Novel Bat Coronaviruses, Brazil and Mexico (original) (raw)
To the Editor: Bats are now recognized as natural reservoirs for many families of viruses that can cross species barriers and cause emerging diseases of humans and animals. Protecting humans against emerging diseases relies on identifying natural reservoirs for such viruses and surveillance for host-jumping events. The emergence of the Middle East respiratory syndrome coronavirus (MERS-CoV) on the Arabian Peninsula (1) further justifies increased surveillance for coronaviruses (CoVs) in bats. MERS-CoV most likely is a zoonotic virus from a bat reservoir and is associated with high case-fatality rates among humans. The existence of a diverse array of alphacoronaviruses in bats in the United States, Canada, and Trinidad has been reported (2–6). Recently, a possible new alphacoronavirus was detected in an urban roost of bats in southern Brazil (7), and a survey of bats in southern Mexico reported 8 novel alphacoronaviruses and 4 novel betacoronaviruses, 1 with 96% similarity to MERS-CoV (8). These findings expand the diversity and range of known bat coronaviruses and increase the known reservoir for potential emerging zoonotic CoVs.
Expanding on our previous work (2,3), we analyzed samples from 97 bats from Brazil and 75 bats from Mexico (Technical Appendix). During 2007–2010, intestinal samples were collected from bats of 10 species in northwest São Paulo state in southeastern Brazil. These bats had been submitted to the University Estadual Paulista for rabies testing as a result of epidemiologic surveillance or, in some cases, because of possible or known contact with humans. During 2011–2012, as part of an ongoing rabies surveillance project, intestinal samples were collected from bats of 12 species in their usual habitats in Jalisco state in midwestern Mexico. Bats from a variety of species, including insectivorous, nectarivorous, frugivorous, and hematophagous bats, were included in this study for the purpose of obtaining a diverse array of potential exposures. Intestines were collected and stored, and RNA was purified as described (2). CoV RNA was detected by using a pancoronavirus PCR selective for the RNA-dependent RNA polymerase gene, and amplicons were sequenced as described (3). Virus isolation was not attempted as part of this study.
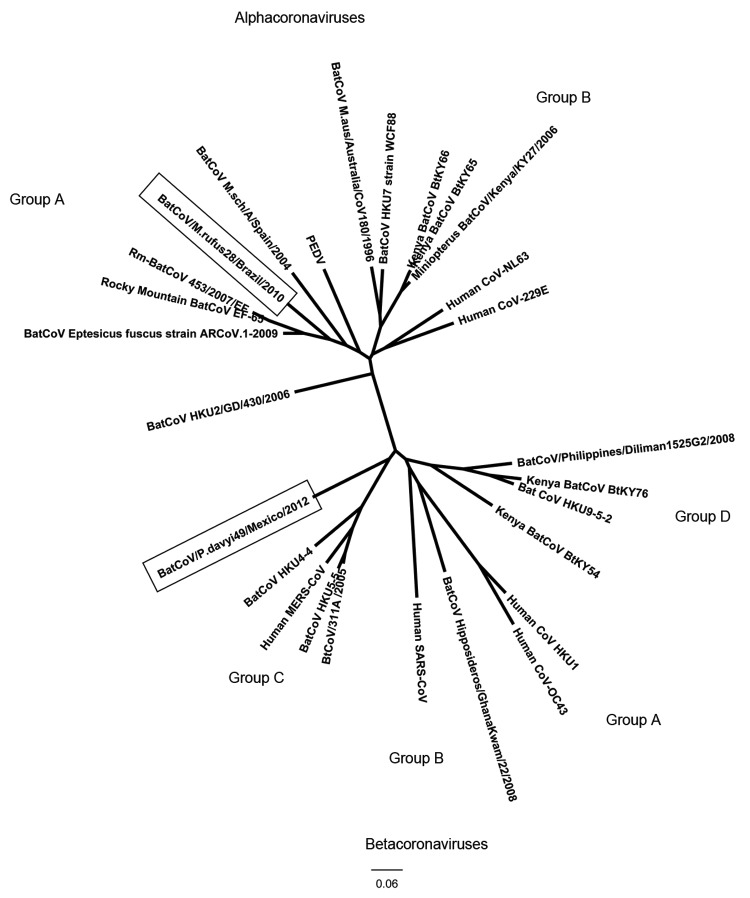
From 1 of 17 Molossus rufus bats and 1 of 8 Molossus molossus bats, an identical novel alphacoronavirus was detected (BatCoV-_M.rufus_28/Brazil/2010, GenBank accession no. KC886321). Both specimens were collected in Brazil during 2010 from adult male bats that had been found in urban areas on residential property. The 412-nt sequence of this virus was most closely related to alphacoronaviruses detected in Eptesicus fuscus bats in North America (82% nt identity), Myotis australis bats in Australia (77% nt identity), Miniopterus bats in Kenya (77% nt identity), and Rhinolophus bats in Hong Kong (77% nt identity) (Figure). Bats of the genus Molossus are insectivorous; their geographic range is restricted to the New World, from northern Mexico to northern Argentina.
Figure.
Phylogenetic tree showing relationships based on 412-nt and 439-nt sequences of a conserved region of gene 1b of BatCoV/Molossus rufus28/Brazil/2010 (alphacoronavirus) and BatCoV/Pteronotus davyi49/Mexico/2012 (betacoronavirus) to other known coronaviruses. Sequences were aligned by using ClustalW (www.clustal.org/), phylogenetic analyses were conducted by using the neighbor-joining method and BioEdit (www.mbio.ncsu.edu/BioEdit/BioEdit.html), and trees were constructed by using FigTree version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). Boxes surround the novel alphacoronavirus detected in Molossus rufus and M. molossus bat specimens from São Paulo state in southeastern Brazil, and the novel betacoronavirus detected in a specimen from a Pteronotus davyi bat from Jalisco state in midwestern Mexico. GenBank accession numbers aree AB539081.1, DQ648808.1, GU065420.1, HM211099.1, EF065512.1, EF065512.1, JX869059.2, GU065398.1, EF507794.1, FJ710054.1, JX537914.1, EF544566.1, EU834956.1, EF203064.1, GU065410.1, HQ728484.1, GU065409.1, HQ184049.1, DQ666339.1, HQ336974.1, DQ445911.1, KC210147.1, AY278741, NC_002645.1, and NC_005147.1. PEDV, porcine epidemic diarrhea virus. Scale bar indicates nucleotide substitutions per site.
A novel betacoronavirus (presumably group C) was detected in a specimen from 1 of 4 Pteronotus davyi bats (BatCoV-_P.davyi_49/Mexico/2012, GenBank accession no. KC886322). This specimen was collected in 2012 from an adult male bat roosting in a cave in La Huerta, Mexico. The 439-nt sequence of this virus has 71% nt identity to the novel human group C betacoronavirus MERS-CoV and 72% nt identity to various group D betacoronaviruses detected in Rousettus, Pipistrellus, and Tylonycteris bats in the Philippines, China, and Kenya (Figure). Bats of the species P. davyi (Davy’s naked-backed bat) are insectivorous and are found from southern Mexico to the northern parts of South America. They prefer to roost in caves and man-made structures, such as mines.
In summary, we found a novel alphacoronavirus in bats from Brazil and a novel betacoronavirus in a bat from Mexico. Both viruses were detected in bats with known or potential contact with humans. Because the bats we sampled were mostly adult males, the prevalence of CoVs that we identified is probably an underestimation of the true incidence of CoVs in these bat populations. For bats of other species, incidence of CoVs among juvenile and female bats is higher (2,9). Furthermore, we used a non-nested, broadly conserved CoV PCR, which might have limited the sensitivity of CoV RNA detection. The finding of a novel betacoronavirus in insectivorous bats in the New World is noteworthy. Three human CoVs (229E, SARS-CoV, and MERS-CoV) all have animal reservoirs of closely related viruses in Old World insectivorous bats (10) from which they most likely emerged, either directly or indirectly, into the human population. Ongoing surveillance for CoVs in wildlife and increased research efforts to better understand the factors associated with CoV host-switching events are warranted.
Technical Appendix
Results of reverse transcription PCR analysis of coronaviruses in bats from Brazil and Mexico.
Acknowledgments
We thank Anaís Ruesga, Heriberto Verduzco M., Adrian Gutierrez A., Edgar Gómez, Carol Alexis, Jazmín Osorio, and Jansen de Araujo for help with capturing and dissecting bats. We thank Kathryn Holmes and Thomas M. Yuill for critically reviewing this manuscript and advising and facilitating this collaboration.
This study was funded by National Institutes of Health grant no. K08 AI-073525 (S.R.D.), by a fellowship from the Consejo Nacional para Ciencia y Tecnología of the Mexican federal government (S.G.R.), and by a fellowship from the Brazilian government agencies Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Conselho Nacional de Desenvolvimento Científico e Tecnológico (L.G.B.G.).
Suggested citation for this article: Góes LGB, Ruvalcaba SG, Campos AA, Queiroz LH, de Carvalho C, Jerez JA, et al. Novel bat coronaviruses, Brazil and Mexico [letter]. Emerg Infect Dis [Internet]. 2013 Oct [_date cited_]. http://dx.doi.org/10.3201/eid1910.130525
1
These authors contributed equally to this article.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20 . 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 2.Osborne C, Cryan PM, O'Shea TJ, Oko LM, Ndaluka C, Calisher CH, et al. Alphacoronaviruses in New World bats: prevalence, persistence, phylogeny, and potential for interaction with humans. PLoS ONE. 2011;6:e19156 . 10.1371/journal.pone.0019156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis. 2007;13:1295–300. 10.3201/eid1309.070491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington CV, Foster JE, Zhu HC, Zhang JX, Smith GJ, Thompson N, et al. Detection and phylogenetic analysis of group 1 coronaviruses in South American bats. Emerg Infect Dis. 2008;14:1890–3. 10.3201/eid1412.080642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. Metagenomic analysis of the virome of three North American bat species: viral diversity between different bat species that share a common habitat. J Virol. 2010;84:13004–18. 10.1128/JVI.01255-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misra V, Dumonceaux T, Dubois J, Willis C, Nadin-Davis S, Severini A, et al. Detection of polyoma and corona viruses in bats of Canada. J Gen Virol. 2009;90:2015–22. 10.1099/vir.0.010694-0 [DOI] [PubMed] [Google Scholar]
- 7.Lima FE, Campos FS, Filho H, Batista HB, Junior P, Cibulski SP, et al. Detection of Alphacoronavirus in velvety free-tailed bats (Molossus molossus) and Brazilian free-tailed bats (Tadarida brasiliensis) from urban areas of southern Brazil. Virus Genes. 2013. Mar 16. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 8.Anthony SJ, Ojeda-Flores R, Rico-Chavez O, Navarrete-Macias I, Zambrana-Torrelio C, Rostal MK, et al. Coronaviruses in bats from Mexico. J Gen Virol. 2013;94:1028–38. 10.1099/vir.0.049759-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gloza-Rausch F, Ipsen A, Seebens A, Gottsche M, Panning M, Felix Drexler J, et al. Detection and prevalence patterns of group I coronaviruses in bats, northern Germany. Emerg Infect Dis. 2008;14:626–31. 10.3201/eid1404.071439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Annan A, Baldwin H, Corman V, Klose S, Owusu M, Enkrumah E, et al. Human betacoronavirus 2C EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–9 . 10.3201/eid1903.121503 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Technical Appendix
Results of reverse transcription PCR analysis of coronaviruses in bats from Brazil and Mexico.