Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy (original) (raw)
Significance
β-Amyloid aggregates are often found in the brains of Alzheimer’s patients. We have previously reported that β-amyloid levels can be down-regulated through activation of autophagy, a process involving the degradation of unnecessary or harmful proteins. However, the underlying mechanism of β-amyloid autophagy-mediated degradation is unknown. Here we demonstrate that the complex adaptor protein 2/phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (AP2/PICALM), via an interaction with a β-amyloid precursor, bridges β-amyloid degradation and autophagy. This work reveals mechanistic steps for the targeting of the β-amyloid precursor for degradation via autophagy and supports a genome-wide association study identifying PICALM as a risk factor for Alzheimer’s disease (AD). Altogether, these findings support the notion that activating autophagy is a valid approach for the AD field, which urgently needs novel therapeutic strategies.
Keywords: endocytosis, trafficking, aggregate removal
Abstract
The hallmarks of Alzheimer’s disease (AD) are the aggregates of amyloid-β (Aβ) peptides and tau protein. Autophagy is a major cellular pathway leading to the removal of aggregated proteins. We have reported recently that autophagy was responsible for amyloid precursor protein cleaved C-terminal fragment (APP-CTF) degradation and amyloid β clearance in an Atg5-dependent manner. Here we aimed to elucidate the molecular mechanism by which autophagy mediates the degradation of APP-CTF and the clearance of amyloid β. Through affinity purification followed by mass spectrum analysis, we identified adaptor protein (AP) 2 together with phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) as binding proteins of microtubule-associated protein 1 light chain 3 (LC3). Further analysis showed that AP2 regulated the cellular levels of APP-CTF. Knockdown of AP2 reduced autophagy-mediated APP-CTF degradation. Immunoprecipitation and live imaging analysis demonstrated that AP2 and PICALM cross-link LC3 with APP-CTF. These data suggest that the AP-2/PICALM complex functions as an autophagic cargo receptor for the recognition and shipment of APP-CTF from the endocytic pathway to the LC3-marked autophagic degradation pathway. This molecular mechanism linking AP2/PICALM and AD is consistent with genetic evidence indicating a role for PICALM as a risk factor for AD.
A hallmark pathological feature of Alzheimer’s disease (AD) is the amyloid plaque made of aggregated amyloid β (Aβ) peptides. Among genetic mutations identified in familial AD patients, large numbers are associated with accumulation of Aβ peptides. These peptides are generated via sequential proteolysis of amyloid precursor protein (APP) during the course of its trafficking along the secretory pathway. APP is sequentially matured in the endoplasmic reticulum (ER) and Golgi apparatus and then delivered to the plasma membrane through the trans-Golgi networks. Within minutes of arrival at the cell surface, APP is internalized in clathrin-coated vesicles through endocytosis (1) via a tetrapeptide motif, YENP, located at the carboxyl terminus of APP (2). The internalized APP is then delivered to endosomes, where it is processed first by β-secretase, also known as BACE1, and then by γ-secretase, to generate Aβ peptides (1, 3).
Adaptor protein 2 (AP2) is a well-characterized complex involved in clathrin-mediated endocytosis. It is heterotetrameric and consists of four subunits AP2A1, AP2B1, AP2M1, and AP2S1 (α, β, μ, and σ). It was suggested that the α subunit, AP2A1, mediates the binding to the plasma membrane and acts as a scaffold for endocytic accessory proteins. The μ subunit, AP2M1, is responsible for cargo selection through the recognition of a YxxФ signal (where Ф is a bulky hydrophobic residue) in the cytosolic region of type I transmembrane proteins during endocytosis (4–6). Phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) is a cytoplasmic adaptor protein that also plays a critical role in clathrin-mediated endocytosis. It binds to clathrin, phosphatidylinositol, and AP2 to aid in the formation of clathrin-coated pits (7–9). PICALM was also identified by genomewide association studies (GWAS) as a significant risk factor for AD (10). However, the pathogenic mechanism for PICALM as an AD risk factor is unclear.
Macroautophagy, hereafter referred to as autophagy, is a highly conserved catabolic process in which proteins and organelles are engulfed in double-membraned vacuoles called autophagosomes and then transported to lysosomes for degradation. The membrane origins of autophagosomes are unclear but may include ER (11–13), mitochondria (14), and plasma membrane (15). During the formation of autophagosomes, the small cytosolic ubiquitin-like microtubule-associated protein 1 light chain 3 (LC3-I) is processed and conjugated to phosphatidylethanolamine (PE) to form lipidated LC3 (LC3-II). LC3-II is specifically targeted to elongating preautophagosomal structures and remains on mature autophagosomes until it is degraded by the fusion to lysosomes (16). Lipidated LC3-II serves as a docking site for specific cargo receptors, such as SQSTM1/p62, NBR1, Nix, NDP52, and OPTN (17–21). They all bind to LC3 through an LC3-interacting region (LIR) to recruit specific cargos for degradation.
We and others have reported that autophagy regulates the levels of Aβ peptides (22–24). Furthermore, we showed that the clearance of Aβ peptides results from the degradation of the precursor amyloid precursor protein cleaved C-terminal fragment (APP-CTF) via autophagy. However, the molecular mechanism by which autophagy leads to the down-regulation of the membrane-bound APP-CTF and, in turn, of Aβ is not known. We report here that AP2 functions as an LC3 receptor, which shuttles APP-CTF from the endocytic pathway to autophagosomes for degradation.
Results
Identification of AP2 as an Interacting Partner of LC3.
We have previously shown that APP-CTF is degraded through autophagy. To understand the underlying mechanism, we sought to identify factors that interact with LC3 and, therefore, might mediate the targeting of APP-CTFs to autophagosomes. Affinity purification followed by mass spectrometry (MS) analysis was performed in HeLa cells stably expressing LC3 fused with eGFP (eGFP-LC3). As shown by the heatmap of MS analysis in Fig. 1_A_, various polypeptides were shown to be associated with LC3. Interestingly, a large number of the LC3 interacting factors identified are involved in intracellular trafficking, such as clathrin heavy chain 1 (CLTC), clathrin heavy chain like 1 (CLTCL1), clathrin interactor 1 (CLINT1), adaptor protein α and β subunit (AP2A1/A2/B1), phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (PIK3C2A), FYVE and coiled-coil domain containing 1 (FYCO1), and protein transport protein Sec16A (SEC16A). Sequestosome 1 (SQSTM1, also named p62), a previously known LC3 partner, was also recovered by our MS analysis (Fig. 1_A_).
Fig. 1.
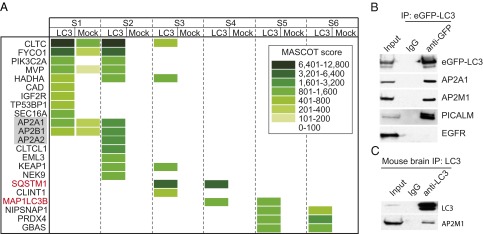
Identification of AP2 as an LC3 binding protein. (A) HeLa cells stably expressing eGFP-LC3 were lysed and immunoprecipitated (IP) with mouse IgG or monoclonal GFP antibody. Eluates were resolved by SDS/PAGE and stained by colonial blue. Six bands (S1 to S6) from top to bottom of each IP were subjected to liquid chromatography-mass spectrometry. Proteins associated with LC3 were displayed as a heatmap based on their MASCOT score. The presence of the same polypeptides found in mock IP is also shown. (B) IP experiments with monoclonal GFP antibody were carried out as in A. (C) IP experiments with LC3 antibody using brain extracts from AD double transgenic mice, followed by immunoblotting using antibodies as indicated.
Given the well-established role of the AP2 complex in endocytosis, a critical event for the processing of APP, we chose to focus on this complex. The association of AP2A1 with LC3 was confirmed through immunoprecipitation (IP) experiments by using an anti-GFP antibody and HeLa cells expressing eGFP-LC3 lysates (Fig. 1_B_). AP2M1, another subunit of AP2, as well as PICALM, a protein reported to interact with AP2 and play a role in AP2-mediated endocytosis, were also confirmed to bind to LC3 (Fig. 1_B_). Epidermal growth factor receptor (EGFR), which also undergoes endocytosis like APP, was used as a negative control and did not coimmunoprecipitate with LC3 (Fig. 1_B_). The interaction of AP2M1 and LC3 was further confirmed through IP with an antibody against endogenous LC3 using the same AD double transgenic mice (APPswe/PS1ΔE9) (Fig. 1_C_) as the ones used for APP-CTF IPs (see Fig. 3_C_).
Fig. 3.
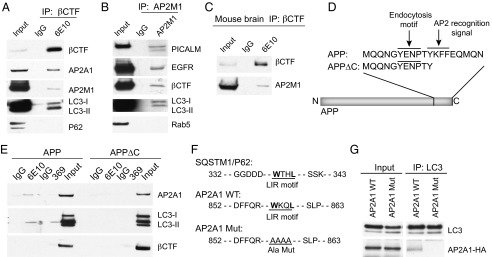
AP2 interacts with APP-βCTF and LC3. (A) N2a cells stably expressing βCTF were lysed and immunoprecipitated with mouse IgG or 6E10 antibody and then analyzed by immunoblotting with corresponding antibodies. (B) IP was performed on the same cell lysates by using AP2M1 antibody. (C) IP experiments using brain extracts from AD double transgenic mice, using 6E10 antibody (N terminus of βCTF). (D) Schematic representation of the YxxФ motif deletion in APP C-terminal region. (E) N2a cells transiently transfected with APP or APP∆C were lysed and immunoprecipitations carried out with 6E10 or RU-369 antibodies, followed by SDS/PAGE and immunoblotting using AP2A1, LC3, and 6E10 antibodies. (F) Alignment of the LIR motifs of SQSTM1/P62 and AP2A1 is presented. The putative LIR motif of AP2A1 was mutated to alanine repeats in the AP2A1 Mut construct. (G) N2a cells were transiently transfected with HA-tagged AP2A1-WT or AP2A1 Mut constructs. Cell lysates were immunoprecipitated with LC3 antibody and immunoblotted with HA antibody.
AP2 Is Required for the Cellular Turnover of APP-CTF.
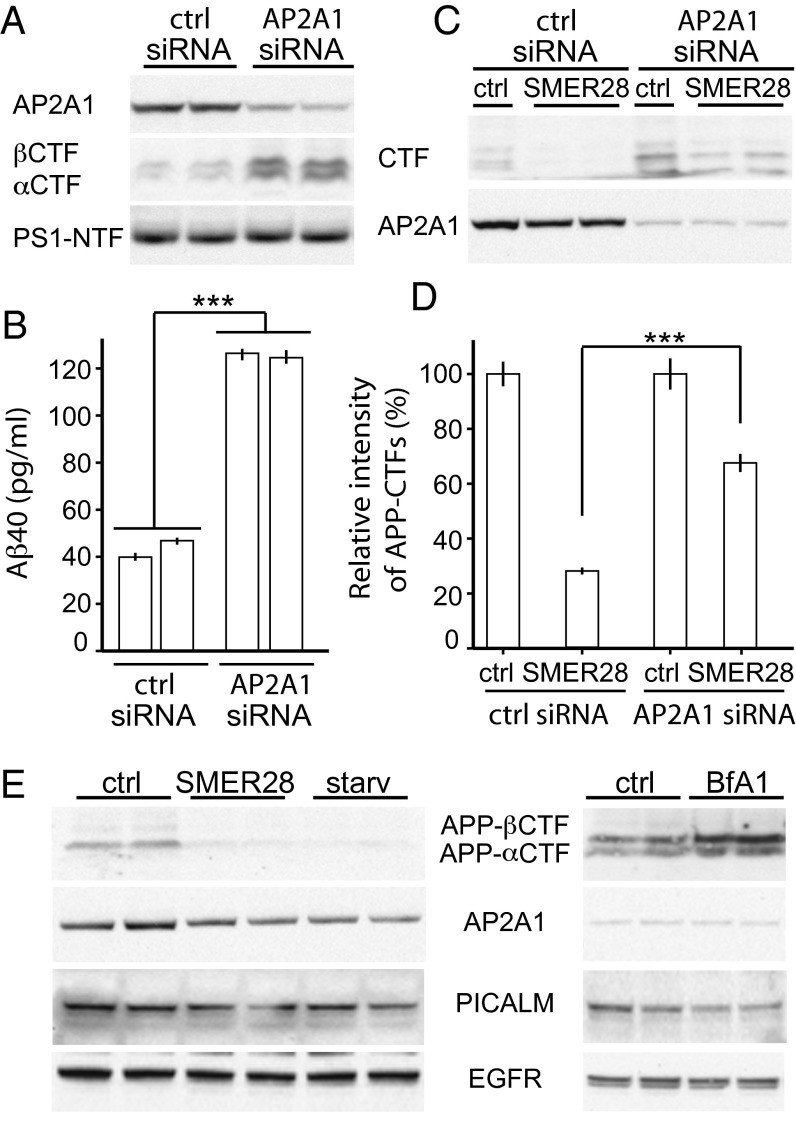
Prompted by the importance of AP2 in endocytosis and the role that endocytosis plays in the regulation of the proteolytic processing of APP, we next asked whether AP2 is involved in the regulation of APP-CTF levels. To this end, using siRNA, we silenced AP2A1 in N2a cells stably expressing APP at full length, and found that knockdown of AP2A1 led to a dramatic increase in the levels of APP-CTF (both APP-βCTF and APP-αCTF) (Fig. 2_A_). The accumulation of APP-CTF was unlikely mediated by γ-secretase, because no change in the level of PS1-NTF, the catalytic subunit of γ-secretase, was detected upon AP2A1 knockdown (Fig. 2_A_). A similar up-regulation of Aβ40 peptide production was seen in the AP2A1 knocked-down cells (Fig. 2_B_). It has been reported that inhibition of APP endocytosis by mutation of its internalization signal “YENP” increased soluble sAPPα fragment secretion and reduced Aβ production (3). This accumulation of APP-CTF and Aβ40 peptide upon knockdown of AP2A1 strongly suggested that AP2 not only functioned upstream at the endocytosis level, but also downstream of the production of APP-βCTF. With its ability to interact with LC3 (Fig. 1), we asked whether AP2 is involved in autophagy-mediated degradation of APP-CTF. Upon treatment with small molecule enhancer of rapamycin 28 (SMER28), APP-CTF was dramatically diminished in N2a APP cells (Fig. 2_C_), consistent with our previous finding (24). However, when AP2A1 expression was reduced by siRNA, the down-regulation of APP-CTFs by SMER28 was largely inhibited (Fig. 2_C_). Quantification of the level of APP-CTF showed that, although SMER28 treatment resulted in an approximate 80% reduction in APP-CTF in cells treated with control siRNA, it led to a lesser decrease after AP2A1 knockdown (approximately 40%) (Fig. 2_D_). These results suggest that AP2 function is required for the degradation of APP-CTF by autophagy. We next tested whether AP2 and PICALM themselves are regulated by autophagy. The levels of APP-CTFs were greatly reduced upon autophagy induction by using SMER28 or starvation, but not the levels of EGFR, AP2, or PICALM (Fig. 2_E_). Furthermore, Bafilomycin A1 treatment, which inhibits autophagy, led to an accumulation of APP-CTFs but not EGFR, AP2A1, or PICALM (Fig. 2_E_). Taken together, these results indicate that although AP2 and PICALM bind to LC3, they do not seem to be direct substrates of autophagy.
Fig. 2.
AP2 regulates protein levels of APP-CTF. N2a cells stably expressing APP were used and transfected with either control or AP2A1 siRNA for 48 h. (A) Cell lysates were analyzed by SDS/PAGE and Western blotting using antibodies as indicated. APP-CTFs were detected by using RU-369 antibody (C terminus of APP). (B) Aβ40 production was measured by ELISA using the supernatant. (C) Cells were then incubated with SMER28 (50 µM) for 16 h. Cell lysates were then analyzed by immunoblotting using RU-396 and AP2A1 antibodies. (D) Quantification of APP-CTF levels from repeated experiments as in C (n ≥ 3; ***P < 0.001; error bars represent SEM). (E) Cells were starved for 2 h or treated with SMER28 (50 μM) for 16, or treated with Bafilomycin (BfA1) (200 ng/mL) for 2 h. Cell lysates were analyzed by SDS/PAGE and Western blotting using antibodies as indicated.
AP2 Cross-Links LC3 with APP-CTF.
Previous studies have shown that several factors, including SQSTM1/p62, function as bridging molecules to deliver certain cargos to autophagosomes for degradation via LC3. We next addressed the possibility that AP2 plays a similar role in the autophagy-mediated degradation of APP-CTFs by targeting APP-CTF–containing vacuoles to LC3-marked autophagosomes. We first examined whether AP2 interacts with APP-CTFs. As shown in Fig. 3_A_, IP of βCTF using 6E10 antibody in N2a cells stably expressing βCTF recovered AP2A1 and AP2M1, two subunits of AP2, together with LC3, but not SQSTM1/p62 (Fig. 3_A_). In addition, using AP2M1 antibody to IP endogenous AP2M1, we found it interacted with PICALM, EGFR, βCTF, and LC3 (Fig. 3_B_). Furthermore, the interaction of βCTF and AP2M1 was also detected in AD double transgenic mouse brain extracts (Fig. 3_C_).
The ability of AP2 to interact with βCTF and with LC3 suggests the possibility that these proteins may form a complex. In fact, this hypothesis is supported by subcellular fractionation of N2a APP cells, which showed that AP2A1, LC3-II, and APP-CTF localized in the same subcellular compartment (Fig. S1_A_). In addition, glycerol gradient analysis of the FLAG affinity purified LC3 complexes showed colocalization of AP2A1 and AP2M1 with a portion of LC3 as well as APP-CTFs (Fig. S1_B_). To further confirm the interaction of AP2, βCTF and LC3, we took advantage of the existence of a conserved tetra-peptide motif, “YKFF,” as an AP2 recognition signal on the C terminus of APP (Fig. 3_D_). We generated constructs with a deletion of a short C-terminal region containing this motif, named APPΔC. IP experiments were then performed in cells transfected with APPΔC or full-length APP (referred as APP). Whereas APP was able to coimmunoprecipitate with AP2A1 by using either 6E10 antibody or RU-369 antibody, which respectively recognize the _N_- and C-terminal regions of APP-βCTF, APPΔC failed to do so (Fig. 3_E_), indicating that this YKFF motif is required for the interaction between APP-CTFs and AP2. More importantly, APPΔC lost its ability to interact with LC3 in contrast to APP (Fig. 3_E_), further emphasizing the requirement of AP2 in complex formation to link APP-CTF to LC3. To rule out the possibility that deletion of the YKFF motif affected APPΔC endocytosis and, therefore, its binding to LC3, we performed cell-surface biotinylation experiments of APP and APPΔC. When APP and APPΔC were expressed at the same levels, the same amounts of proteins were biotinylated on the cell surface (Fig. S2), indicating that deletion of the YKFF motif did not interfere with endocytosis of APPΔC.
Most of the known autophagy receptors and adaptors bind to LC3 via an LIR containing a consensus sequence “W/Y/F-X-X-I/L” (17–21). For example, a “WTHL” sequence is present in SQSTM1/p62 (Fig. 3_F_). We found that AP2A1 contains a similar LIR motif, which is “WKQL” (Fig. 3_F_). Once this motif in AP2A1 was mutated to alanine repeats (Fig. 3_E_, referred to as AP2A1 Mut), LC3 could no longer coimmunoprecipitate with HA-tagged AP2A1 (Fig. 3_G_). Taken together, our biochemical analysis delineated a sequence in AP2 that serves as an intermediate to link APP-CTFs and LC3, therefore potentially facilitating the fusion of APP-CTF–residing vesicles with LC3-containing autophagosomes.
AP2 Mediates Endosome–Autophagosome Fusion.
Having established AP2 as a bridging molecule to mediate the interaction between LC3 and APP-CTF by IP, we next aimed to understand how the interaction of LC3 with AP2M1 and PICALM was affected by starvation-induced autophagy. HeLa cells were analyzed by IP experiments using anti-GFP antibody. The interaction between LC3 and AP2M1 as well as PICALM showed a significant increase triggered by starvation (Fig. 4_A_). Thus, AP2 appears to function as an adaptor to tether APP-CTF–containing vesicles and LC3-containing autophagosomes, promoting their fusion and, in turn, the degradation of APP-CTF.
Fig. 4.
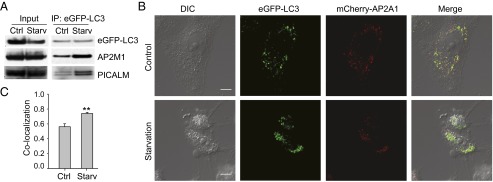
Starvation enhances LC3 and AP2 interaction (A) and colocalization (B and C). (A) HeLa cells stably expressing eGFP-LC3 were used. Cells were starved or not for 2 h, and lysates were then used for IP (eGFP antibody). Immunoblots are presented as indicated. (B) Cells were transiently transfected with mCherry-AP2A1 and treated with complete medium or starvation medium for 2 h. Images were taken during the second hour of starvation by live-cell confocal microscopy. The micrographs presented are representative from three independent experiments. Two additional sets of images are shown in Fig. S4. (Scale bar: 10 μm.) (C) Quantification of Pearson's colocalization coefficient showed statistically significant difference in GFP-LC3 and mCherry-AP2A1 colocalization with or without serum starvation (n = 15–18; error bars represent SEM; **P < 0.001, Mann–Whitney u test).
We next investigated the intracellular localization of LC3 and AP2M1 in living cells as described (25). Confocal images of HeLa cells stably expressing eGFP-LC3 (Fig. S3, Upper) or transiently transfected with mCherry-tagged AP2M1 (Fig. S3, Lower) displayed punctate structures throughout the cytoplasm. Importantly, we could observe efficient cytosolic localization in both eGFP-LC3 and mCherry-AP2A1 overexpression cases, demonstrating that eGFP-LC3 and mCherry-AP2A1 were delivered to the appropriate subcellular localization where autophagy is active.
To further validate that starvation-induced autophagy modulates the binding between AP2 and LC3 in living cells, colocalization of mCherry-AP2A1 and eGFP-LC3 were investigated by dual-channel live-cell confocal microscopy (Fig. 4_B_). Visual inspection of HeLa cells coexpressing mCherry-AP2A1 and eGFP-LC3 without starvation showed moderate levels of colocalization (Fig. 4_B, Upper_). Colocalization was also confirmed by quantification of Pearson’s correlation coefficient (PCC = 0.56 ± 0.04, mean ± SEM). Remarkably, as shown in Fig. 4_B, Lower_, and consistent with our biochemical data (Fig. 4_A_), starvation yielded a significant increase (Fig. 4_C_) in mCherry-AP2A1 and eGFP-LC3 colocalization (PCC = 0.74 ± 0.02, mean ± SEM). An additional two sets of images yielding similar results are shown in Fig. S4. To ensure that the increased colocalization is not random but a synchronized movement as one would expect in autophagosome trafficking, we performed time-lapse imaging (Fig. 5 A and B). Indeed, we were able to follow the synchronized movement of mCherry-AP2A1 and eGFP-LC3 for at least 200 s (Fig. 5_C_ and Movie S1). Taken together, the enhanced interaction between LC3 and AP2 as well as PICALM upon induction of autophagy by starvation provides a mechanistic explanation as to how autophagy leads to down-regulation of APP-CTFs (Fig. S5).
Fig. 5.
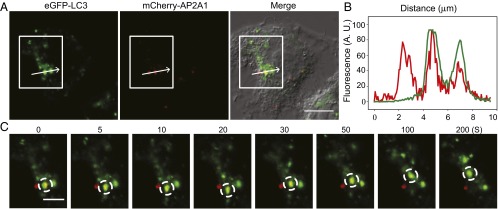
Starvation-induced autophagy mediates LC3 and AP2 colocalization with time-scale on the order of a few hundred seconds. (A) Representative images of live HeLa cells expressing eGFP-LC3 and mCherry-AP2A1 upon 1-h serum starvation treatment. The white box highlights the colocalization of eGFP-LC3 and mCherry-AP2A1. (Scale bar: 10 μm.) (B) Fluorescent intensity profiles of the arrow in the white box indicate the position of a line scan, where two of three mCherry-AP2A1 (red line) punctate structures displayed high colocalization with eGFP-LC3 (green line). (C) Time-lapse images of the white box (A) showing that eGFP-LC3 and mCherry-AP2A1 present an increased colocalization following starvation. Note that the synchronized movement of eGFP-LC3 and mCherry-AP2A1 (dashed circles) lasted for at least 200 s. (Scale bar: 5 μm.)
Discussion
Our previous work showed that APP-CTF and Aβ peptides can be targeted for removal by autophagy in an Atg5-dependent manner (24). A small molecule enhancer of autophagy (SMER28) promotes this process and reduces the levels of Aβ (24). Autophagy is known to degrade intracellular aggregation-prone proteins. How autophagy reduces membrane-bound APP-CTF and secreted Aβ peptides is largely unknown. The present work identified AP2 as a mediator that bridges the APP endocytic pathway with the autophagic pathway. AP2 binds to the APP C terminus during endocytosis and brings APP-CTF to autophagosomes via direct binding to LC3 through LIR. Therefore, AP2, functioning as an LC3 receptor, specifically targets APP-CTF to autophagy for degradation. Most APP-βCTF is produced in the endocytic pathway (1). When endocytic APP-βCTF is degraded by autophagy, Aβ levels are greatly reduced. It has been known that AP complexes are important for vesicular transport and cargo selection (26). Therefore, it is not surprising to find that AP2 is used as an LC3 cargo receptor.
Recent mounting evidence has indicated that autophagy degradation is more selective than initially thought (27). The supporting evidence includes the continuing discovery of specific autophagy receptors that are responsible for recruiting specific cargos to the site of autophagosomes. Thus far, several autophagy receptors have been identified, such as SQSTM1/p62, NBR1, Nix, NDP52, and OPTN (17–21). Indeed, it has been shown that they regulate the selective degradation of damaged organelles, protein aggregates, and pathogens. Our discovery of AP2 as another autophagy receptor, which selectively mediates the degradation of APP-CTF, further supports the notion of targeted elimination of unwanted components by autophagy. This study also proposes a mechanism by which autophagy is involved in Aβ removal. Impaired or disabled autophagy has been linked to various human pathologies, including neurodegenerative diseases (28). In our study, we found that PICALM, a known binding partner of AP2 involved in clathrin-mediated endocytosis, was also recruited to LC3 marked autophagosomes along with AP2 and APP-CTF. Because enhanced autophagy increases the binding of PICALM to autophagosomes, we speculate that PICALM might have an important function in the clearance of APP-βCTF and, in turn, in the clearance of Aβ via autophagy. However, many questions remain to be resolved, including the precise role of PICALM in the bridging of APP-CTF to autophagy. This work, together with the fact that PICALM was identified as a risk factor for AD by GWAS (10), highlights the crucial role of PICALM in APP metabolism and opens a therapeutic avenue for AD intervention.
Materials and Methods
Reagents.
Antibodies were diluted 1:1,000 in 5% (wt/vol) milk unless specified. Commercially available antibodies are listed in SI Materials and Methods. RU-369, a rabbit polyclonal antibody that recognizes the C-terminal of APP695 (29); Ab14 antiserum targeting residues 1–25 of PS1-NTF (30); and monoclonal GFP antibody was produced by the monoclonal antibody core facility at Memorial Sloan–Kettering Cancer Center. Compound SMER28 was purchased from EMD Chemicals.
Cell Culture and siRNA.
N2a cells stably expressing APP were maintained in medium containing 50% DMEM and 50% Opti-MEM, supplemented with 5% FBS (Invitrogen) plus 400 μg/mL geneticin. HeLa cells stably expressing eGFP-LC3 were maintained in DMEM with 10% FBS plus 400 μg/mL geneticin. The siRNA for AP2A1 was purchased from Dharmacon (On-TARGETplus Set of 4 siRNAs J-055895-05). The control siRNA was purchased from Dharmacon (On-TARGET plus GAPD Control siRNA D-001830-02-05).
Coimmunoprecipitation.
All coimmunoprecipitation experiments using cell lysates or brain lysates were performed by using the co-Immunoprecipitation Kit from Invitrogen (Invitrogen) according to manufacturer’s protocol. Briefly, antibodies conjugated to Dynabeads were incubated with lysates solubilized in a lysis buffer [0.5% Triton (Sigma-Aldrich, St. Louis)] for 30 min.
Subcellular Fractionation.
For sucrose density gradient fractionation, cells were prepared as described (31). Briefly, cells were homogenized by using a stainless steel ball-bearing homogenizer. Homogenates were loaded on top of a step sucrose gradient (1 mL at 2 M, 4 mL at 1.3 M, 3.5 mL at 1.16 M, and 2.0 mL at 0.8 M). Gradients were centrifuged (2.5 h at 390,000 × g), and 1-mL fractions were collected and assayed by Western blot.
Microscopy.
Live-cell confocal images were obtained on a Leica TCS SP8 confocal imaging system equipped with a 63×/1.4 numerical aperture oil-immersion objective lens. The temperature-controlled stage of the confocal microscope was maintained at 32–35 °C. Cells for live-cell imaging were seeded on poly-d-lysine–coated MatTek glass bottom culture dishes. Microscopy setup and imaging acquisition were performed as described (32). Time-lapse fluorescent images were acquired at 5-s intervals, following the movement of eGFP-LC3 and mCherry-AP2A1. To minimize photodamage and photobleaching, we restricted time-lapsed confocal imaging to a period of 300 s.
Statistical Analyses for Colocalization.
A minimum of 15 sets of micrographs from three independent experiments were used for colocalization analyses. Pearson's colocalization coefficient was calculated by using Image-Pro Plus (Version 6.0). Mann–Whitney test was performed to determine statistical significance. Data were analyzed in SigmaPlot (Version 11.0).
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. Kaye Thomas and Alison North for technical assistance; Drs. Aviva Tolkovsky and Zhengyu Yue for providing the HeLa eGFP-LC3 cell line; and Drs. Victor Bustos, John Steele, and William Netzer for critical discussion. This work was supported by the Fisher Center for Alzheimer’s Research Foundation and National Institutes of Health Grant AG09464 (to P.G. and M.F.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2012;2(5):a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez RG, et al. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274(27):18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- 3.Koo EH, Squazzo SL, Selkoe DJ, Koo CH. Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J Cell Sci. 1996;109(Pt 5):991–998. doi: 10.1242/jcs.109.5.991. [DOI] [PubMed] [Google Scholar]
- 4.Canfield WM, Johnson KF, Ye RD, Gregory W, Kornfeld S. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24-29 of the cytoplasmic tail. J Biol Chem. 1991;266(9):5682–5688. [PubMed] [Google Scholar]
- 5.Jadot M, Canfield WM, Gregory W, Kornfeld S. Characterization of the signal for rapid internalization of the bovine mannose 6-phosphate/insulin-like growth factor-II receptor. J Biol Chem. 1992;267(16):11069–11077. [PubMed] [Google Scholar]
- 6.Ohno H, et al. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269(5232):1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 7.Dreyling MH, et al. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA. 1996;93(10):4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tebar F, Bohlander SK, Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: Localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell. 1999;10(8):2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyerholz A, et al. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6(12):1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 10.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi-Nishino M, et al. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2010;6(2):301–303. doi: 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- 13.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5(8):1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 14.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15(2):231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Mostowy S, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286(30):26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11(1):45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 21.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vingtdeux V, et al. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-β peptide degradation. FASEB J. 2011;25(1):219–231. doi: 10.1096/fj.10-167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spilman P, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS ONE. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Y, Bustos V, Flajolet M, Greengard P. A small-molecule enhancer of autophagy decreases levels of Abeta and APP-CTF via Atg5-dependent autophagy pathway. FASEB J. 2011;25(6):1934–1942. doi: 10.1096/fj.10-175158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: From autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1(1):23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 26.Nakatsu F, Ohno H. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct Funct. 2003;28(5):419–429. doi: 10.1247/csf.28.419. [DOI] [PubMed] [Google Scholar]
- 27.Fimia GM, Kroemer G, Piacentini M. Molecular mechanisms of selective autophagy. Cell Death Differ. 2013;20(1):1–2. doi: 10.1038/cdd.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe DM, et al. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37(12):1949–1961. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Netzer WJ, et al. Lowering beta-amyloid levels rescues learning and memory in a Down syndrome mouse model. PLoS ONE. 2010;5(6):e10943. doi: 10.1371/journal.pone.0010943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thinakaran G, et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17(1):181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, et al. Presenilins and gamma-secretase inhibitors affect intracellular trafficking and cell surface localization of the gamma-secretase complex components. J Biol Chem. 2004;279(39):40560–40566. doi: 10.1074/jbc.M404345200. [DOI] [PubMed] [Google Scholar]
- 32.Chang JC, et al. Single molecule analysis of serotonin transporter regulation using antagonist-conjugated quantum dots reveals restricted, p38 MAPK-dependent mobilization underlying uptake activation. J Neurosci. 2012;32(26):8919–8929. doi: 10.1523/JNEUROSCI.0048-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information