spindle checkpoint and the establishment of chromosome bi-orientation.
Figure 6. Association of PP1 and the motor activity of Klp5-Klp6 play distinct roles in microtubule dynamics and the timing of anaphase onset.
(A) Log phase cultures of cdc13-gfp or Δklp5 Δklp6 cdc13-gfp cells, that were either wild type (mad3+) or lacking mad3 (Δmad3), and expressing either nothing (+ Empty + Empty) or wild type copies of both genes (+ klp5 + klp6), or both genes mutated in their respective PP1-binding sites (+ klp5(PP1mut) + klp6(PP1mut)) or both genes mutated in their respective motor domains (+ klp5(SwII) + klp6(SwII)) were fixed and the percentage of cells with Cdc13 (Cyclin B) on separated spindle poles and spindles assessed. All error bars correspond to standard deviation.
(B) Log phase cultures of fta3-gfp sid4-tdtomato cells or Δklp5 Δklp6 fta3-gfp sid4-tdtomato cells expressing the same combination of wild type or mutant alleles of klp5 and klp6 as in (A) were fixed and the percentage in pro-metaphase and metaphase (PM + M) assessed.
(C) Pre-anaphase spindle length of individual cells in log phase populations of strains in (B) is plotted. Horizontal bars represent the mean length. Representative images of pre-anaphase cells (right panels) showing SPBs (red), kinetochores (green) and chromatin (DAPI, blue). Bar, 2μm.
(D) Log phase cultures of wild type or Δklp5 Δklp6 cells expressing the same combination of wild type or mutant alleles of klp5 and klp6 as in (A) were spotted in ten-fold serial dilutions onto YEA plates containing various concentrations of thiabendazole (TBZ) and incubated for 3 days at 33°C.
Table 1.
Association of PP1 to Klp5 and Klp6 is required for accurate bi-orientation
| Mini-chromosomeloss per division | Viability | ||
|---|---|---|---|
| Δbub3 | Δdam1 | ||
| wild type | 0.08% (± 0.03%) | ++++ | ++++ |
| Δklp6 | 0.5% (± 0.1%) | − | − |
| _Δklp5 Δklp6_+ Empty + Empty | 0.4% (± 0.3%) | − | − |
| _Δklp5 Δklp6_+ klp5 + klp6 | 0.1% (± 0.01%) | ++++ | ++++ |
| _Δklp5 Δklp6_+ _klp5(PP1mut)_+ klp6(PP1mut) | 2.1% (± 1.2%) | + | +++ |
| _Δklp5 Δklp6_+ _klp5(SwII)_+ klp6(SwII) | N.D. | − | − |
Spc7 and Klp5-Klp6 bound pools of PP1 co-ordinate spindle checkpoint silencing
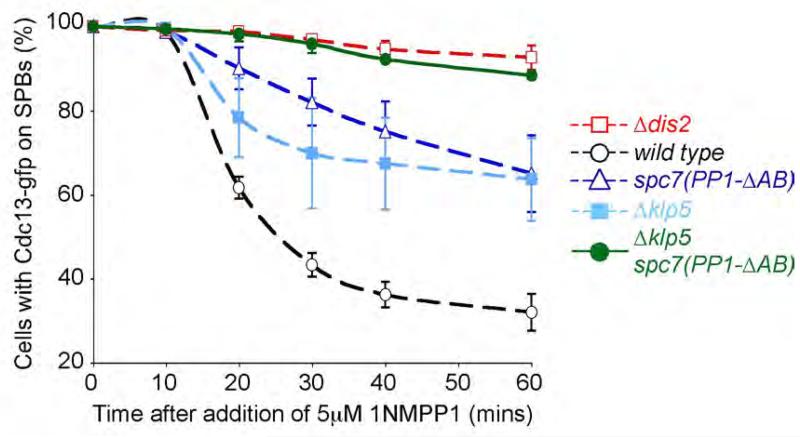
To examine the relationship between Spc7 and Klp5/Klp6 complex in checkpoint silencing, we constructed a nda3-KM311 ark1-as3 cdc13-GFP spc7(PP1-ΔAB) Δklp5 double mutant. These cells arrested normally at the restrictive temperature indicating that spindle checkpoint activation was intact. However, on addition of 1NMPP1 we observed little destruction of Cdc13 in spc7(PP1-ΔAB) klp5Δ cells over the course of the experiment, similar to that seen in the absence of Dis2 (Figure 7), suggesting that recruitment of PP1 to both Spc7 and kinesin-8 motors is necessary to silence the spindle checkpoint.
Figure 7. Asssociation of PP1 to both Klp5-Klp6 and Spc7 is required to efficiently silence the spindle checkpoint.
Spc7 and Klp5-Klp6 bound pools of PP1 combine to efficiently silence the spindle checkpoint. nda3-KM311 ark1-as3 cdc13-gfp spc7(PP1-ΔAB) Δklp5 cells were arrested in mitosis and the percentage of cells with spindle pole associated Cdc13 (Cyclin B) assessed after the addition of 1NMPP1. Dashed lines indicate data from control cultures reproduced from a previous figure. Error bars correspond to standard deviation.
Discussion
In this paper we provide direct and compelling evidence that association of PP1 to the N-terminus of Spc7 is required for efficient spindle checkpoint silencing. We define two binding motifs for PP1 in the Spc7 kinetochore protein and show that deletion of both motifs abolishes binding of PP1Dis2 to Spc7 and slows destruction of Cyclin B (Cdc13) following chemical inactivation of Aurora B kinase (Ark1) in mitotically arrested nda3-KM311 cells. The phenotype of these spc7-PP1 binding mutants can only be studied in cells that contain an additional C-terminally truncated allele of spc7 that cannot contact kinetochores. This is because deletion of either the RRVSF or KGILK motifs by themselves, or in combination, is lethal. Strikingly, the lethality caused by deletion of the RRVSF motif in Spc7 is rescued by deletion of either mad2 or mad3. This is consistent with a role for this pool of PP1Dis2 in spindle checkpoint inactivation. However, the lethality caused by deletion of the KGILK motif or both motifs in Spc7 is not rescued by deletion of either mad2 or mad3, suggesting that association of PP1Dis2 to the KGILK binding site plays other essential mitotic functions, such as stabilising microtubule-kinetochore interactions. Further work will be necessary to determine whether association of PP1Dis2 to both the KGILK and RRVSF motifs co-ordinately stabilises microtubule-kinetochore interaction and inactivates checkpoint silencing or whether these two binding sites differentially control these activities.
Since association of PP1Dis2 to Spc7 is not sufficient to fully silence the spindle checkpoint in our assay, we employed a genetic screen to identify additional silencing factors. Our data strongly indicate that the Klp5-Klp6 kinesin heterodimer acts in conjunction with Spc7-PP1 to efficiently silence the spindle checkpoint in fission yeast (Figure 7). Importantly, both Klp5 and Klp6 bind PP1Dis2 in vitro and contain several consensus PP1-binding sites that are necessary for the function of the Klp5-Klp6 complex in checkpoint silencing and the timing of anaphase onset in vivo. These data demonstrate for the first time that a kinesin-8 family member binds PP1 and, moreover, that at least two different pools of PP1 are required for efficient spindle checkpoint silencing. Additionally, we have identified PP1-binding motifs in many other members of the kinesin-8 family, including budding yeast KIP3 and human KIF18A. Notably, KIF18A interacts with the catalytic subunit of PP1-α by two-hybrid analysis, suggesting that the spindle checkpoint silencing function of kinesin-8 motors may be conserved (Colland et al., 2004). We are currently testing this possibility in human cells.
Members of the kinesin-8 family are plus-end directed molecular motors that concentrate at kinetochores during early mitosis. Deletion or knockdown of kinesin-8 function causes increased spindle length, resistance to microtubule depolymerising drugs (at least in yeast), altered kinetochore dynamics, defective chromosome congression and a delay or block in anaphase onset (Mayr et al., 2007; Stumpff et al., 2008). Although budding yeast KIP3 is a microtubule depolymerase in vitro (Gupta et al., 2006; Varga et al., 2006; Varga et al., 2009), this activity cannot be detected for Klp5 and Klp6 (Grissom et al., 2009) and remains unclear for human KIF18A (Du et al., 2010; Peters et al., 2010). Nevertheless, we find that cells expressing a motor defective Klp5-Klp6 complex display most of the phenotypes associated with Δklp5 and Δklp6 cells, indicating that motor activity is important for kinesin-8 function. Despite this we find that the motor activity of Klp5-Klp6 is not required for spindle checkpoint silencing in the absence of microtubules. In stark contrast, cells expressing a Klp5-Klp6 complex that is unable to bind PP1 are highly sensitive to microtubule depolymerising agents, display a profound defect in the establishment of chromosome bi-orientation and are also delayed in anaphase onset, but have normal metaphase spindle length. This suggests that the kinesin-8-PP1 complex contains two distinct activities that co-ordinate chromosome bi-orientation and spindle checkpoint signalling.
During early mitosis, association of PP1γ to the N-terminus of KNL-1/Blinkin is prevented by phosphorylation of the serine residue in the RRVSF motif by Aurora B kinase (Liu et al., 2010). The application of tension between sister chromatids pulls the outer kinetochore away from Aurora B kinase, which is located at the inner centromere. This allows dephosphorylation of this residue and the association of PP1γ. It is tempting to speculate, based on the results presented in this paper, that association of PP1 to the Spc7/Blinkin/KNL-1 family of kinetochore proteins co-ordinately stabilises tension bearing microtubule-kinetochore attachments and inactivates the spindle checkpoint. Indeed, our preliminary evidence suggests that mimicking phosphorylation in the RRVSF motif of Spc7 abrogates binding of PP1 to this site and hinders checkpoint silencing (JCM and JBAM, unpublished data). However, mutation of this residue to alanine does not override the spindle checkpoint in nda3-KM311 cells, suggesting that Aurora B kinase does not prevent silencing of the checkpoint simply by preventing association of PP1 to this motif (JCM and JBAM, unpublished data). Our data raise the possibility that de-regulation of both Spc7-PP1 and Klp5-Klp6-PP1 pathways is needed to override the spindle checkpoint. Association of PP1Dis2 to the Klp5-Klp6 complex or association of the Klp5-Klp6-PP1 complex to kinetochores could be regulated by Aurora B kinase. However, the regions surrounding the PP1 binding motifs in Klp5 and Klp6 do not contain consensus Aurora B sites and, although Klp5 binds the kinetochore during mitosis, it is unclear whether this is important for checkpoint silencing, since association of Klp5 to kinetochores is microtubule dependent (Garcia et al., 2002). Indeed, we note that the kinetics of checkpoint silencing by the Spc7-PP1 and Klp5-Klp6-PP1 pathways is quite different. Whereas Spc7-PP1 is required at early time points, the Klp5-Klp6-PP1 pathway is only required at later time points. One possibility is that, in the absence of microtubules, the Spc7-PP1 pathway acts rapidly at the kinetochore, whereas Klp5-Klp6-PP1 may act more slowly to dephosphorylate key targets, such as Mad3, in the nucleoplasm. Regardless, we contend that the Klp5-Klp6-PP1 complex is normally likely to silence the spindle checkpoint at the microtubule-kinetochore interface since this is where it concentrates during early mitosis.
We previously demonstrated that dissociation of the MCC from APC/C requires PP1 activity (Vanoosthuyse and Hardwick, 2009). The key targets of PP1 in triggering inactivation of the spindle checkpoint, however, remain unknown in both fission yeast and other systems. In principle these substrates could include core components of the kinetochore (such as Ndc80), spindle checkpoint proteins (such as Mad1, Mad2 or Mad3) or the APC/C itself. It is particularly interesting in this regard that Bub1 and BubR1 checkpoint proteins associate with the N-terminus of Blinkin/KNL-1 in human cells and this is thought to be required for spindle checkpoint activation (Kiyomitsu et al., 2011; Kiyomitsu et al., 2007). However, we found that association of Bub1 to the fission yeast kinetochore is dependent on Bub3 and that Δbub3 mutants have a robust spindle checkpoint (Vanoosthuyse et al., 2009). Instead Δbub3 mutants are partially defective in spindle checkpoint silencing, arguing that kinetochore association of Bub1 is important for spindle checkpoint inactivation. One possibility is that Spc7-bound Bub1 and/or Bub3 may provide a docking site for the MCC at kinetochores to allow it to be dephosphorylated by Spc7-PP1 and/or the Klp5-Klp6-PP1 complex. Clearly, in the future it will be important to determine whether the Spc7-bound and Klp5-Klp6-bound pools of PP1 dephosphorylate the same or different targets and what influence this has on the checkpoint silencing process.
It is necessary to point out that the checkpoint silencing assay we have employed is performed in the absence of microtubules so we cannot assess the contribution of microtubule-dependent checkpoint silencing pathways, should they exist in fission yeast. Although dynein is not required for checkpoint silencing in fission yeast (Courtheoux et al., 2007), dynein-dependent stripping of Mad2 from kinetochores plays a key role in spindle checkpoint silencing in animal cells (Howell et al., 2001). However, since binding of dynein to the mammalian kinetochore is dependent on phosphorylation (Whyte et al., 2008), it is possible that dynein-dependent stripping of checkpoint proteins is triggered by the tension-dependent recruitment of PP1 to kinetochores. Additionally, metazoa may employ checkpoint silencing pathways that are not present in fungi. For example, an Aurora/PP1 phosphorylation switch modulates CENP-E motor activity in mammalian cells where it is required for both chromosome congression and stable microtubule capture by those chromosomes (Kim et al., 2010). Since CENP-E binds the kinetochore through BubR1 (Mao et al., 2005) binding of CENP-E to PP1 may also play a direct role in spindle checkpoint inactivation. Regardless, we propose that association of PP1 to both Spc7 kinetochore protein and kinesin-8 motors is the primary and evolutionarily conserved mechanism that inactivates the spindle checkpoint in all eukaryotes.
Experimental Procedures
Checkpoint silencing assay
The checkpoint silencing assays were performed as previously described (Vanoosthuyse and Hardwick, 2009) any modifications are detailed in Supplemental Information.
MCC-APC/C association assay
This experiment was performed as previously described (Vanoosthuyse and Hardwick, 2009), more information is provided in Supplemental Information.
ChIP analysis of PP1Dis2
ChIP analysis was performed as previously described (Vanoosthuyse et al., 2009). See Supplemental Information for further details.
Plasmid and strain construction
Construction of spc7, klp5 and klp6 mutant alleles and GST-fusion proteins are detailed in Supplemental Information.
Biochemistry
Details of protein expression, purification, in vitro binding assays and western blotting are in Supplemental Information.
Yeast two-hybrid analysis
The Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was used to conduct yeast two-hybrid experiments. See Supplemental Information for further experimental details.
Fluorescence microscopy
Fluorescence imaging of cells expressing Gfp or Tdtomato-tagged proteins was performed on a Nikon TE-2000 inverted microscope with a 100× 1.49 N.A. objective lens equipped with a Photometrics Coolsnap-HQ2 liquid cooled CCD camera (Photometrics, Tucson, AZ). Images were collected and analysed using Metamorph (version 7.5.2.0 MAG Biosystems Software). Exposure times of 1 second were used for both GFP and tdtomato and 0.25 seconds for DAPI.
Supplementary Material
01
Includes Supplementary Figures 1-6 and Supplementary Tables 1 and 2.
Highlights.
* Spc7 bound PP1 is important for silencing the spindle checkpoint
* Kinesin-8 motors bind PP1 to silence the spindle checkpoint by a motor-independent mechanism
* Both Spc7 and kinesin-8 bound pools of PP1 are needed to silence the spindle checkpoint
Acknowledgements
We thank U. Fleig, I. Hagan, S. Hauf, J. Javerzat, D. McIntosh, P. Russell, T. Toda and M. Yanagida for fission yeast strains. Individual author contributions are detailed in Supplemental Information. LAS is funded by a MRC doctoral training fellowship. AMS is funded by SULSA and the University of Edinburgh. This work was supported by an MRC Programme Grant to JBAM and a Wellcome Trust Programme Grant to KGH.
References
- Alvarez-Tabares I, Grallert A, Ortiz JM, Hagan IM. Schizosaccharomyces pombe protein phosphatase 1 in mitosis, endocytosis and a partnership with Wsh3/Tea4 to control polarised growth. J Cell Sci. 2007;120:3589–3601. doi: 10.1242/jcs.007567. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Browning H, Hackney DD, Nurse P. Targeted movement of cell end factors in fission yeast. Nat Cell Biol. 2003;5:812–818. doi: 10.1038/ncb1034. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtheoux T, Gay G, Reyes C, Goldstone S, Gachet Y, Tournier S. Dynein participates in chromosome segregation in fission yeast. Biol Cell. 2007;99:627–637. doi: 10.1042/BC20070047. [DOI] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr., Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hauf S, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 2007;26:4475–4486. doi: 10.1038/sj.emboj.7601880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A, Jakopec V, Fleig U. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol Biol Cell. 2007;18:2441–2454. doi: 10.1091/mbc.E06-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Murakami H, Yanagida M. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol Cell Biol. 2011 doi: 10.1128/MCB.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR, 3rd, He X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 2010;29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y, Niwa O. Schizosaccharomyces pombe Bub3 is dispensable for mitotic arrest following perturbed spindle formation. Genetics. 2008;179:785–792. doi: 10.1534/genetics.107.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol Syst Biol. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth A, Masuda H, Dhut S, Toda T. Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol Biol Cell. 2008;19:5104–5115. doi: 10.1091/mbc.E08-02-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol. 2009;19:1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Meadows JC, van der Sar SJ, Millar JB, Hardwick KG. Bub3p facilitates spindle checkpoint silencing in fission yeast. Mol Biol Cell. 2009;20:5096–5105. doi: 10.1091/mbc.E09-09-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Wakula P, Beullens M, Ceulemans H, Stalmans W, Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR, 3rd, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Whyte J, Bader JR, Tauhata SB, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe EH, Vaughan PS, Vaughan KT. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windecker H, Langegger M, Heinrich S, Hauf S. Bub1 and Bub3 promote the conversion from monopolar to bipolar chromosome attachment independently of shugoshin. EMBO Rep. 2009;10:1022–1028. doi: 10.1038/embor.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Includes Supplementary Figures 1-6 and Supplementary Tables 1 and 2.