ROOT HAIR DEFECTIVE3 Family of Dynamin-Like GTPases Mediates Homotypic Endoplasmic Reticulum Fusion and Is Essential for Arabidopsis Development (original) (raw)
RHD3 proteins mediate ER fusion and are essential for plant development and the formation of the tubular ER network.
Abstract
In all eukaryotic cells, the endoplasmic reticulum (ER) forms a tubular network whose generation requires the fusion of ER membranes. In Arabidopsis (Arabidopsis thaliana), the membrane-bound GTPase ROOT HAIR DEFECTIVE3 (RHD3) is a potential candidate to mediate ER fusion. In addition, Arabidopsis has two tissue-specific isoforms of RHD3, namely RHD3-like (RL) proteins, and their function is not clear. Here, we show that a null allele of RHD3, rhd3-8, causes growth defects and shortened root hairs. A point mutant, rhd3-1, exhibits a more severe growth phenotype than the null mutant, likely because it exerts a dominant-negative effect on the RL proteins. Genetic analysis reveals that the double deletion of RHD3 and RL1 is lethal and that the rhd3 rl2 plants produce no viable pollen, suggesting that the RL proteins are redundant to RHD3. RHD3 family proteins can replace Sey1p, the homolog of RHD3 in yeast (Saccharomyces cerevisiae), in the maintenance of ER morphology, and they are able to fuse membranes both in vivo and in vitro. Our results suggest that RHD3 proteins mediate ER fusion and are essential for plant development and that the formation of the tubular ER network is of general physiological significance.
In all eukaryotic cells, the endoplasmic reticulum (ER) comprises a continuous membrane system of sheets and tubules (Baumann and Walz, 2001; Shibata et al., 2006). ER tubules frequently connect through homotypic membrane fusion to form a reticular network (Lee and Chen, 1988; Prinz et al., 2000; Du et al., 2004). ER fusion in metazoans is mediated by the atlastins (ATLs), a class of dynamin-like, membrane-bound GTPases (Hu et al., 2009; Orso et al., 2009). ATL possesses a cytoplasmic N-terminal GTPase domain, followed by a helical domain, two closely spaced transmembrane domains, and a C-terminal cytosolic tail. ATL proteins localize mostly to ER tubules and they interact with the tubule-shaping proteins, reticulons and DP1 (Hu et al., 2009). A role for the ATLs in ER fusion is suggested by the fact that depletion of ATLs leads to long, nonbranched ER tubules in cultured cells (Hu et al., 2009) and to ER fragmentation in Drosophila melanogaster (Orso et al., 2009), possibly due to insufficient fusion between the tubules. Nonbranched ER tubules are also observed upon the expression of dominant-negative ATL mutants (Hu et al., 2009). In addition, antibodies to ATL inhibit ER network formation in Xenopus laevis egg extracts (Hu et al., 2009). Moreover, proteoliposomes containing purified D. melanogaster ATL undergo GTP-dependent fusion in vitro (Orso et al., 2009; Bian et al., 2011). The physiological significance of ER fusion is supported by the observation that mutations in human ATL1, the dominant isoform in the brain, cause hereditary spastic paraplegia (Zhao et al., 2001), a neurodegenerative disease characterized by axon shortening in corticospinal motor neurons and progressive spasticity and weakness of the lower limbs (Salinas et al., 2008).
Many organisms lack ATL homologs. In yeast (Saccharomyces cerevisiae), another dynamin-like GTPase, Sey1p, has been found to share the same signature motifs and membrane topology as ATL (Hu et al., 2009). Recent work suggests that Sey1p mediates ER membrane fusion both in vivo and in vitro (Anwar et al., 2012). Cells lacking Sey1p grow normally (Hu et al., 2009), but additional mutation of an ER SNARE Ufe1p, which probably represents an alternative ER fusion mechanism in yeast, causes severe growth defects (Anwar et al., 2012). In Arabidopsis (Arabidopsis thaliana), the potential functional ortholog of ATL appears to be ROOT HAIR DEFECTIVE3 (RHD3; Hu et al., 2009), which was initially discovered by a genetic screen of root hair-defective mutants (Schiefelbein and Somerville, 1990). It is sequence related to Sey1p over the entire length (Wang et al., 1997; Brands and Ho, 2002). Mutations of RHD3 cause short and wavy root hairs (Schiefelbein and Somerville, 1990; Wang et al., 1997; Stefano et al., 2012) and defects in cell expansion (Wang et al., 2002).
Despite the sequence homology between Sey1p and RHD3, it was reported that Sey1p could not replace RHD3 in plants and vice versa (Chen et al., 2011). Therefore, it is not clear whether RHD3 can mediate ER fusion. Another complication in plants is that the Arabidopsis RHD3 family also contains two RHD3-like (RL) proteins (Hu et al., 2003): RL1 is expressed only in pollen, whereas RL2 is expressed ubiquitously, but both are present at very low levels. Deletion of either RL protein causes no detectable defects in root hair development or overall growth (Chen et al., 2011). Whether RL proteins support the role of RHD3 in a tissue-specific manner remains to be investigated.
Here, we have analyzed the function of RHD3 and RL proteins in Arabidopsis. We show that RHD3 and the two RL proteins play redundant roles but function during different stages of Arabidopsis development. In addition, we show that RHD3 proteins can functionally replace Sey1p in yeast and mediate ER membrane fusion.
RESULTS
Characterization of an RHD3 Deletion Mutant
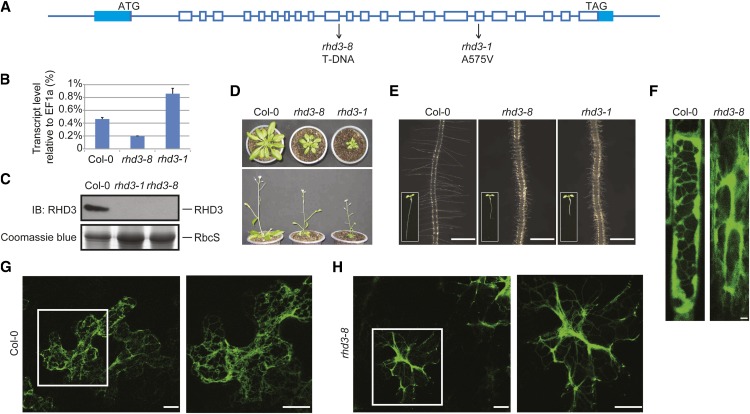
To investigate the precise role of RHD3, we aimed to obtain a null allele of RHD3. Thus, a transfer DNA (T-DNA) insertion line of RHD3 (SALK_025215; rhd3-8; for T-DNA insertion site, see Fig. 1A) was analyzed. The expression of RHD3 in the mutant line was first measured using reverse transcription-PCR and quantitative real-time PCR. As expected, the amount of RHD3 mRNA decreased dramatically in rhd3-8 compared with the wild type (Fig. 1B). To directly measure the levels of RHD3 protein, a polyclonal antibody was raised against RHD3 (Supplemental Fig. S1). Immunoblotting of total cell lysates from plants confirmed a lack of detectable RHD3 protein in rhd3-8 (Fig. 1C). These results indicate that rhd3-8 is a null allele of RHD3.
Figure 1.
Characterization of rhd3-8, a null mutant of RHD3. A, Schematic diagram of the genomic locus of RHD3. Exons and introns are shown in boxes and lines, respectively. Mutation sites for rhd3-8 and rhd3-1 are indicated. B, The transcription levels of RHD3 in wild-type (Columbia [Col-0]), rhd3-8, and rhd3-1 plants were analyzed by reverse transcription and real-time PCR. EF1α was used as an internal control. The data represent means ± sd from three replicates. C, The protein levels of RHD3 in wild-type (Col-0), rhd3-8, and rhd3-1 plants were determined by immunoblotting (IB) with anti-RHD3 antibodies (top panel). Coomassie blue staining of the RuBisCO subunit (RbcS) served as a loading control (bottom panel). D, Mature plants of Col-0, rhd3-8, and rhd3-1 shown in top view (top panel) and side view (bottom panel). E, Root and root hairs of wild-type (Col-0), rhd3-8, and rhd3-1 plants. The corresponding 5-d-old seedlings are shown in insets. Bars = 0.5 mm. F, Wild-type (Col-0) and rhd3-8 root cells in a Q4 line as an ER marker were visualized by fluorescence confocal microscopy. Bars = 2 μm. G, Wild-type (Col-0) and rhd3-8 leaf epidermal cells were transfected with the ER marker ss-GFP-HDEL and were visualized by fluorescence confocal microscopy. The right panels show enlarged views of the highlighted areas. Bars = 10 μm.
Next, we monitored root hair development and overall growth of the RHD3 null mutant. Similar to previously characterized alleles (Schiefelbein and Somerville, 1990; Wang et al., 1997; Stefano et al., 2012), rhd3-8 had short and wavy root hairs, a small rosette, and a dwarf size (Fig. 1, D and E). While rhd3-8 was being characterized, another T-DNA insertion line of RHD3 (SALK_106309; named rhd3-7; Stefano et al., 2012), and likely an RHD3 null allele, was reported. It was shown that rhd3-7 had no detectable RHD3 transcripts and exhibited the typical defects in root hair development as other RHD3 mutants. We obtained consistent results with rhd3-7 and yet another line (SALK_047559; rhd3-9; for T-DNA insertion site, see Supplemental Fig. S2A). Taken together, these findings indicate that complete loss of RHD3 causes defects in root hair and plant development, particularly in cell expansion.
Previous studies have shown that RHD3 mutations cause “cable-like” or “nonbranched” ER tubules (Zheng et al., 2004; Stefano et al., 2012), indicating a lack of fusion between ER tubules. When the ER in rhd3-8 root cells was visualized using Q4 as a marker (Cutler et al., 2000) or leaf epidermal cells transiently transfected with an ER marker (a fusion of a signal sequence with the GFP and HDEL [ss-GFP-HDEL] under the control of the 35S promoter), similar morphology defects were observed (Fig. 1F). These results suggest that RHD3 plays an important role in maintaining ER morphology in plant cells.
To test whether the defects in rhd3-8 can be restored by the expression of RHD3, we transformed the null mutant with a construct expressing RHD3 under the control of its endogenous promoter (Wang et al., 2002). We obtained approximately 100 transformed T1 lines that exhibited normal root hair growth and an improved dwarf phenotype (Supplemental Fig. S3, A and B). Western-blot analysis revealed that the levels of RHD3 in these lines ranged from being barely detectable to being comparable with the wild type (Supplemental Fig. S3C). We also found that the degree of restoration of the wild-type phenotypes correlated with RHD3 abundance. Collectively, these results indicate that the rhd3-8 defects are caused by the loss of RHD3.
To compare the null allele of RHD3 with previously identified point mutants, we obtained rhd3-1 plants, in which Ala-575 in the helical domain is mutated to Val (Fig. 1A; Supplemental Fig. S2B). In contrast to rhd3-8, rhd3-1 exhibited elevated levels of RHD3 transcript (Fig. 1B). Surprisingly, the levels of RHD3 protein were barely detectable in rhd3-1 (Fig. 1C). Thus, the A575V mutation seems to destabilize RHD3. Notably, rhd3-1 exhibited more severe growth defects than that of the null mutant (Fig. 1D). It is likely that residual levels of mutated RHD3 in rhd3-1 have a dominant-negative effect on Arabidopsis development.
RHD3 Proteins Are Essential for Plant Development
Both of the RL proteins have high sequence homology to RHD3 but are expressed in different tissues at low levels. Null alleles of RL1 or RL2 alone have no detectable phenotypes under normal growth conditions (Chen et al., 2011) and exhibit normal ER morphologies (Supplemental Fig. S4, A and B), raising the possibility that despite the similarity to RHD3, the RL proteins may have a redundant or even no role in plant development. To investigate whether the RHD3 family as a whole is essential for Arabidopsis development, we crossed rhd3-8 with T-DNA insertion mutants of RL1 (SALK_048580) or RL2 (SALK_116772). Notably, no rhd3−/− rl1−/− or rhd3−/− rl2−/− plants were recovered in F2 populations, suggesting that double deletion of RHD3 and either RL1 or RL2 leads to lethality.
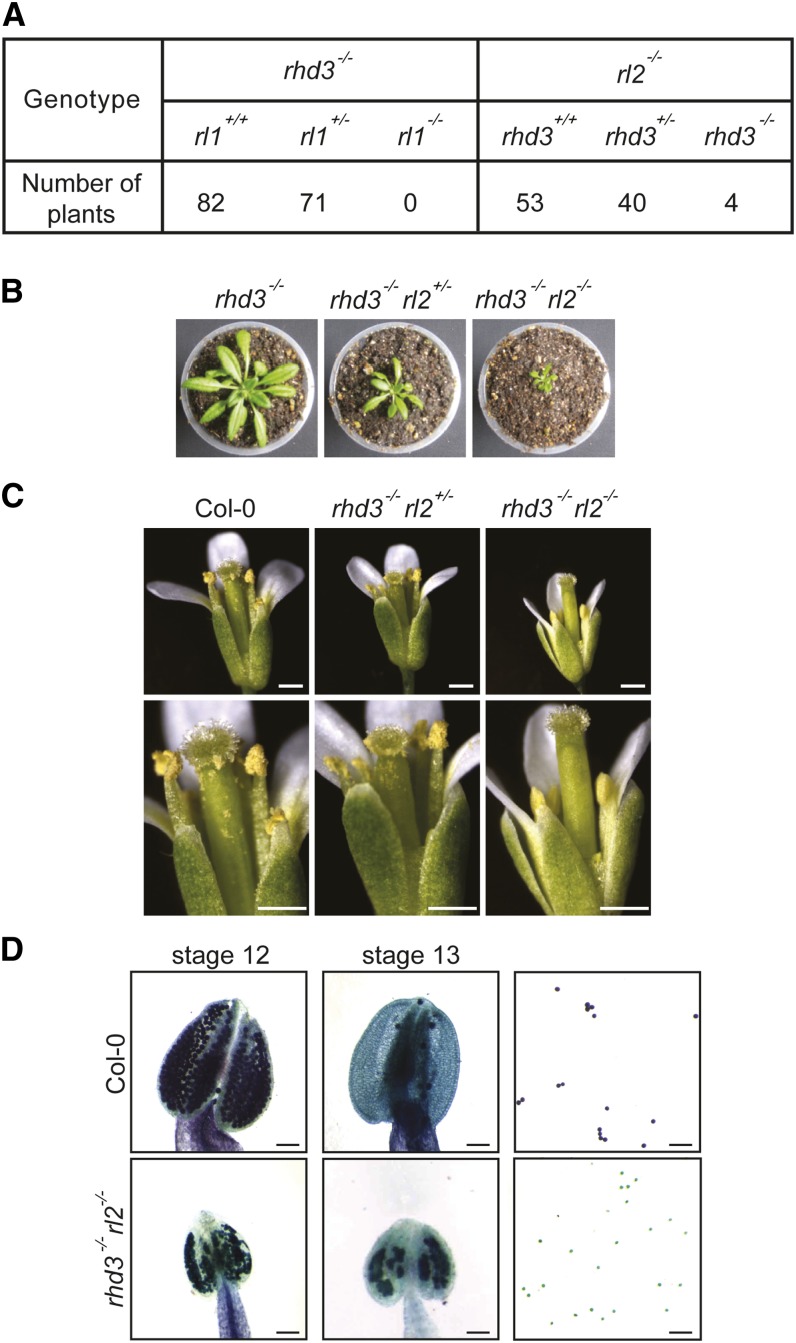
To further test whether rhd3−/− rl1−/− or rhd3−/− rl2−/− is lethal, F2 seeds of rhd3−/− rl1+/− and rhd3+/− rl2−/− plants were collected separately and their F3 seedlings were individually genotyped (Fig. 2A). rhd3−/− rl2−/− lines were recovered only at an abnormally low rate (four out of 97 plants; χ2 = 52.485 and χ test = 5.75E-07), with much more severe growth defects than rhd3−/− plants (Fig. 2B). Further comparison showed that rhd3−/− rl2+/− mutants adopted an intermediate size between rhd3−/− rl2−/− and rhd3−/− (Fig. 2B). These results suggest that RL2 plays a similar role in cell expansion to RHD3 (Wang et al., 2002). Interestingly, we also found that rhd3−/− rl2−/− plants had no seed production. A closer examination of the flowers revealed that their pollen cannot be released from the pollen sac and are not viable (Fig. 2, C and D), even though all floral organs were present (Supplemental Fig. S5A). Pollen production was not affected in either rhd3−/− rl2+/− or rhd3−/− plants (Fig. 2C; Supplemental Fig. S5, B and C). These results suggest that RL2 is required to compensate the loss of RHD3 postembryonically, particularly for cell expansion and pollen development. When seedlings of rhd3−/− rl1+/− were analyzed, we failed to obtain any rhd3−/− rl1−/− (zero out of 153 plants; χ2 = 88.686 and χ test = 9.24E-13). Unlike rhd3−/− rl2+/− plants, rhd3−/− rl1+/− lines were of the same size as rhd3−/− (Supplemental Fig. S5D). Thus, RL1 plays an essential role in supplementing RHD3 function during preembryonic growth. Collectively, our findings suggest that the function of RHD3 family members is required for Arabidopsis development. In addition, RL1 and RL2 both support RHD3’s role in plant growth.
Figure 2.
Genetic interactions between RHD3 and RLs. A, rhd3 was crossed with either rl1 or rl2. F3 seedlings were genotyped and counted. B, Mature plants of rhd3−/−, rhd3−/− rl2+/−, and rhd3−/− rl2−/−. C, The flowers of the wild type (Col-0), rhd3−/− rl2+/−, and rhd3−/− rl2−/− were imaged. The bottom panels show enlarged views. Note that rhd3−/− rl2−/− has no visible pollen grains. Bars = 1 mm. D, The anthers of the wild type (Col-0) and rhd3−/− rl2−/− were collected at stage 12 (left panels) and 13 (middle panels) and were imaged. Pollen viability was analyzed by Alexander staining (right panels). Note that the viable pollen grains are purple and the inactive ones are green. Bars = 100 μm.
To test whether the defects in rhd3 rls mutants are due specifically to the lack of the RHD3/RLs, we performed reciprocal crosses. When mutant stigmas of rhd3 rl2, rhd3 rl1+/−, or rhd3 rl2+/− were pollinated with pollen grains from the wild type, or when they were crossed with line 4 of the rhd3-8 plants expressing RHD3 at an endogenous level, all resulting populations exhibited normal growth and normal pollen development with the expected segregation ratio (Supplemental Fig. S6). These results confirm that the phenotypes of the double mutants are not caused by background effects, but rather, the deficiency of the RHD3 family.
RHD3 Proteins Can Functionally Replace Sey1p
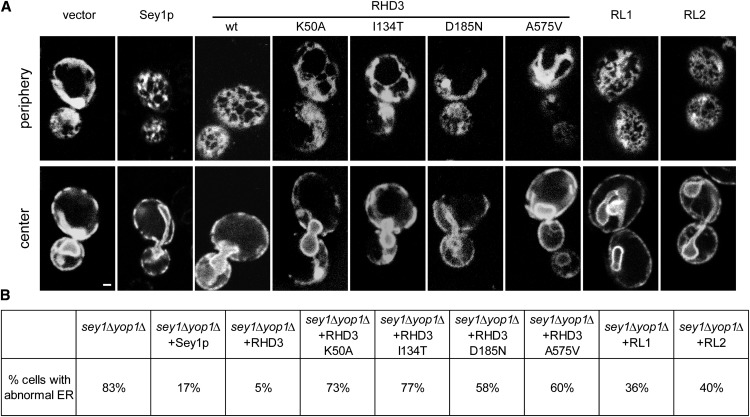
To analyze the function of RHD3, we first tested whether RHD3 can replace Sey1p in S. cerevisiae. We previously showed that yeast cells missing Sey1p and either Yop1p or Rtn1p, proteins that generate high curvature in the membranes of ER tubules (Voeltz et al., 2006; Hu et al., 2008), have abnormal ER morphology with a decreased number of ER tubules, increased number of sheets, and aberrant ER distribution in the cell cortex (Hu et al., 2009). ER morphology can be restored by the expression of wild-type Sey1p or human ATL1 (Hu et al., 2009; Anwar et al., 2012). To test whether RHD3 can replace Sey1p, we expressed wild-type RHD3 in sey1Δyop1Δ cells from a CEN plasmid using the upstream sequence of Sey1p as the promoter (Supplemental Fig. S7). We found that most cells expressing wild-type RHD3 had normal ER morphology (Fig. 3). As expected, when the GTP-binding mutant RHD3 K50A, or the previously identified RHD3 mutants (I134T, rhd3-5; D185N, rhd3-2; A575V, rhd3-1; for mutation sites, see Supplemental Fig. S2B) were expressed, the ER morphology defects were not rescued (Fig. 3). We also found that both RL proteins can partially restore the formation of the tubular ER network in sey1Δyop1Δ cells, even though their expression levels are relatively low (Fig. 3; Supplemental Fig. S8C). These results suggest that members of the RHD3 family and Sey1p have similar functions in the maintenance of ER morphology.
Figure 3.
RHD3 proteins can replace Sey1p in yeast. A, Wild-type (wt) RHD3 proteins or the indicated mutants were expressed under the control of the endogenous SEY1 promoter in S. cerevisiae cells lacking Sey1p and Yop1p (sey1Δ yop1Δ cells). For expression levels, see Supplemental Figure S7. The ER was visualized by expressing Sec63-GFP, focusing the microscope on either the periphery or the center of the cells. sey1Δ yop1Δ cells expressing Sey1p or empty vector were also analyzed for comparison. Bar = 1 μm. B, The percentage of cells with abnormal ER was determined from 80 to 200 cells per mutant.
RHD3 Proteins Can Mediate ER Fusion in Yeast
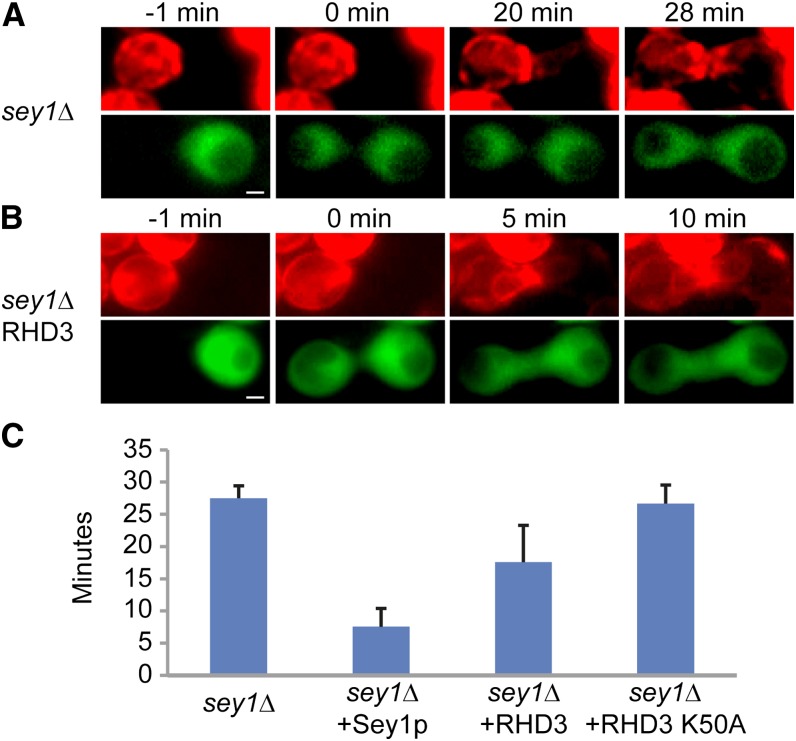
To further test whether RHD3 mediates ER fusion, we used a previously established in vivo assay (Anwar et al., 2012). Haploid yeast cells expressing cytosolic GFP were mated with cells expressing a red fluorescent protein (RFP)-containing ER marker (ss-RFP-HDEL). When cell fusion occurs between two types of cells, the cytosolic GFP of one cell rapidly diffuses to the other cell, marking the starting point for ER fusion. The efficiency of ER fusion was monitored by the equilibration of the RFP signal between two cells. As described previously, cells lacking Sey1p exhibited slow equilibration of the ER marker (an average 27 min after cell fusion; Fig. 4, A and C). The expression of Sey1p in both cell types accelerated the fusion process drastically (Fig. 4C). To test whether RHD3 is capable of mediating ER fusion, we expressed wild-type RHD3 in both GFP- and RFP-labeled cells (Supplemental Fig. S8A). We found that RHD3 significantly enhanced the rate of ER fusion in sey1Δ cells (an average 16 min after cell fusion; Fig. 4, B and C). Such enhancement was not observed when the GTP-binding mutant RHD3 K50A was expressed (Fig. 4C). RL1 and RL2 showed only low activity, likely because of their low expression levels (Supplemental Fig. S8, B and D). Collectively, these results indicate that RHD3 proteins play a similar role in ER fusion to Sey1p.
Figure 4.
ER-ER fusion mediated by RHD3 proteins in yeast. A, sey1Δ cells of opposite mating types expressing either ss-RFP-HDEL or cytosolic GFP were mixed, placed on an agarose pad, and imaged at 1-min intervals. Selected images from the time-lapse video are shown. Time 0 min is the first image taken after cell fusion, as indicated by GFP in both cells. Bar = 2 μm. B, As in A, but with sey1Δ cells expressing wild-type RHD3. C, The average time between cell fusion and ER fusion during mating was determined from eight to 10 cells per sample. Values shown are means ± sd.
RHD3 Proteins Can Mediate Membrane Fusion in Vitro
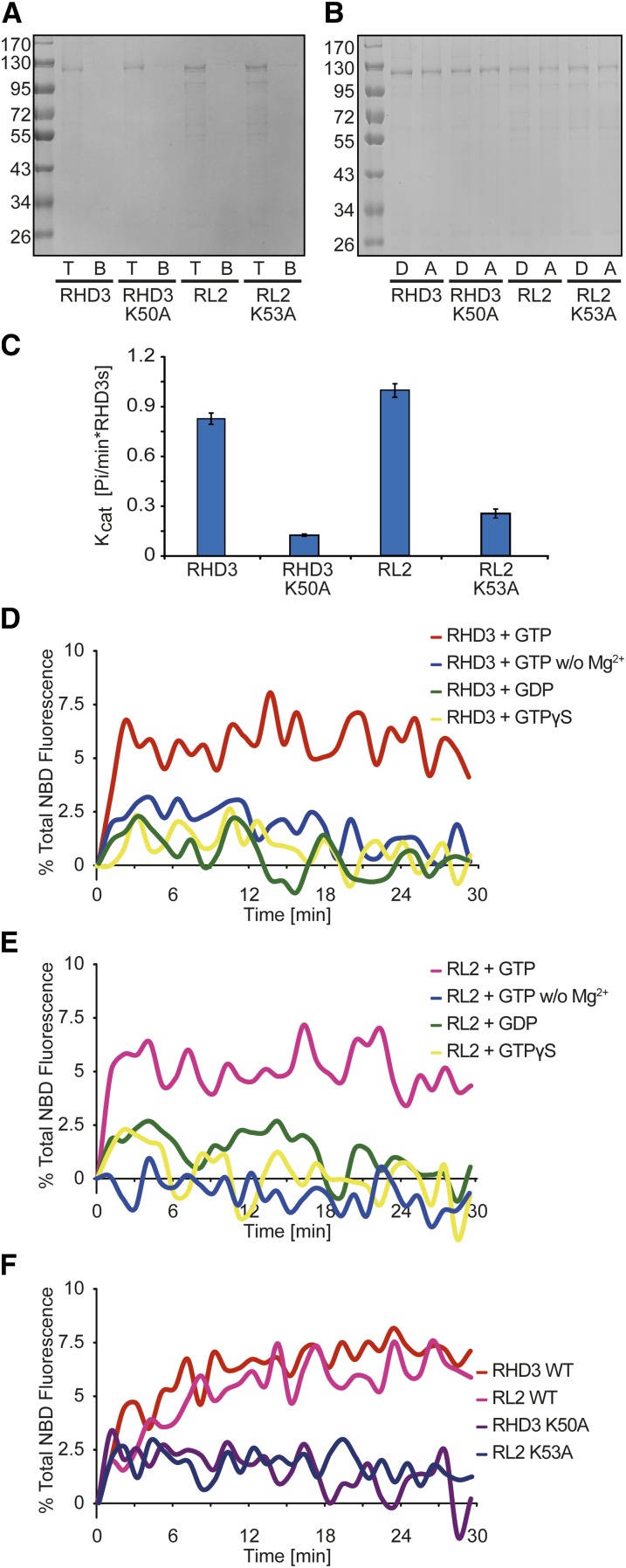
To test whether RHD3 proteins are sufficient to mediate membrane fusion, we performed in vitro lipid-mixing experiments. In this assay, the donor vesicles contained lipids labeled with nitrobenzoxadiazole (NBD) and rhodamine at quenching concentrations; fusion with the unlabeled acceptor vesicles leads to fluorophore dilution and dequenching. Full-length RHD3 or RL2 were expressed as glutathione _S_-transferase fusion proteins in Escherichia coli. After purification, the proteins were reconstituted into donor and acceptor vesicles (Fig. 5A). Efficient reconstitution was confirmed by flotation experiments (Fig. 5B). The GTPase activity of RHD3 proteins was also measured (Fig. 5C). When tested in the lipid-mixing assay, wild-type RHD3 and RL2 caused fusion of the vesicles (Fig. 5, D and E). As shown previously with ATL and Sey1p, no fusion was observed in the absence of magnesium ions or when GTP was replaced by GDP or the nonhydrolyzable analog GTPγS (Fig. 5, D and E). A mutation in the conserved P loop (K50A in RHD3 or K53A in RL2), which affects GTP binding, abolished the fusion activity of the RHD3 proteins (Fig. 5F). Taken together, these results demonstrate that RHD3 proteins are capable of mediating membrane fusion.
Figure 5.
RHD3 proteins mediate membrane fusion in vitro. A, RHD3, RL2, or the indicated mutants were purified and reconstituted into proteoliposomes. Flotation in a sucrose gradient (right) shows efficient reconstitution of the proteins (T, top fraction; B, bottom fraction). B, Donor (D; with NBD- and rhodamine-labeled lipids at quenching concentrations) and acceptor (A; unlabeled) proteoliposomes containing RHD3, RL2, or the indicated mutants were analyzed by SDS-PAGE and Coomassie blue staining. C, GTPase activity of full-length wild-type RHD3, RL2, or the indicated mutants was measured by phosphate release. The data represent means ± sd from six replicates. Kcat, Catalytic constant. D, Full-length RHD3 was reconstituted at equal concentrations into donor and acceptor vesicles. Fusion was monitored by the dequenching of the NBD-labeled lipids present in the donor vesicles and was initiated by the addition of GTP. Control experiments were performed in the absence of Mg2+ or the presence of GDP or GTPγS instead of GTP. E, As in D, but with full-length RL2. F, Fusion with RHD3 or RL2 was compared with that of the indicated GTP-binding mutants. WT, Wild type.
DISCUSSION
Our in vivo and in vitro results strongly support the notion that RHD3 mediates homotypic ER fusion. Morphological defects in the ER of RHD3 mutant cells imply the involvement of RHD3 in ER fusion (Zheng et al., 2004). Surprisingly, a recent report suggested that RHD3 and Sey1p are not interchangeable (Chen et al., 2011). It was shown that RHD3 failed to restore ER morphology in sey1Δyop1Δ cells (Chen et al., 2011), although the expression of RHD3 in yeast was driven by an exogenous promoter and the expression level was not determined. In contrast, we have shown here that RHD3 is able to rescue the ER defects in sey1Δyop1Δ cells. The fusion activity of RHD3 in mating yeast cells and in reconstituted proteoliposomes provides compelling evidence that RHD3 mediates ER fusion and is functionally analogous to the ATLs and to Sey1p. Chen et al. (2011) also showed that Sey1p was not able to complement rhd3-1. It is possible that an excess amount of Sey1p is required to maintain proper development in plants, especially in the presence of mutated RHD3. Alternatively, RHD3 could possess unique features that are essential for its function in plants.
Recent structural and biochemical studies have suggested that ATL-mediated fusion starts with GTP binding to ATL molecules sitting in two apposing membranes, which induces dimerization and membrane tethering; GTP hydrolysis and inorganic phosphate release triggers conformational changes that pull the two membranes together, eventually leading to membrane fusion (Bian et al., 2011; Byrnes and Sondermann, 2011). Additional investigation revealed that the C-terminal tail and transmembrane domains of ATL also play an important role in the fusion process (Liu et al., 2012). The analogous domain structure and signature motifs between RHD3 and ATLs suggest that the fusion process driven by RHD3 is presumably very similar to that of ATLs. In addition, the C-terminal tail of RHD3 is predicted to have an amphipathic helix, a feature in ATLs that appears to be important for fusion activity (Liu et al., 2012). In fact, RHD3 proteins lacking the tail are inactive in vivo (Stefano et al., 2012). However, the helical domain that follows the GTPase domain is much longer in RHD3 than in ATLs, so the exact conformational change between the GTPase and the helical domain in RHD3 remains to be determined.
The RHD3 family in Arabidopsis consists of RHD3 and two RL proteins. RL proteins may function similarly to RHD3, since overexpression of RL2 rescued the RHD3 mutant (Chen et al., 2011). We showed that, like RHD3, RL proteins complement the loss of Sey1p in yeast cells and are able to mediate fusion in yeast cells and in vitro. Importantly, RHD3 is much more abundant than RL proteins; RHD3 deletion causes prominent growth defects, whereas deletion of RLs, individually (Chen et al., 2011) or together (Supplemental Fig. S6), resulted in no detectable phenotype. Thus, RHD3 plays a dominant role, while the RL proteins are auxiliary. In Arabidopsis, it is quite common that GTPases possess several minor isoforms or family members with tissue-specific expression (Vernoud et al., 2003). Our genetic analysis suggests that the presence of both RL proteins is critical in rhd3 cells. These findings suggest that the retained ER fusion detected in rhd3-7 cells (Stefano et al., 2012) is likely mediated by RL proteins. It is also plausible to speculate that the increased growth defects observed in rhd3-1, compared with rhd3-8, are due to a dominant-negative effect by mutated RHD3 toward the RL proteins.
As ATL1 mutations affect mostly the axons of upper motor neurons and rhd3 causes root hair defects, the maintenance of ER integrity by GTP-dependent ER fusogens may be particularly important in cells with long protrusions (Hu et al., 2009, 2011). Our double deletion experiments indicate that other cell types, such as pollen, could also be affected drastically. These findings suggest that the tubular ER network is of general importance.
MATERIALS AND METHODS
Molecular Cloning and Antibodies
RHD3, RL1, and RL2 were amplified from the complementary DNA of wild-type Arabidopsis (Arabidopsis thaliana) plants, and the endogenous promoter of RHD3 was amplified from genomic DNA. For expression in yeast (Saccharomyces cerevisiae), hemagglutinin (HA) tagged-RHD3 or HA-RL was subcloned into pYC2/CT (a URA3/CEN plasmid with the original GAL promoter deleted) along with the endogenous promoter and terminator of SEY1 or into pESC-URA (a 2μ plasmid with the GAL promoter). For expression in plants, HA-RHD3 with the endogenous promoter was subcloned into pCAMBIA 1301. For expression in bacteria, RHD3 or RL2 with a C-terminal Strep(II) tag was inserted into pGEX-6P-1. All point mutations were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and confirmed by DNA sequencing. The purified fragment of RHD3 (residues 273–558) was used to raise polyclonal antibodies in rabbits (GL Biochem).
Plant Materials
All seeds of mutant plants were obtained from the Arabidopsis Biological Resource Center (Ohio State University). rhd3-8 expressing HA-RHD3 was generated by _Agrobacterium tumefaciens_-mediated floral dipping (Clough and Bent, 1998) transformation of corresponding plasmid into rhd3-8 plants. Transgenic plants were selected on one-half-strength Murashige and Skoog medium containing 25 µg L−1 hygromycin. Plants were grown either on one-half-strength Murashige and Skoog medium or in the soil at 20°C to 22°C under 16-h-light/8-h-dark conditions.
Microscopy
Yeast cells expressing Sec63p-GFP and root cells from the plant ER marker line Q4 were analyzed with a Leica TCS SP5 confocal microscope for ER morphology. For alternative ER visualization in Arabidopsis, A. tumefaciens strain GV3101 expressing ss-HDEL-GFP5 or p19 was grown to an optical density at 600 nm (OD600) of 1.0 to 1.2. Cells were collected and resuspended in the infiltration medium (10 mm MES, 10 mm MgCl2, and 200 μm acetosyringone) to an OD600 of 0.4 to 0.6 for ss-HDEL-GFP5 cells and 0.8 to 1.0 for p19 cells. These cultures were then mixed in a 1:1 ratio (v/v) and used to infiltrate leaves of 5-week-old healthy plants by 1-mL syringes without a needle. Following infiltration, plants were cultivated for another 5 to 7 d before confocal analysis. For cytochemical analysis, pollen were stained by dipping flowers several times in 4′,6-diamino-phenylindole or Alexander solution (Alexander, 1969) or directly on microscope slides with 0.3 μg mL−1 propidium iodide and 0.5 μg mL−1 fluorescein diacetate (Regan and Moffatt, 1990). A Leica M165 FC was used for bright-field imaging of root hairs, flowers, and pollen. In vivo fusion assay was performed with a Zeiss Axio imager Z1. All changes were made linearly across the entire image using Adobe Photoshop.
In Vivo Fusion Assay
Yeast in vivo fusion assays were performed as described previously (Anwar et al., 2012). In brief, cells of opposite mating types expressing either ss-RFP-HDEL or cytosolic GFP were grown to an OD600 of 0.1 to 0.4 and mixed to allow mating. They were then placed on an agarose pad and imaged at 1-min intervals.
Recombinant RHD3 and RL2 Production
Glutathione _S_-transferase- and Strep(II)-tagged full-length RHD3 or RL2 was expressed in the Escherichia coli strain Rosetta DE3 (Novagen). Cells were grown in superbroth medium at 37°C. When cultures reached an OD600 of 0.6, protein expression was induced by 0.5 mm isopropylthio-β-galactoside for 12 h at 24°C. The cells were harvested and lysed by sonication in buffer containing 50 mm Tris, pH 8.0, 500 mm NaCl, 2 mm β-mercaptoethanol, and protease inhibitor cocktail (Roche). Total membranes were sedimented by ultracentrifugation at 40,000 rpm for 30 min, and the pellet was homogenized and solubilized in the buffer containing 2% (v/v) Triton X-100 for 2 h. The Strep(II)-tagged proteins were then isolated from the cell extracts by StrepTactin Sepharose beads (GE Healthcare), and bound recombinant proteins were washed with buffer containing 0.1% (v/v) Triton X-100 and eluted with 2.5 mm _d_-desthiobiotin. The _d_-desthiobiotin was removed using a desalting column (GE Healthcare), and the purified protein was concentrated to 1 mg mL−1 for subsequent analysis.
GTPase Activity Assay
GTPase activity was determined using the EnzChek phosphate assay kit (Invitrogen). Reactions were performed in a 100-μL volume and initiated by the addition of 0.5 mm GTP. Optical density at 360 nm was measured at 1-min intervals for 30 min at 37°C. The catalytic constant was determined using three different protein concentrations (1, 2, and 5 μm).
Lipid-Mixing Assay
The in vitro lipid-mixing assay was performed as described previously (Bian et al., 2011). In brief, labeled donor proteoliposomes were mixed with unlabeled acceptor proteoliposomes in a total volume of 100 μL per reaction. NBD fluorescence was measured at 1-min intervals at 37°C. After 30 min, 5 μL of 10% (w/v) _n_-Dodecyl β-D-Maltopyranoside was added to determine total NBD fluorescence. Fusion is expressed as the percentage of total fluorescence.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RHD3, At3g13870; RL1, At1g72960; RL2, At5g45160.
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure S1. A polyclonal antibody against RHD3.
- Supplemental Figure S2. Sites of the RHD3 mutations.
- Supplemental Figure S3. Expression of RHD3 rescues rhd3-8.
- Supplemental Figure S4. ER morphology of RLs mutants.
- Supplemental Figure S5. Plants of rhd3 crossed with rl1 or rl2.
- Supplemental Figure S6. Reciprocal crosses for RHD3 RLs double mutants.
- Supplemental Figure S7. Protein expression levels in the assay shown in Fig. 3.
- Supplemental Figure S8. Expression and in vivo fusion by RHD3 proteins.
Acknowledgments
We thank W. Prinz for materials, S. Men and T. Zhang for technical assistance, the Arabidopsis Biological Resource Center for the Arabidopsis mutants, and T. Rapoport and T. Liu for critically reading the manuscript.
Glossary
ER
endoplasmic reticulum
ATL
atlastin
T-DNA
transfer DNA
RFP
red fluorescent protein
HA
hemagglutinin
OD600
optical density at 600 nm
Col-0
Columbia
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA. (2012) The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol 197: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Walz B. (2001) Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205: 149–214 [DOI] [PubMed] [Google Scholar]
- Bian X, Klemm RW, Liu TY, Zhang M, Sun S, Sui X, Liu X, Rapoport TA, Hu J. (2011) Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA 108: 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands A, Ho TH. (2002) Function of a plant stress-induced gene, HVA22: synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol 130: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes LJ, Sondermann H. (2011) Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci USA 108: 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Stefano G, Brandizzi F, Zheng H. (2011) Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J Cell Sci 124: 2241–2252 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Ferro-Novick S, Novick P. (2004) Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci 117: 2871–2878 [DOI] [PubMed] [Google Scholar]
- Hu J, Prinz WA, Rapoport TA. (2011) Weaving the web of ER tubules. Cell 147: 1226–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. (2008) Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. (2009) A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhong R, Morrison WH, III, Ye ZH. (2003) The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217: 912–921 [DOI] [PubMed] [Google Scholar]
- Lee C, Chen LB. (1988) Dynamic behavior of endoplasmic reticulum in living cells. Cell 54: 37–46 [DOI] [PubMed] [Google Scholar]
- Liu TY, Bian X, Sun S, Hu X, Klemm RW, Prinz WA, Rapoport TA, Hu J. (2012) Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc Natl Acad Sci USA 109: E2146–E2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA**, et al** (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460: 978–983 [DOI] [PubMed] [Google Scholar]
- Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. (2000) Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol 150: 461–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan SM, Moffatt BA. (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas S, Proukakis C, Crosby A, Warner TT. (2008) Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol 7: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA. (2006) Rough sheets and smooth tubules. Cell 126: 435–439 [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Moss T, McNew JA, Brandizzi F. (2012) In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J 69: 957–966 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Wang H, Lee MM, Schiefelbein JW. (2002) Regulation of the cell expansion gene RHD3 during Arabidopsis development. Plant Physiol 129: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. (1997) The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev 11: 799–811 [DOI] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L**, et al** (2001) Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 29: 326–331 [DOI] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I. (2004) A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J 37: 398–414 [DOI] [PubMed] [Google Scholar]