S6K1−/−/S6K2−/− Mice Exhibit Perinatal Lethality and Rapamycin-Sensitive 5′-Terminal Oligopyrimidine mRNA Translation and Reveal a Mitogen-Activated Protein Kinase-Dependent S6 Kinase Pathway (original) (raw)
Abstract
Activation of 40S ribosomal protein S6 kinases (S6Ks) is mediated by anabolic signals triggered by hormones, growth factors, and nutrients. Stimulation by any of these agents is inhibited by the bacterial macrolide rapamycin, which binds to and inactivates the mammalian target of rapamycin, an S6K kinase. In mammals, two genes encoding homologous S6Ks, S6K1 and S6K2, have been identified. Here we show that mice deficient for S6K1 or S6K2 are born at the expected Mendelian ratio. Compared to wild-type mice, S6K1−/− mice are significantly smaller, whereas _S6K2_−/− mice tend to be slightly larger. However, mice lacking both genes showed a sharp reduction in viability due to perinatal lethality. Analysis of S6 phosphorylation in the cytoplasm and nucleoli of cells derived from the distinct S6K genotypes suggests that both kinases are required for full S6 phosphorylation but that S6K2 may be more prevalent in contributing to this response. Despite the impairment of S6 phosphorylation in cells from _S6K1_−/−/_S6K2_−/− mice, cell cycle progression and the translation of 5′-terminal oligopyrimidine mRNAs were still modulated by mitogens in a rapamycin-dependent manner. Thus, the absence of S6K1 and S6K2 profoundly impairs animal viability but does not seem to affect the proliferative responses of these cell types. Unexpectedly, in _S6K1_−/−/_S6K2_−/− cells, S6 phosphorylation persisted at serines 235 and 236, the first two sites phosphorylated in response to mitogens. In these cells, as well as in rapamycin-treated wild-type, _S6K1_−/−, and _S6K2_−/− cells, this step was catalyzed by a mitogen-activated protein kinase (MAPK)-dependent kinase, most likely p90rsk. These data reveal a redundancy between the S6K and the MAPK pathways in mediating early S6 phosphorylation in response to mitogens.
Recent studies showed that the 40S ribosomal protein S6 kinase (S6K) p70S6K/p85S6K, termed S6K1 (51), is a major effector of cell growth. This conclusion stems from gene deletion studies with Drosophila (39) and with mice (51) as well as recent studies with cell cultures (11). The loss of the Drosophila S6K (dS6K) gene is semilethal, with the few surviving adults having a severely reduced body size. The larvae of such flies exhibit a long developmental delay, consistent with a twofold increase in cell cycle doubling times. The few surviving adults are quite lethargic, living no longer than 2 weeks, and females are sterile. Surprisingly, the reduction in mass is strictly due to a decrease in cell size rather than to a decrease in cell number (39).
In mice, removal of this kinase is not lethal, but the mice are approximately 20% smaller at birth (51). Such mice exhibit normal fasting glucose levels but are mildly glucose intolerant due to markedly reduced levels of circulating insulin (42). Reduced insulin levels are caused by a reduction in pancreatic endocrine mass and an impairment of insulin secretion, which can be traced to a selective reduction in β-cell size. Unexpectedly, the effects on body mass and hypoinsulinemia do not appear to be attributable to a reduction in S6 phosphorylation, as this response proved to be largely intact in S6K1-deficient animals (51). However, S6 phosphorylation in such animals was still sensitive to the bacterial macrolide rapamycin (51), which inhibits the mammalian target of rapamycin (mTOR) (1, 7, 16, 48), the upstream S6K1 kinase (4, 8, 18), suggesting the existence of a second S6K. Subsequent searches of expressed sequence tag databases and biochemical studies led to the identification of S6K2, which exhibited overall homology of over 80% with S6K1 in the highly conserved kinase and linker domains (17, 47, 51). In all tissues examined from S6K1-deficient mice, S6K2 transcripts were upregulated (51). From this observation, it was reasoned that S6K1 and S6K2 functions were redundant and that a deletion of the S6K1 gene led to a compensatory increase in the expression of S6K2.
In parallel studies, it was demonstrated that rapamycin suppressed the serum-induced translational upregulation of a family of mRNAs which contain a polypyrimidine tract at their 5′ end (5′-terminal oligopyrimidine [5′TOP] mRNAs) (20, 55). These mRNAs largely code for components of the translational apparatus, most notably, ribosomal proteins (37). Earlier studies had shown that the translation of such transcripts is under selective translational control (22) and requires an intact 5′TOP tract (19, 49). In addition, a dominant interfering allele of S6K1 inhibited the mitogen-induced translational upregulation of 5′TOP mRNAs to the same extent as rapamycin, whereas an activated allele of S6K1, which exhibits a substantial degree of rapamycin resistance, largely protected these transcripts from the inhibitory effects of rapamycin (19, 49).
Seemingly consistent with these arguments, in embryonic stem (ES) cells from which S6K1 had been homologously deleted by selection with high doses of G418, serum no longer had an effect on the upregulation of 5′TOP mRNAs, nor was there a redistribution of 5′TOP mRNAs from polysomes to nonpolysomes in the presence of rapamycin (24). However, S6 phosphorylation was initially reported to be abolished in these cells (24), despite the fact that it was largely intact in cells and tissues derived from S6K1−/− mice (51). This difference seemed to be resolved in subsequent studies, where S6 phosphorylation was detected in these same _S6K1_−/− ES cells and S6K2 was present and active (31, 60). Despite these observations, it was again recently reported that S6 phosphorylation was absent from these same cells (53). Furthermore, it was also claimed in the latter study that S6K activation, S6 phosphorylation, and rapamycin had little impact on 5′TOP mRNA translation in PC12 cells (53), although others working with these same cells had reported earlier that rapamycin treatment abolished the selective recruitment of these transcripts from small to large polysomes (44).
Obviously, cells lacking both S6K1 and S6K2 would facilitate such studies. Therefore, we set out to delete the S6K2 gene from mice and to determine whether we could generate _S6K1_−/−/_S6K2_−/− mice. Here we report on the deletion of the S6K2 gene and the effects of deleting both S6K1 and S6K2 on animal growth and viability as well as on S6 phosphorylation, cell proliferation, and 5′TOP mRNA translation.
MATERIALS AND METHODS
Generation of an S6K2-targeted allele.
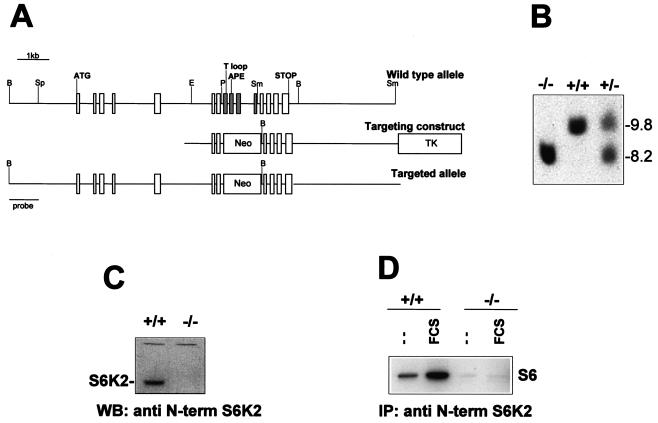
A P1 129/Ola mouse ES cell library (Genome Systems Inc., St. Louis, Mo.) was screened by PCR with the following S6K2-specific primers: 5′-CCTTTGAGGGGTTCCGG and 5′-TTCTCACAGCTGCCCTCTCTTCCTCTATTCTCCTAACG. A positive clone containing a genomic DNA fragment larger than 70 kb encompassed all of the S6K2 coding exons. The 1.3-kb EcoRI-PstI fragment of the S6K2 gene was ligated to the ClaI-NotI sites of a targeting vector containing a neomycin resistance gene and a thymidine kinase gene for negative selection and positive selection, respectively (2). The 5.5-kb SmaI fragment of the S6K2 gene was ligated to the HpaI site of the targeting vector (Fig. 1A). The S6K2 targeting vector was linearized at the ClaI site and electroporated into E14 129/Ola ES cells as described previously (51). Homologous recombination events were identified by Southern blot analysis of ES cell DNA after digestion with BamHI and hybridization with the probe corresponding to the 1-kb BamHI-SpeI fragment of the S6K2 gene (Fig. 1A). Three clones were found to be positive for the homologous recombination event. Hybridization with a probe for the neomycin resistance gene revealed a single integration site.
FIG. 1.
Generation of an _S6K2_-null allele. (A) Murine S6K2 gene map and targeting vector. Rectangles represent the coding exons of the S6K2 gene as well as the neomycin resistance (Neo) and thymidine kinase (TK) genes. The exons containing the T loop and APE sequences and the initiation and stop codons are indicated. The four exons deleted following the homologous recombination event are shown in grey. Restriction sites: B, BamHI; Sp, SpeI; E, EcoRI; P, PstI; Sm, SmaI. (B) Genotypes of littermates from S6K2+/− crosses. Tail DNA was digested with BamHI, analyzed by Southern blotting, and hybridized with the probe depicted in panel A. The corresponding BamHI fragments of genomic DNA are 9.8 kb in the wild-type allele and 8.2 kb in the targeted allele. (C and D) Loss of S6K2 protein and kinase activity in S6K2 homozygous mutant cells. (C) Western blot (WB) analysis of fibroblast cell extracts from wild-type or _S6K2_−/− embryos with a MAb against the N-terminal (N-term) region of S6K2. The apparent molecular mass of S6K2 is 68 kDa. (D) MEFs from wild-type or _S6K2_−/− embryos were starved overnight and either stimulated with 10% FCS for 30 min or left untreated. Equal amounts of protein extracts were immunoprecipitated (IP) with the anti-S6K2 antibody, and S6K activity was measured with an immune complex assay (see Materials and Methods).
Animals and cell cultures.
S6K2-targeted ES cells from clone 36 were aggregated with morula-stage embryos as described previously (51). Chimeric mice were crossed with C57BL/6 mice, and germ line transmission was assessed by Southern blot analysis. _S6K2_−/− mice were crossed with S6K1−/− mice (51) to generate mice carrying the combined deletions. Wild-type and mutant mice initially were kept in a hybrid C56BL/6-129/Ola background and subsequently were backcrossed in a pure C57BL/6 background for 10 generations. Animals were maintained on a 12-h light-dark cycle and were allowed free access to food. Mouse embryonic fibroblasts (MEFs) were prepared from embryos at embryonic day 13.5 and analyzed for cell proliferation and mRNA translation as previously described (51).
Primary hepatocytes from 12- to 14-week-old male mice were isolated by liver perfusion by the method of Seglen (50) as modified by Klaunig et al. (25). After cannulation in the subhepatic vena cava, the liver was perfused at a flow rate of 10 ml/min with calcium-free HEPES buffer (0.33 mM, pH 7.6) for 3 min and then with HEPES buffer containing 7 μg of collagenase (Liberase; Roche)/ml and 5 mM calcium chloride for 5 min. The solutions were allowed to run through a cut made in the portal vein. After enzymatic digestion, cells were collected in L-15 medium (Invitrogen Corporation) supplemented with 1 mg of bovine serum albumin (BSA)/ml, filtered, and centrifuged for 2 min at 100 × g. Viable hepatocytes were purified by centrifugation on a 45% Percoll gradient for 10 min at 100 × g. The pellet was collected and washed twice in L-15 medium containing 1 mg of BSA/ml. Hepatocytes were plated at 12 × 104 cells/cm2 in Primaria dishes (Falcon) or on coverslips coated with collagen I (Sigma) in M-199 medium (Invitrogen) supplemented with 10% fetal calf serum (FCS), 1 mg of BSA/ml, and 100 nM dexamethasone in a 37°C incubator with a 5% CO2 atmosphere. After 3 h of adhesion, cells were incubated in serum-free M-199 medium containing 1 mg of BSA/ml. On the next day, hepatocytes were treated as indicated above. All of the media contained penicillin (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (Fungizone) (250 ng/ml).
Antibody production.
S6K2 cDNA sequences coding for amino acids 1 to 43 or 440 to 484 were cloned into vector pMAL (New England Biolabs) to bacterially express N-terminal or C-terminal S6K2 peptides fused to maltose binding protein. Fusion proteins were purified according to the manufacturer's protocol. Monoclonal antibodies (MAbs) were generated by immunization of BALB/c mice with the following protocol. Two mice were immunized by intrasplenic injection (52) of 13.5 μg of purified antigen, 10 μg of adjuvant peptide (_N_-acetylmuramyl-l-alanyl-d-isoglutamine; Sigma A5919), and 10 mg of liposomes, with encapsulated muramyl tripeptide phosphatidylethanolamine as an additional adjuvant (mixed in phosphate-buffered saline [PBS]; total volume, 70 μl). Booster injections with mixtures of 15 μg of antigen, 10 μg of adjuvant peptide, 550 U of interleukin 2, and 560 U of gamma interferon with 10 mg of liposomes in PBS were administered at days 17 (subcutaneously [s.c.]), 33 (s.c. and intrperitoneally [i.p.]), 49 (s.c.), and 67 (s.c. and i.p.).
Serum samples were taken on days 40 and 73 and tested for the presence of specific antibodies by using an enzyme-linked immunosorbent assay with full-length S6K2 protein fused to glutathione _S_-transferase (GST) (N terminus free and C terminus free, respectively). A final booster injection (same composition) was administered i.p. at day 86, followed by fusion of the spleen cells (30) at day 89. The fusion partner cell line was mouse myeloma line SP2/0-AG-14. After hypoxanthine-aminopterin-thymidine selection, hybridomas were grown in HB101 hybridoma medium containing 10% CLEX serum supplement, and hybridoma supernatants were screened for the presence of antibodies to the GST-S6K2 fusion protein. Positive hybridoma cells were subcloned, and clones producing specific MAbs were selected (enzyme-linked immunosorbent assay, Western blotting, and immunocytochemical analysis). MAbs to the S6K2 N terminus (140.9.11 and 140.8.1) and to the S6K2 C terminus (141.8.5 and 141.5.23) were identified, and milligram amounts were produced by using a Miniperm culture system (Heraeus). Clones were screened for binding to the full-length S6K2 protein fused to GST.
2D PAGE, immunoprecipitation, immunoblotting, kinase assays, and nuclear fractionation.
Total 80S ribosomal proteins were isolated from either MEFs or mouse liver, and the level of in vivo S6 phosphorylation was examined by two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) as previously described (36, 41). For the preparation of total protein extracts, MEFs and hepatocytes were washed twice with cold PBS, scraped from the culture dish into egg lysis buffer, and homogenized with 10 strokes in a Teflon-glass homogenizer. To remove cell debris, homogenates were spun at 8,000 × g for 10 min. For the preparation of nuclear and cytosolic extracts, cells were scraped into hypotonic buffer (10 mM morpholinepropanesulfonic acid [MOPS] [pH 7.4], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 5 mM dithiothreitol [DTT]), homogenized with 10 strokes in a glass-glass homogenizer, and spun at 8,000 × g for 3 min. The supernatants represented the cytosolic extracts. To purify nuclear proteins, pellets were resuspended in buffer I (0.32 mM sucrose, 3 mM CaCl2, 2 mM Mg acetate, 0.1 mM EDTA, 0.1% Triton X-100, 1 mM DTT, 10 mM Tris [pH 8]), layered onto a sucrose cushion (2 M sucrose, 5 mM Mg acetate, 0.1 mM EDTA, 1 mM DTT, 10 mM Tris [pH 8]), and spun at 30,000 × g for 45 min. Protein concentrations were measured by using a Bio-Rad D/C protein assay.
For immunoblot analysis, protein extracts were resolved by sodium dodecyl sulfate-PAGE before transfer to Immobilon membranes and incubation with the following primary antibodies: mouse anti-N-terminal or anti-C-terminal S6K2, rabbit anti-ribosomal protein L7a (62), rabbit anti-phosphorylated ribosomal protein S6 (Ser235 and Ser236 [Ser235/236] and Ser240 and Ser244 [Ser240/244]), and anti-phosphorylated ERK1/ERK2 (Cell Signaling Technology). The specificity of the anti-phoshorylated S6 antibodies was demonstrated by competition with differentially phosphorylated S6 peptides (V. L. Goss and R. Polakiewicz, unpublished data). For immunoprecipitation, 300 μg of protein extract was incubated with 4 μg of MAb precoupled to protein G-Sepharose beads. S6K activity was measured by using an immune complex assay with 40S ribosomal subunits as a substrate as previously described (45).
Immunofluorescence analysis.
Primary hepatocytes were plated at 12 × 104 cells/cm2 on coverslips coated with collagen I and incubated overnight in M-199 medium containing 1 mg of BSA/ml. Hepatocytes were stimulated with 1 μM insulin, 25 ng of epidermal growth factor (EGF)/ml, and 10% FCS for 1 h in the presence or absence of 20 nM rapamycin. Cells were washed twice with Tris-buffered saline (TBS) (50 mM Tris [pH 7.4], 150 mM NaCl), fixed with 4% paraformaldehyde for 10 min at room temperature, and washed once again with TBS. Cells were permeabilized with methanol for 5 min at −20°C and washed three times for 5 min each time with TBS. Incubation in fresh 0.1% sodium borohydride in TBS for 5 min quenched the activity of the cells. Cells were rinsed once in TBS, blocked for 1 h at room temperature with PBS containing 10% goat serum, 1% BSA, and 0.02% sodium azide, and washed for 5 min with TBS. Prior to use, anti-phosphorylated ribosomal protein S6 (Ser235/236) primary antibodies or anti-60S ribosomal protein L7a primary antibodies were centrifuged at 13,000 × g for 5 min at 4°C, diluted 1:200 in TBS containing 0.1% BSA, and applied to the cells during overnight incubation at 4°C. After three washes for 5 min each time with TBS, the cells were incubated with a secondary fluorescent antibody (anti-rabbit Alexa 488; Molecular Probes) diluted 1:200 in TBS containing 0.1% BSA for 45 min at room temperature. After the cells were washed three times for 5 min each time with TBS in the dark, coverslips were mounted with a ProLong antifade kit (Molecular Probes) and observed under a confocal microscope.
Histological analysis.
Newborn mice were left for several minutes on ice before being anaesthetized and sacrificed. Tissues were fixed by injecting at several points under the skin 10% formalin in 0.1 M phosphate buffer (pH 7.4) (Baker) and incubating overnight at 4°C. Bodies were cut into three parts: the head, the thorax, and the abdomen. Samples were embedded in paraffin, and five consecutive transverse sections 5 μm thick were obtained at different levels (100 μm apart) of each mouse pup with a Leica RM2135 microtome. Sections were mounted, and one out of five consecutive slides was stained with hematoxylin and eosin for histological examination.
RESULTS
Deletion of the S6K2 gene in mice.
The coding sequence of the murine S6K2 gene spans a 7-kb region of genomic DNA containing 15 exons (Fig. 1A). The overall structure of the S6K2 gene was similar to that of the homologous S6K1 gene, as most of the intron-exon boundaries were conserved between the two genes (data not shown). To generate a targeted deletion of the S6K2 gene by homologous recombination in ES cells (6), we used a vector in which a neomycin resistance cassette replaced four exons of the catalytic domain, including the activation loop and the conserved APE motif (Fig. 1A). Heterozygous mice carrying the targeted allele were identified by Southern blot analysis and mated to obtain homozygous mutant offspring (Fig. 1B). The resulting _S6K2_−/− homozygous mice were viable and showed no obvious phenotypic abnormalities. That the S6K2 gene had been correctly targeted was demonstrated by the absence of S6K2 protein on Western blots of extracts from _S6K2_−/− MEFs compared to MEFs from wild-type mice with monoclonal antibodies directed against either the N terminus or the C terminus of the protein (Fig. 1C and data not shown). Moreover, with either of the two antibodies, S6K2 activity was undetectable by immunoprecipitation in extracts derived from either quiescent or serum-stimulated MEFs of _S6K2_−/− mice compared to those of wild-type mice (Fig. 1D and data not shown). These results indicated that the gene had been correctly deleted without causing any obvious phenotype.
Phenotypes of _S6K1_−/−/_S6K2_−/− mice.
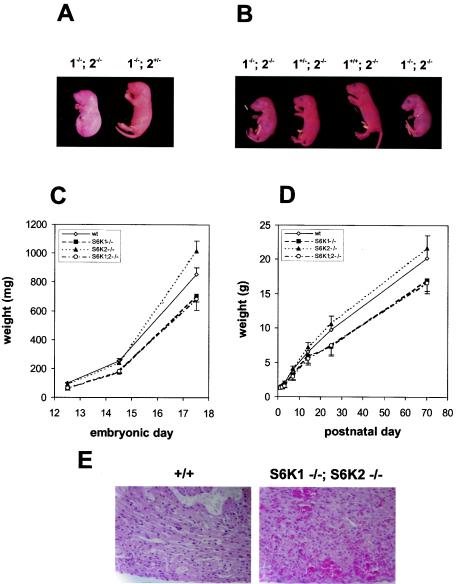
As the S6K1 and S6K2 genes share a high degree of homology and as S6K2 mRNA is upregulated in S6K1−/− mice (51), the two genes might compensate functionally for one another in vivo. To fully evaluate the impact of the S6K pathway on mouse development, we set out to generate mice lacking both protein kinases. We intercrossed mice null for the S6K1 and S6K2 genes to obtain mice with combined heterozygous mutations in both alleles. The S6K1+/−/S6K2+/− mice were bred, and the genotypes of the progeny were identified by Southern blot analysis. As shown in Table 1, all possible genotypic combinations were represented, although _S6K1_−/−/S6K2+/− and _S6K1_−/−/_S6K2_−/− mice were born at lower frequencies than expected, 65 and 30%, respectively (Table 1). Indeed, only 10 _S6K1_−/−/_S6K2_−/− mice were viable from among 526 offspring. However, analysis of embryos at 18.5 days of gestation indicated a normal Mendelian distribution of genotypes (data not shown). These data suggested that the reduced viability of _S6K1_−/−/S6K2+/− and _S6K1_−/−/_S6K2_−/− mice was due to wastage between delivery and the time of weaning. Close observation of the litters revealed that approximately one-third of the _S6K1_−/−/_S6K2_−/− mice were born dead. The corpses were found in a fetal position, still lying in the yolk sac and attached to the placenta (Fig. 2A). Among the _S6K1_−/−/_S6K2_−/− mice that were born alive and emerged from the yolk sac, the majority developed signs of cyanosis, and approximately half of them died shortly after birth. The mice surviving the first day after delivery usually reached adulthood and were fertile. However, the litters from the viable _S6K1_−/−/_S6K2_−/− parents were small and showed a high incidence of perinatal lethality (data not shown). Caesarean delivery appeared to rescue neonatal viability, although the _S6K1_−/−/_S6K2_−/− mice were more likely to develop transient signs of hypoxia, as indicated by a bluish discoloration of the skin (Fig. 2B).
TABLE 1.
Genotype analysis of littermates from crosses of S6K1+/− and S6K2+/− mice at postnatal day 21
| Genotype | % of mice | |
|---|---|---|
| Expected | Obtained | |
| S6K1+/+/S6K2+/+ | 6.25 | 10.0 |
| S6K1+/+/S6K2+/− | 12.5 | 18.1 |
| S6K1+/−/S6K2+/+ | 12.5 | 12.4 |
| S6K1+/−/S6K2+/− | 25 | 24.7 |
| _S6K1_−/−/S6K2+/+ | 6.25 | 6.4 |
| S6K1+/+/_S6K2_−/− | 6.25 | 5.9 |
| S6K1+/−/_S6K2_−/− | 12.5 | 12.6 |
| _S6K1_−/−/S6K2+/− | 12.5 | 8.1 |
| _S6K1_−/−/_S6K2_−/− | 6.25 | 1.9 |
FIG. 2.
Phenotypes of _S6K1_−/−/_S6K2_−/− mice. (A) Littermates from an _S6K1_−/−/S6K2+/− cross a few minutes after natural delivery. The genotypes of the mice are indicated. The _S6K1_−/−/_S6K2_−/− mouse was born dead and left in the yolk sac. (B) Littermates from an S6K1+/−/_S6K2_−/− cross at embryonic day 19.5 a few minutes after caesarean delivery. The genotypes of the mice are indicated. _S6K1_−/−/_S6K2_−/− mice were more prone to develop transient signs of cyanosis. (C and D) Fetal (C) and postnatal (D) growth curves for mice from homozygous crosses. Values for mice with the same genotype did not differ from those for mice derived from heterozygous crosses. Data represent the average and standard deviation for at least 10 mice per genotype. wt, wild type. (E) Heart histologic findings for wild-type and _S6K1_−/−/_S6K2_−/− mice a few minutes after delivery. The _S6K1_−/−/_S6K2_−/− mouse was born dead. Note hemorrhages in the myocardium.
To analyze the rates of growth of _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− mice during development, mice homozygous for each genotype were inbred, and the weights of their offspring were compared to those of wild-type mice during embryonic development and early postnatal life (Fig. 2C and D). As previously reported, at birth, S6K1−/− mice were about 15% smaller than wild-type mice (51). This growth defect was more evident in embryos at midgestation, when the difference in body weight was approximately 30% at embryonic day 12.5 (Fig. 2C). During postnatal growth, the S6K1−/− mice failed to attain the same body size as wild-type mice and remained about 20% smaller at 2 months of age (Fig. 2D). Conversely, the _S6K2_−/− mice tended to be larger at later stages of embryonic development and during the postnatal period; however, their weights did not differ significantly from those of wild-type mice throughout the entire period of observation (Fig. 2C and D). Moreover, the growth defect of the S6K1−/− mice was not reduced further in an _S6K2_−/− background, as S6K1−/− mice and the surviving _S6K1_−/−/_S6K2_−/− mice had similar growth rates (Fig. 2C and D). Growth rates and newborn viability were initially assessed with mice in a hybrid C57BL/6-129Ola genetic background and were confirmed following backcrossing for 10 generations in a C57BL/6 genetic background (data not shown). Thus, the impairment of body growth appears to be caused selectively by deletion of the S6K1 gene, whereas perinatal lethality is revealed only when the S6K1 and S6K2 gene products are both absent.
To assess the possible causes of reduced viability of the _S6K1_−/−/_S6K2_−/− mice, histopathological analyses of mice born dead, viable mice with signs of cyanosis, and viable mice with no signs of cyanosis were compared to those of wild-type mice at postnatal day 0. Such analyses failed to reveal any gross anatomical defects in _S6K1_−/−/_S6K2_−/− neonates. Moreover, in all viable neonates, the lungs were inflated and properly developed. However, in the nonviable neonates and in the cyanotic neonates, the internal organs were hyperemic and the heart chambers tended to be more dilated. In addition, the nonviable neonates showed several hemorrhagic sites, as revealed by diapedesis of red blood cells throughout the entire myocardium (Fig. 2E).
S6 phosphorylation in S6K-deficient mice.
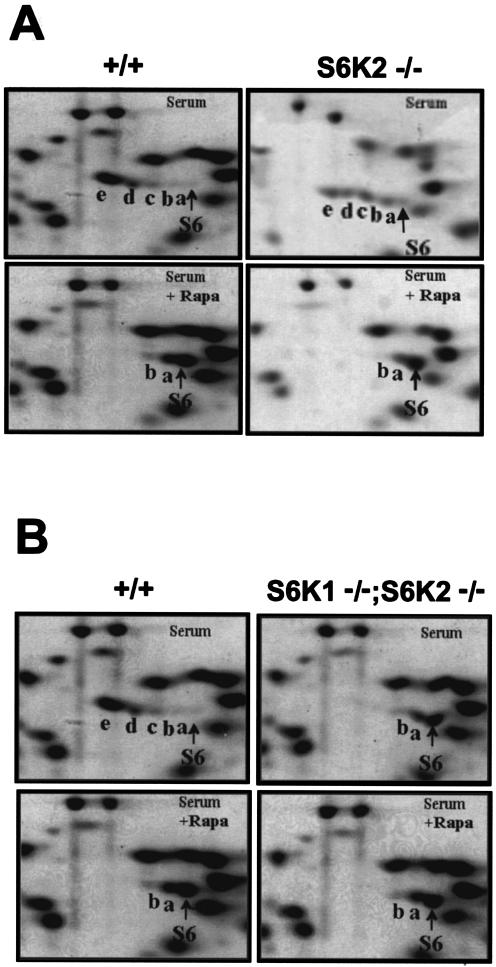
Upon growth factor stimulation, ribosomal protein S6 is multiply phosphorylated at the carboxy terminus on five serine residues in an ordered fashion beginning with Ser236, followed sequentially by Ser235, Ser240, Ser244, and Ser247 (27, 36). This response is readily measured by the decreased electrophoretic mobility of S6 following the separation of its differentially phosphorylated derivatives by 2D PAGE. To determine whether the deletion of the S6K2 gene affected the mitogen-induced phosphorylation of ribosomal protein S6, total ribosomal proteins from serum-stimulated MEFs were analyzed by 2D PAGE. As shown in Fig. 3A, in serum-stimulated wild-type MEFs, the majority of S6 migrated in highly phosphorylated derivatives d and e, corresponding to proteins phosphorylated on four and five serine residues, respectively (36, 41). Under similar experimental conditions, Shima et al. previously demonstrated that the extent of S6 phosphorylation in S6K1−/− cells was largely comparable to that in wild-type MEFs (51). In _S6K2_−/− MEFs, S6 was still highly phosphorylated in response to serum, although to a lesser extent than in wild-type and S6K1−/− MEFs; a considerable amount of S6 was present in phosphorylated derivatives a, b, and c, representing phosphorylation on one, two, and three serine residues, respectively (Fig. 3A). Pretreatment with rapamycin drastically decreased S6 phosphorylation in all three genotypes, resulting in the presence of the unphosphorylated, monophosphorylated, or biphosphorylated forms of S6 (Fig. 3A) (51). These data suggest that in _S6K2_−/− cells, S6 phosphorylation was catalyzed by a second rapamycin-sensitive kinase, most likely S6K1, although somewhat less efficiently. To test this hypothesis, we assessed S6 phosphorylation in MEFs from _S6K1_−/−/_S6K2_−/− mice. As shown in Fig. 3B, the combined deletion of the S6K1 and S6K2 genes severely suppressed S6 phosphorylation compared to that in wild-type cells. Moreover, rapamycin treatment attenuated S6 phosphorylation in wild-type MEFs, bringing it to the same level as that in serum-stimulated _S6K1_−/−/_S6K2_−/− MEFs, where rapamycin had no effect on S6 phosphorylation (Fig. 3B). S6 phosphorylation was also attenuated in the liver of _S6K1_−/−/_S6K2_−/− mice following refeeding of starved animals (data not shown). These data suggested that S6K1 and S6K2 are the sole kinases responsible for mediating rapamycin-sensitive phosphorylation.
FIG. 3.
Regulation of S6 phosphorylation. (A) In _S6K2_−/− cells, S6 phosphorylation was reduced yet regulated by mitogens and inhibited by rapamycin (Rapa). 2D PAGE was carried out with 80S ribosomal proteins from wild-type and _S6K2_−/− MEFs. Cells were stimulated with 10% FCS for 1 h, with or without pretreatment with 20 nM rapamycin. (B) S6 phosphorylation in _S6K1_−/−/_S6K2_−/− cells was reduced to the same extent as in rapamycin-treated cells. 2D PAGE was carried out with 80S ribosomal proteins from wild-type and _S6K1_−/−/_S6K2_−/− MEFs. Cells were treated as described for panel A. See the text for an explanation of a to e on the panels.
Intracellular localization of S6K activity.
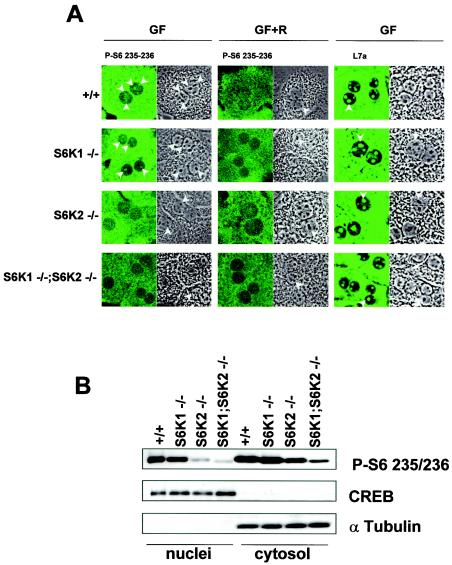
The majority of S6 is found in the cytoplasm as an integral component of the 40S ribosomal subunit. However, S6 is also present in nucleoli, the site of ribosome biogenesis, and in perinucleolar structures called coiled bodies (or Cajall bodies). Given the large relative abundance of S6 in the cytoplasm compared to the nucleus, the quantitation of S6 phosphorylation by 2D PAGE would largely reflect the status of S6 phosphorylation in the cytoplasm. However, as the p85S6K isoform of S6K1 is targeted to the nucleus (46) and S6K2 also contains a nuclear targeting sequence (17, 47, 51), it is possible that one of the two kinases is responsible for mediating selective phosphorylation of S6 in the nucleolus. Consistent with this idea, previous studies demonstrated that S6K1 and S6K2 are differentially targeted within the cell (26, 46, 57). In wild-type hepatocytes, growth factors predominantly stimulated S6 Ser235/236 phosphorylation in the cytosol and nucleoli (Fig. 4A). This staining matched the localization of the ribosomal subunits, as shown by immunostaining with antibodies against 60S ribosomal protein L7a (Fig. 4A) (62). In agreement with the 2D PAGE analysis (Fig. 3A), the deletion of S6K2 caused a sharper reduction of S6 phosphorylation in the cytosol than did the deletion of S6K1 (Fig. 4A). Interestingly, this also appeared to be the case in nucleoli (Fig. 4A). The overall intensity of S6 phosphorylation was further reduced by combining the deletion of S6K1 with the deletion of S6K2. In addition to the immunofluorescence analysis, nuclear fractionation of hepatocytes was performed following growth factor stimulation (Fig. 4B). The purity of the fractions was assessed with specific nuclear and cytosolic protein markers, CREB and α-tubulin, respectively (Fig. 4B). Consistent with the results of the immunofluorescence experiments, the deletion of S6K2 significantly affected S6 phosphorylation in both cell compartments, and the combination of the two gene deletions further reduced the levels of the phosphorylated protein. Moreover, in contrast to the results obtained with the other genotypes, the residual S6 phosphorylation staining observed with the anti-phosphorlyated S6 (Ser235/236) antibody in the cytosol and the nuclei of _S6K1_−/−/_S6K2_−/− hepatocytes was not inhibited by rapamycin treatment (Fig. 4A) (also see below).
FIG. 4.
Intracellular localization of S6K activity. (A) Immunostaining of wild-type, _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− hepatocytes with anti-phosphorylated S6 (Ser235/236) and anti-L7a antibodies (left panels). The right panels represent phase-contrast images of the same fields. Cells were growth factor deprived for 1 day and stimulated with 4 nM EGF (25 ng/ml) and 1 μM insulin for 1 h (GF), with or without pretreatment with 20 nM rapamycin (R). Arrowheads indicate nucleoli. (B) Immunoblot analysis of nuclear and cytosolic extracts from wild-type, _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− hepatocytes with anti-phosphorylated S6 (Ser235/236), anti-α-tubulin, and anti-CREB antibodies. The latter two were used as cytosolic and nuclear markers, respectively.
5′TOP mRNA translation and cell cycle progression of _S6K1_−/−/_S6K2_−/− cells.
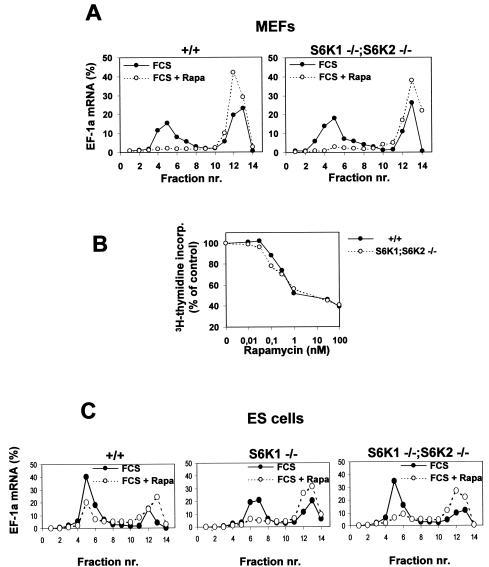
It was previously demonstrated that rapamycin inhibits the selective upregulation of 5′TOP mRNAs following mitogen stimulation (20, 55) and that this effect requires intact 5′TOP (19, 49) and may be mediated by S6K1 (19, 49). However, recently it was claimed that neither the S6Ks nor rapamycin appears to be involved in this response (53). As 5′TOP mRNAs code for ribosomal proteins, their increased expression is essential to sustaining translation during the phases of cell growth (56). To examine the roles of S6K1, S6K2, and rapamycin in this response, we used cells derived from _S6K1_−/−/_S6K2_−/− mice. Initially, we examined the distribution of 5′TOP elongation factor 1α (EF1α) mRNA on polysomes in MEFs derived from wild-type and _S6K1_−/−/_S6K2_−/− mice. As shown in Fig. 5A, serum induced the recruitment of EF1α mRNA into polysomes in both cell types to almost the same extent, and this effect was largely abolished by rapamycin in both _S6K1_−/−/_S6K2_−/− and wild-type cells. Furthermore, the combined deletion of S6K1 and S6K2 did not alter the cell doubling times or the saturation densities compared to that of wild-type MEFs (data not shown). Rapamycin inhibited the S-phase entry of cells from both genotypes with similar potencies and 50% inhibitory concentrations (Fig. 5B).
FIG. 5.
Regulation of 5′TOP mRNA translation and cell cycle progression is preserved in _S6K1_−/−/_S6K2_−/− MEFs. (A and C) 5′TOP mRNA translation is modulated by serum and rapamycin (Rapa), although S6 phosphorylation is resistant to rapamycin treatment. Cytoplasmic extracts were prepared from wild-type (+/+), _S6K1_−/−, and _S6K1_−/−/_S6K2_−/− MEFs (A) or ES cells (C) that had been stimulated with 10% serum (FCS) in the presence (empty circles) or absence (filled circles) of 20 nM rapamycin after serum deprivation for 48 h. Extracts were centrifuged on a sucrose gradient prior to fractionation as previously described (22). RNA from the different fractions was analyzed by Northern blotting with a probe specific for EF1α (EF-1a) mRNA. The relative amount of mRNA present in each fraction is expressed as a percentage. Fractions 1 to 7 contain polysomes, whereas fractions 8 to 14 are enriched in monosomes, ribosomal subunits, and mRNPs. nr., number. (B) Inhibitory effect of rapamycin on the proliferation of S6K-null cells. Wild-type MEFs (filled circles) and _S6K1_−/−/_S6K2_−/− MEFs (empty circles) were seeded in a 24-well plate at a density of 2 × 104 cells per well. Cells then were treated with 0.5% serum for 48 h and stimulated for 20 h with 10% serum in the absence or presence of rapamycin concentrations ranging from 0 to 100 nM. Cells were labeled with [3H]thymidine (1 μCi/ml) throughout the entire stimulation period Radioactivity incorporated (incorp.) into the DNA was measured, and the average values for triplicate samples were determined. Data are expressed as the percent inhibition of [3H]thymidine incorporation by rapamycin-treated cells compared to untreated cells.
The analysis of 5′TOP mRNA translation was extended to ES cells, as Kawasome et al. (24) previously reported that 5′TOP mRNAs are constitutively upregulated in S6K1−/− ES cells and resistant to the effects of rapamycin, although these findings were not consistent with those of a later study with the same ES cells (53). To examine this possibility, we analyzed the distribution of EF1α mRNA on polysomes in ES cells derived from embryos of _S6K1_−/−/_S6K2_−/−, _S6K1_−/−, and wild-type mice. Similar to what was observed for MEFs, the deletion of S6K1 alone or the deletion of both S6K1 and S6K2 in ES cells did not alter the upregulation of 5′TOP mRNA or sensitivity to rapamycin (Fig. 5C). Thus, despite a pronounced effect on the development and growth of the animal, in two distinct cell types, neither the reduction of S6 phosphorylation nor the loss of S6K1 and S6K2 function is sufficient to deregulate 5′TOP mRNA translation; these findings suggest that other rapamycin-sensitive events are required.
Evidence for functional redundancy with the mitogen-activated protein kinase (MAPK) pathway.
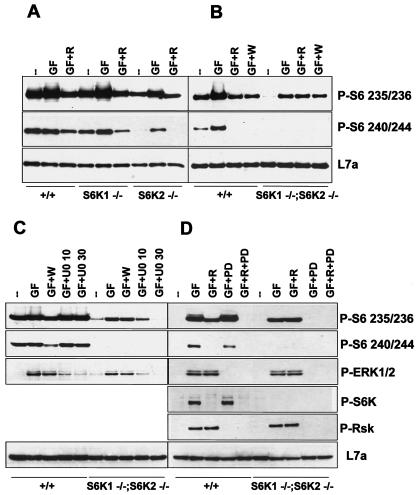
The findings described above raised the possibility of either a compensatory or a redundant S6K pathway. On closer analysis of S6 phosphorylation, a small amount of partially phosphorylated S6 was still detected in MEFs from _S6K1_−/−/_S6K2_−/− mice as well as in rapamycin-treated wild-type MEFs, as evidenced by the presence of derivatives a and b (Fig. 3B). The residual S6 phosphorylation in wild-type MEFs could have been due to the relatively short treatment in the presence of rapamycin, as longer times were reported for complete S6 dephosphorylation (20). However, this finding was unexpected for _S6K1_−/−/_S6K2_−/− MEFs, as the level of S6 phosphorylation was low in these cells and was unaffected by rapamycin (Fig. 3B). To examine this situation further, we monitored S6 phosphorylation in primary hepatocytes stimulated with EGF and insulin by using phosphospecific antibodies directed against the first and second phosphorylation sites, Ser236 and Ser235, or the third and fourth phosphorylation sites, Ser240 and Ser244. Following insulin stimulation, phosphorylation at both Ser235/236 and Ser240/244 increased in wild-type, _S6K1_−/−, and _S6K2_−/− hepatocytes, whereas only Ser235/236 phosphorylation was detectable in _S6K1_−/−/_S6K2_−/− hepatocytes (Fig. 6A and B); these results were consistent with the order and extent of S6 phosphorylation (Fig. 3B) (3). In wild-type hepatocytes as well as in _S6K1_−/− and _S6K2_−/− hepatocytes, the increase in S6 phosphorylation was sensitive to rapamycin (Fig. 6A). However, in _S6K1_−/−/_S6K2_−/− hepatocytes, the increase in S6 phosphorylation was resistant to rapamycin (Fig. 6B), suggesting that a rapamycin-resistant S6K was operating in the absence of S6K1 and S6K2.
FIG. 6.
Evidence for a rapamycin-insensitive S6K. Western blot analysis of protein extracts from wild-type, _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− hepatocytes was carried out with anti-phosphorylated S6 (Ser235/236 and Ser240/244), anti-phosphorylated ERK1/ERK2, anti-phosphorylated S6K (Thr389), anti-phosphorylated Rsk (Thr359 and Ser363), and anti-L7a antibodies, the latter being used as a loading control. Cells were stimulated with 4 nM EGF (25 ng/ml) and 1 μM insulin for 30 min (GF), with or without pretreatment with 20 nM rapamycin (R), 1 μM wortmannin (W), 10 or 30 μM U0126 (U0 10 or U0 30, respectively), and 3 μM PD184352 (PD). Extracts in panels A and C were derived from cells deprived of GFs for 1 day, whereas those in panels B and D were derived from cells deprived of GFs for 2 days. In panel D, cells were also deprived of nutrients (all of the amino acids and glucose) for 3 h.
It is known that Akt has a substrate recognition motif similar to that of S6K1 and S6K2 (32) and phosphorylates S6 in vitro (23). However, the phosphatidylinositide-3OH kinase inhibitor wortmannin, which blocks Akt activation, had no effect on insulin-induced S6 phosphorylation in _S6K1_−/−/_S6K2_−/− hepatocytes, whereas it was as potent as rapamycin in blocking S6 phosphorylation in wild-type hepatocytes (Fig. 6B). As p90rsk appears to be the rapamycin-resistant kinase responsible for regulating S6 phosphorylation during progesterone-induced meiotic maturation of Xenopus oocytes (9, 49), we next tested the effect of the MEK inhibitor U0126 on S6 phosphorylation in _S6K1_−/−/_S6K2_−/− hepatocytes. MEK mediates MAPK activation, which in turn phosphorylates and activates p90rsk (13, 54). Such treatment had no measurable effect on S6 phosphorylation in wild-type cells; however, it inhibited Ser235/236 and MAPK phosphorylation in a dose-dependent manner in _S6K1_−/−/_S6K2_−/− cells (Fig. 6C).
The MAPK-dependent phosphorylation of S6 could represent either the upregulation of a compensatory pathway or the presence of a pathway that normally operates in wild-type cells but whose activity is masked by S6Ks. It was difficult to distinguish between these possibilities, given the high level of basal S6 phosphorylation displayed by wild-type hepatocytes. To circumvent this problem, hepatocytes were deprived of nutrients as well as serum, a step which resulted in the complete dephosphorylation of S6 (Fig. 6D). Under these conditions, rapamycin abolished the insulin-induced increase in Ser240/244 phosphorylation but only attenuated the phosphorylation of Ser235/236 (Fig. 6D). In contrast, PD184352, an MKK1/ERK1/ERK2 pathway inhibitor that is more specific than U0126 (38), had little effect on S6 phosphorylation when used alone but, when used together with rapamycin, totally abolished the insulin-induced response. In contrast, in _S6K1_−/−/_S6K2_−/− hepatocytes, rapamycin had no effect on Ser235/236 phosphorylation, whereas PD184352 abolished this response (Fig. 6C). Thus, in the absence of S6K1 and S6K2 activity, an MAPK pathway which cooperates in the regulation of S6 phosphorylation was unveiled.
DISCUSSION
Studies with gene deletions or RNA interference can be instructive in revealing biological functions. For S6Ks, such studies have been carried out by double-stranded RNA interference with Caenorhabditis elegans (33) and by gene deletion with Drosophila melanogaster (39) and mice (42). In C. elegans, disruption of the gene encoding the S6K1/S6K2 orthologue, Cep70, by double-stranded RNA interference led to only minor growth deficits (33). In contrast, deletion of the Drosophila gene encoding the S6K1/S6K2 orthologue, the dS6K gene, resulted in semilethality, with most flies dying during the late third larval instar or early pupation (39). In addition, the surviving flies were strikingly reduced in size and lived only a few weeks, and the females were sterile (39). However, S6K1-deficient mice are viable and fertile, although they are 15 to 20% smaller than wild-type mice at birth and are hypoinsulinemic (42, 51). Furthermore, these mice, unlike Drosophila, have a second S6K, S6K2, which is upregulated in all tissues examined in S6K1-deficient mice, suggesting a compensatory response for the loss of S6K1. Consistent with this argument, we show here that deletion of S6K1 and S6K2 in mice, as for dS6K in flies, is also semilethal, with pups dying at the time of delivery or shortly thereafter. A potential difference between the findings for C. elegans or D. melanogaster and mice is that small interfering RNAs reduce gene expression, whereas it is abolished by gene deletion. It should also be noted that the combined inactivation of two other elements of the phosphatidylinositide-3OH kinase pathway, Akt1 and Akt2, also leads to perinatal lethality of unknown cause (43). The results of macroscopic and histological analyses of newborns lacking S6K1 and S6K2 were compatible with the mutant mice experiencing a period of hypoxic stress. This finding might have been partly due to heart failure or a placentation defect, a possibility which is presently being addressed.
One of the early intracellular responses to growth factor stimulation in both invertebrates and vertebrates is multiple phosphorylation of 40S ribosomal protein S6 (14, 58). Although many kinases have been shown to mediate this response in vitro, only two have been demonstrated to phosphorylate S6 in the same specific order and extent as those observed in vivo: S6K1 (10) and p90rsk from Xenopus (61). Although a role for p90rsk in this response has been controversial, studies of progesterone-induced meiotic maturation of Xenopus oocytes support such a function (29, 49). That S6K1 is not involved in this response is further supported by the fact that rapamycin, which selectively inhibits S6K1 but not p90rsk activation (28), abolished S6K1 activity, with little effect on progesterone-induced S6 phosphorylation (49). In contrast to the findings in progesterone-treated Xenopus oocytes, rapamycin eliminates serum-induced S6 phosphorylation in mammalian cells (20), an inhibitory effect that is reversed by the use of an S6K1 variant that is largely rapamycin resistant (19, 59). In seeming agreement with the findings in mammalian cells, _S6K1_−/− ES cells were reported to be devoid of phosphorylated S6 (24, 53). However, in tissues derived from _S6K1_−/− mice, S6 phosphorylation was shown to be intact but still rapamycin sensitive (51), leading to the discovery of S6K2 (17, 47, 51). Although studies with ES cells have been difficult to rationalize, some investigators recently demonstrated that growth factor stimulation of these same cells leads to S6K2 activation and increased S6 phosphorylation in a rapamycin-sensitive manner (31, 60). These conflicting results may be attributable to different stimulation conditions or detection methods.
The studies presented here reveal two novel aspects of S6 phosphorylation in mammalian cells in vivo, i.e., the major role of S6K2 in this response and the involvement of an MAPK pathway, whose function is detectable when S6K1 and S6K2 are inactive. S6K2 appears to be the dominant S6K in embryo fibroblasts as well as in adult hepatocytes and skeletal and heart muscle, as _S6K2_−/− mice showed a more dramatic reduction in the amount of phosphorylated S6 in these tissues than did _S6K1_−/− mice (Fig. 3 and 6 and data not shown). In addition, the nucleolar phosphorylation of S6 was greatly affected in _S6K2_−/− hepatocytes (Fig. 4), implying that the nucleus-targeted isoform of S6K1, p85S6K (46), may not efficiently mediate phosphorylation in nucleoli. Ectopically expressed S6K2 predominantly resides in the nucleus due to a nuclear localization signal in the C-terminal region of the protein (26, 57). Upon mitogen stimulation, protein kinase C-dependent phosphorylation of S6K2 in the C-terminal region masks the nuclear localization signal, causing the retention of the kinase in the cytosol (57). Our data are compatible with this model and suggest the presence of active S6K2 in the cytosol and nucleoli, where it can phosphorylate S6. However, S6K1 clearly contributes to the overall amount of S6 phosphorylation, as deletion of both kinases was required to reduce S6 phosphorylation to the same extent as rapamycin (Fig. 3). Indeed, with our experimental model, we cannot conclusively determine to what extent each kinase contributes to this response under physiological conditions, as it was previously reported that in _S6K1_−/− mice, S6K2 mRNA transcripts are upregulated in all tissues examined (51). Such upregulation of S6K2 could explain its dominance as an S6K and the reliance of complete S6 phosphorylation on S6K1. That S6K1 and S6K2 are the only genes coding for rapamycin-sensitive S6Ks is consistent with two other S6K genes identified by the human genome project representing pseudogenes (34). However, the combined inactivation of S6K1 and S6K2 revealed the presence of another S6K, most likely p90rsk, which is rapamycin resistant and phosphorylates at least the first two Ser residues of S6 (Fig. 3 and 6). p90rsk appears to be the physiological S6K during Xenopus (49) and mouse (15) meiotic maturation. The murine p90rsk family includes several members which are directly phosphorylated and activated by ERK1 and ERK2 (12). p90rsk family members have two kinase domains; the N-terminal one shows approximately 60% sequence identity with S6K1 and S6K2 and has a similar consensus substrate recognition motif (32). The functional redundancy among the S6K pathways indicates the importance of the phosphorylation of S6 or another common substrate at one or more steps of embryonic development.
The functional compensation between S6K1 and S6K2 was initially proposed based on the upregulation of S6K2 in _S6K1_−/− mice and could represent a mechanism to ensure the survival of the animal (51). This suggestion is supported by the data presented here, showing semilethality in both S6K1−/−/S6K2−/− and S6K1−/−/S6K2+/− mice. Interestingly, Wang et al. (60) showed that S6 phosphorylation was reduced in the _S6K1_−/− ES cells described above (24) but found no difference in the level of S6K2 protein. These ES cells were obtained by selection in vitro with high doses of G418 (24), rather than from crossing heterozygous founders. Therefore, they may not have upregulated S6K2 compensatorily, as they have not undergone the stress associated with embryonic development. Following a similar reasoning, the MAPK-dependent S6K activity described here could alleviate the phenotype of the double-mutant mice. Surprisingly, the body size phenotype of the single-mutant mice did not appear to correlate with the extent of S6 phosphorylation. For _S6K1_−/− mice, body size was clearly reduced, with little effect on S6 phosphorylation, whereas _S6K2_−/− mice exhibited normal body size but had reduced levels of S6 phosphorylation. Similarly, the defect in β-cell growth and insulin levels was associated mainly with the deletion of S6K1, which has a minor effect on S6 phosphorylation in β cells (data not shown). These data raise the interesting possibility that S6K1 and S6K2 also exert specific functions through distinct substrates. Consistent with such a model, the two kinases have been reported to be differentially localized and regulated (5, 31, 35). Despite these observations, it should be noted that recent studies with phosphospecific site mutations in Drosophila S6 suggested that Drosophila S6 is epistatic to dS6K, the single S6K gene in Drosophila, arguing that it may be required to elicit the dS6K growth response (T. Radimerski and G. Thomas, unpublished data).
Initial studies showed that rapamycin selectively inhibited the translation of 5′TOP mRNAs (20, 55), with the extent of inhibition varying between cell types (21). This finding led to the conclusion that some cell types use both rapamycin-sensitive and rapamycin-resistant pathways to control this response, whereas other cell types are largely reliant on the rapamycin-sensitive pathway (20). Such an explanation is consistent with the ability of rapamycin to inhibit cell proliferation in certain cell types, such as endothelial, smooth muscle, and naive T cells, while having a less dramatic impact on other cell types. Here we show, as previously demonstrated (51), that the inhibitory effect of rapamycin on 5′TOP mRNA translation is complete in MEFs derived from wild-type cells (Fig. 5). Earlier it was also demonstrated that the effect of rapamycin requires an intact polypyrimidine tract, that dominant interfering alleles of S6K1 were as effective in blocking the upregulation of 5′TOP mRNAs as rapamycin, and that the inhibitory effects of rapamycin could be largely prevented by the use of a rapamycin-resistant allele of S6K1 (19, 49). Unexpectedly, we found here that rapamycin still inhibited the translation of 5′TOP mRNAs despite the absence of S6K1 and S6K2 (Fig. 5). The effect of the dominant-negative S6K1 allele can be rationalized by its ability to titrate mTOR, potentially through Raptor (40), and block mTOR from phosphorylating other key downstream targets (19, 59). However, in view of the findings presented here, it is more difficult to understand the ability of the rapamycin-resistant allele of S6K1 to protect 5′TOP mRNA translation from the inhibitory effects of rapamycin (19, 49). One possibility is that S6K1 is not involved in this response but that ectopic expression of the rapamycin-resistant allele can lead to the phosphorylation of a target protein that is involved in 5′TOP mRNA translation and whose phosphorylation is not mediated by endogenous S6K1. It is also possible that in the absence of S6K1 and S6K2, a rapamycin-sensitive, compensatory pathway is activated during development. Stolovich et al. (53) recently reported that S6 phosphorylation, S6K1, and rapamycin had little impact on 5′TOP mRNA translation. However, they used ES cells in which the S6K pathway was not completely inhibited, as S6K2 was still expressed (31, 60). To resolve this issue and the discrepancies concerning the inhibitory effect of rapamycin on 5′TOP mRNA translation in nerve growth factor-treated PC12 cells (42, 51), an understanding of the molecular mechanisms which control 5′TOP mRNA translation is required.
In summary, these studies provide a first comparative analysis of the distinct phenotypes of S6K1- and S6K2-deficient mice and should serve as a basis to search for other specific cellular targets of these enzymes. They also reveal an alternative mechanism leading to S6 phosphorylation in the absence of S6K1 and S6K2, i.e, an MAPK pathway; this function should be taken into account when the impact of this pathway on growth and development is evaluated.
Acknowledgments
We thank P. Kopp and J.-F. Spetz for expert technical assistance in generating _S6K2_−/−mice. We are grateful to R. Polakiewicz, A. Sobering, A. Sotiropoulos, and T. Nobukuni for critical reading of the manuscript and for helpful discussions. We also thank A. Ziemiecki for providing L7a antibodies and Paul Schofield and M. Stumm for help with histological studies.
M.P. is a recipient of a stipend from the Foundation pour la Recherche Medicale. This work was supported in part by a grant to M.P. from the INSERM Avenir Program (R01131KS) and a grant to S.C.K. and G.T. from the EEC (EU BIO4-CT97-2071) and the Novartis Research Foundation.
REFERENCES
- 1.Abraham, R. T. 2002. Identification of TOR signaling complexes: more TORC for the cell growth engine. Cell 111**:**9-12. [DOI] [PubMed] [Google Scholar]
- 2.Arber, S., J. J. Hunter, J. Ross, Jr., M. Hongo, G. Sansig, J. Borg, J. C. Perriard, K. R. Chien, and P. Caroni. 1997. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88**:**393-403. [DOI] [PubMed] [Google Scholar]
- 3.Bandi, H. R., S. Ferrari, J. Krieg, H. E. Meyer, and G. Thomas. 1993. Identification of 40 S ribosomal protein S6 phosphorylation sites in Swiss mouse 3T3 fibroblasts stimulated with serum. J. Biol. Chem. 268**:**4530-4533. [PubMed] [Google Scholar]
- 4.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95**:**1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett, P. E., S. Blackshaw, M. M. Lai, I. A. Qureshi, A. F. Burnett, D. M. Sabatini, and S. H. Snyder. 1998. Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc. Natl. Acad. Sci. USA 95**:**8351-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capecchi, M. R. 1989. Altering the genome by homologous recombination. Science 244**:**1288-1292. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, P. B., S. Fumagalli, and G. Thomas. 1999. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 9**:**49-54. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294**:**1102-1105. [DOI] [PubMed] [Google Scholar]
- 9.Erikson, E., and J. L. Maller. 1989. In vivo phosphorylation and activation of ribosomal protein S6 kinases during Xenopus oocyte maturation. J. Biol. Chem. 264**:**13711-13717. [PubMed] [Google Scholar]
- 10.Ferrari, S., H. R. Bandi, B. M. Bussian, and G. Thomas. 1991. Mitogen-activated 70K S6 kinase. J. Biol. Chem. 266**:**22770-22775. [PubMed] [Google Scholar]
- 11.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16**:**1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frodin, M., and S. Gammeltoft. 1999. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol. 151**:**65-77. [DOI] [PubMed] [Google Scholar]
- 13.Frodin, M., C. J. Jensen, K. Merienne, and S. Gammeltoft. 2000. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 19**:**2924-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fumagalli, S., and G. Thomas. 2000. S6 phosphorylation and signal transduction, p. 695-717. In M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Gavin, A. C., and S. Schorderet-Slatkine. 1997. Ribosomal S6 kinase p90rsk and mRNA cap-binding protein eIF4E phosphorylations correlate with MAP kinase activation during meiotic reinitiation of mouse oocytes. Mol. Reprod. Dev. 46**:**383-391. [DOI] [PubMed] [Google Scholar]
- 16.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15**:**807-826. [DOI] [PubMed] [Google Scholar]
- 17.Gout, I., T. Minami, K. Hara, Y. Tsujishita, V. Filonenko, M. D. Waterfield, and K. Yonezawa. 1998. Molecular cloning and characterization of a novel p70 S6 kinase, p70 S6 kinase b, containing a proline-rich region. J. Biol. Chem. 273**:**30061-30064. [DOI] [PubMed] [Google Scholar]
- 18.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274**:**34493-34498. [DOI] [PubMed] [Google Scholar]
- 19.Jefferies, H. B. J., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 12**:**3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferies, H. B. J., C. Reinhard, S. C. Kozma, and G. Thomas. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA 91**:**4441-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jefferies, H. B. J., and G. Thomas. 1996. Ribosomal protein S6 phosphorylation and signal transduction, p. 389-409. In N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Jefferies, H. B. J., G. Thomas, and G. Thomas. 1994. Elongation factor-1a mRNA is selectively translated following mitogenic stimulation. J. Biol. Chem. 269**:**4367-4372. [PubMed] [Google Scholar]
- 23.Jones, P. F., T. Jakubowicz, F. J. Pitossi, F. Maurer, and B. A. Hemmings. 1991. Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc. Natl. Acad. Sci. USA 88**:**4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasome, H., P. Papst, S. Webb, G. M. Keller, G. L. Johnson, E. W. Gelfand, and N. Terada. 1998. Targeted disruption of p70s6k defines its role in protein synthesis and rapamycin sensitivity. Proc. Natl. Acad. Sci. USA 95**:**5033-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaunig, J. E., P. J. Goldblatt, D. E. Hinton, M. M. Lipsky, J. Chacko, and B. F. Trump. 1981. Mouse liver cell culture. I. Hepatocyte isolation. In Vitro 17**:**913-925. [DOI] [PubMed] [Google Scholar]
- 26.Koh, H., K. Jee, B. Lee, J. Kim, D. Kim, Y. H. Yun, J. W. Kim, H. S. Choi, and J. Chung. 1999. Cloning and characterization of a nuclear S6 kinase, S6 kinase-related kinase (SRK): a novel nuclear target of Akt. Oncogene 18**:**5115-5119. [DOI] [PubMed] [Google Scholar]
- 27.Krieg, J., J. Hofsteenge, and G. Thomas. 1988. Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J. Biol. Chem. 263**:**11473-11477. [PubMed] [Google Scholar]
- 28.Kuo, C. J., J. Chung, D. F. Fiorentino, W. M. Flanagan, J. Blenis, and G. R. Crabtree. 1992. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature 358**:**70-73. [DOI] [PubMed] [Google Scholar]
- 29.Lane, H. A., S. J. Morley, M. Doree, S. C. Kozma, and G. Thomas. 1992. Identification and early activation of a Xenopus laevis p70s6k following progesterone-induced meiotic maturation. EMBO J. 11**:**1743-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, R. D., R. S. Crissman, and S. Ginn. 1986. High efficiency fusion procedure for producing monoclonal antibodies against weak immunogens. Methods Enzymol. 121**:**183-192. [DOI] [PubMed] [Google Scholar]
- 31.Lee-Fruman, K. K., C. J. Kuo, J. Lippincott, N. Terada, and J. Blenis. 1999. Characterization of S6K2, a novel kinase homologous to S6K1. Oncogene 18**:**5108-5114. [DOI] [PubMed] [Google Scholar]
- 32.Leighton, I. A., K. N. Dalby, F. B. Caudwell, P. T. Cohen, and P. Cohen. 1995. Comparison of the specificities of p70 S6 kinase and MAPKAP kinase-1 identifies a relatively specific substrate for p70 S6 kinase: the N-terminal kinase domain of MAPKAP kinase-1 is essential for peptide phosphorylation. FEBS Lett. 375**:**289-293. [DOI] [PubMed] [Google Scholar]
- 33.Long, X., C. Spycher, Z. S. Han, A. M. Rose, F. Muller, and J. Avruch. 2002. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 12**:**1448-1461. [DOI] [PubMed] [Google Scholar]
- 34.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298**:**1912-1934. [DOI] [PubMed] [Google Scholar]
- 35.Martin, K. A., S. S. Schalm, C. Richardson, A. Romanelli, K. L. Keon, and J. Blenis. 2001. Regulation of ribosomal S6 kinase 2 by effectors of the phosphoinositide 3-kinase pathway. J. Biol. Chem. 276**:**7884-7891. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Pèrez, J., and G. Thomas. 1983. Ordered phosphorylation of 40S ribosomal protein S6 after serum stimulation of quiescent 3T3 cells. Proc. Natl. Acad. Sci. USA 80**:**926-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyuhas, O., D. Avni, and S. Shama. 1996. Translational control of ribosomal protein mRNAs in eukaryotes, p. 363-388. In N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Mody, N., J. Leitch, C. Armstrong, J. Dixon, and P. Cohen. 2001. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 502**:**21-24. [DOI] [PubMed] [Google Scholar]
- 39.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285**:**2126-2129. [DOI] [PubMed] [Google Scholar]
- 40.**Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. I. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa.2003. The mammalian target of rapamycin (mTOR)partner, raptor, binds the mTOR substratesp70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 78:**5461-15464. [DOI] [PubMed] [Google Scholar]
- 41.Olivier, A. R., L. M. Ballou, and G. Thomas. 1988. Differential regulation of S6 phosphorylation by insulin and epidermal growth factor in Swiss mouse 3T3 cells: insulin activation of type 1 phophatase. Proc. Natl. Acad. Sci. USA 85**:**4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pende, M., S. C. Kozma, M. Jaquet, V. Oorschot, R. Burcelin, Y. Le Marchand-Brustel, J. Klumperman, B. Thorens, and G. Thomas. 2000. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature 48**:**994-997. [DOI] [PubMed] [Google Scholar]
- 43.Peng, X. D., P. Z. Xu, M. L. Chen, A. Hahn-Windgassen, J. Skeen, J. Jacobs, D. Sundararajan, W. S. Chen, S. E. Crawford, K. G. Coleman, and N. Hay. 2003. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 17**:**1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petroulakis, E., and E. Wang. 2002. Nerve growth factor specifically stimulates translation of eukaryotic elongation factor 1A-1 (eEF1A-1) mRNA by recruitment to polyribosomes in PC12 cells. J. Biol. Chem. 277**:**18718-18727. [DOI] [PubMed] [Google Scholar]
- 45.Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma, B. A. Hemmings, and G. Thomas. 1998. Phosphorylation and activation of p70s6k by PDK1. Science 279**:**707-710. [DOI] [PubMed] [Google Scholar]
- 46.Reinhard, C., A. Fernandez, N. J. C. Lamb, and G. Thomas. 1994. Nuclear localization of p85s6k: functional requirement for entry into S phase. EMBO J. 13**:**1557-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitoh, M., P. ten Dijke, K. Miyazono, and H. Ichijo. 1998. Cloning and characterization of p70s6kb defines a novel family of p70 S6 kinases. Biochem. Biophys. Res. Commun. 253**:**470-476. [DOI] [PubMed] [Google Scholar]
- 48.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 13:193-200. [DOI] [PubMed] [Google Scholar]
- 49.Schwab, M. S., S. H. Kim, N. Terada, C. Edfjall, S. C. Kozma, G. Thomas, and J. L. Maller. 1999. p70(S6K) controls selective mRNA translation during oocyte maturation and early embryogenesis in Xenopus laevis. Mol. Cell. Biol. 19**:**2485-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seglen, P. O. 1976. Preparation of isolated rat liver cells. Methods Cell Biol. 13**:**29-83. [DOI] [PubMed] [Google Scholar]
- 51.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17**:**6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spitz, M. 1986. “Single-shot” intrasplenic immunization for the production of monoclonal antibodies. Methods Enzymol. 121**:**33-41. [DOI] [PubMed] [Google Scholar]
- 53.Stolovich, M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22**:**8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sturgill, T. W., L. B. Ray, and J. L. Maller. 1988. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinaseII. Nature 334**:**715-718. [DOI] [PubMed] [Google Scholar]
- 55.Terada, N., H. R. Patel, K. Takase, K. Kohno, A. C. Narin, and E. W. Gelfand. 1994. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. USA 91**:**11477-11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, G. 2000. An “encore” for ribosome biogenesis in cell proliferation. Nat. Cell Biol. 2**:**E71-E72. [DOI] [PubMed] [Google Scholar]
- 57.Valovka, T., F. Verdier, R. Cramer, A. Zhyvoloup, T. Fenton, H. Rebholz, M. L. Wang, M. Gzhegotsky, A. Lutsyk, G. Matsuka, V. Filonenko, L. Wang, C. G. Proud, P. J. Parker, and I. T. Gout. 2003. Protein kinase C phosphorylates ribosomal protein S6 kinase βII and regulates its subcellular localization. Mol. Cell. Biol. 23**:**852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volarevic, S., and G. Thomas. 2001. Role of S6 phosphorylation and S6 kinase in cell growth. Prog. Nucleic Acids Res. Mol. Biol. 65**:**101-127. [DOI] [PubMed] [Google Scholar]
- 59.von Manteuffel, S. R., P. B. Dennis, N. Pullen, A. C. Gingras, N. Sonenberg, and G. Thomas. 1997. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol. Cell. Biol. 17**:**5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20**:**4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wettenhall, R. E., E. Erikson, and J. L. Maller. 1992. Ordered multisite phosphorylation of Xenopus ribosomal protein S6 by S6 kinase II. J. Biol. Chem. 267**:**9021-9027. [PubMed] [Google Scholar]
- 62.Ziemiecki, A., R. G. Müller, F. Xiao-Chang, N. E. Hynes, and S. Kozma. 1990. Oncogenic activation of the human trk proto-oncogene by recombination with the ribosomal large subunit protein L7a. EMBO J. 9**:**191-196. [DOI] [PMC free article] [PubMed] [Google Scholar]