Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis (original) (raw)
Nudel/NudE facilitates the binding of Lis1 to dynein, which subsequently enhances the recruitment of dynactin to dynein, and dynactin antagonizes Lis1 to relieve Lis1-induced dynein stall on microtubules.
Abstract
Lis1, Nudel/NudE, and dynactin are regulators of cytoplasmic dynein, a minus end–directed, microtubule (MT)-based motor required for proper spindle assembly and orientation. In vitro studies have shown that dynactin promotes processive movement of dynein on MTs, whereas Lis1 causes dynein to enter a persistent force-generating state (referred to here as dynein stall). Yet how the activities of Lis1, Nudel/NudE, and dynactin are coordinated to regulate dynein remains poorly understood in vivo. Working in Xenopus egg extracts, we show that Nudel/NudE facilitates the binding of Lis1 to dynein, which enhances the recruitment of dynactin to dynein. We further report a novel Lis1-dependent dynein–dynactin interaction that is essential for the organization of mitotic spindle poles. Finally, using assays for MT gliding and spindle assembly, we demonstrate an antagonistic relationship between Lis1 and dynactin that allows dynactin to relieve Lis1-induced dynein stall on MTs. Our findings suggest the interesting possibility that Lis1 and dynactin could alternately engage with dynein to allow the motor to promote spindle assembly.
INTRODUCTION
Cytoplasmic dynein is a 1.2-MDa microtubule (MT)-based motor complex. The dynein heavy chain (DHC) is responsible for ATP-dependent motility toward the minus ends of MTs. The motor domain of DHC is composed of a ring of six AAA ATPase modules, a projecting stalk that binds MTs, and a linker arm that extends from the ring, whereas the tail domain supports DHC homodimerization and serves as a scaffold to anchor the noncatalytic subunits, the dynein IC (DIC), light IC (DLIC), and light chains (DLCs). The linker arm is believed to serve as a mechanical lever to generate dynein's power stroke as a result of ATP hydrolysis at the ring. ATP hydrolysis may also lead to changes in MT-binding affinity at the tip of the stalk via long-range allosteric communication through the stalk (Burgess et al., 2003; Gibbons et al., 2005; Kon et al., 2009, 2012; Carter et al., 2011; Roberts et al., 2012). Dynein drives a wide range of functions in eukaryotic cells, including vesicular transport, spindle morphogenesis, spindle orientation, and the inactivation of the spindle assembly checkpoint. It is therefore not surprising that a large number of regulators influence its motor activity by binding to different parts of the complex.
Nudel/NudE, Lis1, and dynactin are major regulators of dynein (Kardon and Vale, 2009; Allan, 2011; Vallee et al., 2012). Lis1, originally identified as a gene responsible for the human disease lissencephaly (Reiner et al., 1993), was later found to have a role in regulating dynein-mediated nuclear migration in Aspergillus, as NudF (Xiang et al., 1995). Initially identified as a multicopy suppressor of NudF (Efimov and Morris, 2000), NudE and its two mammalian homologues (NudE and Nudel, or Nde1 and Ndel1) have since been shown to bind to both dynein and Lis1 (Feng et al., 2000; Niethammer et al., 2000; Sasaki et al., 2000). Although essential roles for Lis1 and NudE/Nudel in dynein function are now firmly established (Smith et al., 2000; Lee et al., 2003; Sheeman et al., 2003; Stehman et al., 2007; Wynshaw-Boris, 2007; Zhang et al., 2010; Pandey and Smith, 2011; Egan et al., 2012, Lam et al., 2010), their mechanisms of action remain unclear.
Several studies have shown that Lis1 inhibits dynein-mediated MT gliding in vitro, while promoting both MT binding and ATP hydrolysis (Mesngon et al., 2006; Yamada et al., 2008; McKenney et al., 2010; Torisawa et al., 2011; Huang et al., 2012). Single-molecule analysis further revealed that Lis1, either alone or together with NudE, enhances dynein binding to MTs under high load conditions (McKenney et al., 2010). Lis1 was recently shown to bind the dynein motor domain at the interface between AAA3 and AAA4, a site that is directly in the path of the allosteric communication that couples ATP hydrolysis to changes in MT-binding affinity. This has led to the proposal that Lis1 acts as a “clutch” that decouples dynein's chemical and mechanical cycles, explaining its ability to induce a persistent dynein stall on MTs (Huang et al., 2012). Together these studies suggests that Lis1 may facilitate dynein-based transport of heavy cargoes on MTs in vivo by enhancing persistent attachment in the face of ongoing ATP hydrolysis (Yi et al., 2011).
Both in vivo and in vitro work suggests that Nudel/NudE, the binding partners of Lis1, serve as scaffold proteins that facilitate Lis1 recruitment to the dynein motor domain. Nudel/NudE become dispensable for MT aster and spindle assembly in Xenopus egg extracts when the Lis1 concentration is doubled (Wang and Zheng, 2011; Zylkiewicz et al., 2011). In keeping with this, NudE is dispensable for nuclear migration in Aspergillus nidulans when NudF (Lis1) is overexpressed (Efimov, 2003). The Lis1-binding site in Nudel/NudE has been mapped to the C-terminal portion of the coiled-coil domain (aa 103–153; Derewenda et al., 2007). Nudel/NudE may have as many as three dynein-binding sites: one for DIC in the N-terminal portion of the coiled-coil domain (Nudel aa 1–80; Wang and Zheng, 2011; Zylkiewicz et al., 2011), one for DHC in Nudel's unstructured C-terminus (Sasaki et al., 2000; Liang et al., 2004), and a third, for DLC (LC8), also in the unstructured C-terminus (McKenney et al., 2011). Although studies suggest that Nudel/NudE could recruit Lis1 to dynein and relieve Lis1-induced dynein stall in vitro using purified Lis1, Nudel/NudE, and dynein (Yamada et al., 2008; McKenney et al., 2010), the observation that increasing the amount of Lis1 renders Nudel/NudE dispensable in egg extracts and in A. nidulans (Efimov, 2003; Wang and Zheng, 2011; Zylkiewicz et al., 2011) suggests that Nudel/NudE are unlikely to be essential in releasing Lis1-induced dynein stall on MTs and subsequent dynein activity in vivo. Interestingly, it was recently reported that NudE and p150Glued compete with one another for binding to DIC on dynein (McKenney et al., 2011). Although the significance of this competitive binding is unknown, it suggests that Nudel/NudE and dynactin might coordinate with one another to regulate dynein-based motility.
Dynactin is another important dynein regulator involved in all aspects of dynein function examined thus far (Schroer, 2004). Discovered as a factor that is sufficient to allow dynein-mediated transport of membrane vesicles in vitro, dynactin is a complex of 11 distinct subunits that comprise a short polymer of actin-family proteins and a shoulder structure that binds dynein via the p150Glued subunit (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; King et al., 2003). A number of studies have shown that dynactin regulates dynein by targeting it to different cellular locations or cargoes (Schroer, 2004; Zhang et al., 2011; Yeh et al., 2012) and by increasing its processivity (King and Schroer, 2000; Ross et al., 2006; Kardon et al., 2009). A recent RNA interference (RNAi) screen in HeLa cells found that dynactin, surprisingly, was dispensable for dynein-dependent spindle pole organization (Raaijmakers et al., 2013). This finding is in direct conflict with a similar RNAi analysis in Cos7 cells (Yeh et al., 2012) and contradicts a large body of previous in vitro work showing that formation of HeLa mitotic asters or Xenopus meiotic spindles is prevented when dynactin is immunodepleted or the dynein–dynactin interaction is inhibited by antibody addition (Gaglio et al., 1996, 1997; Heald et al., 1996, 1997). The discrepancy is most likely due to insufficient depletion of dynactin in the HeLa RNAi screen (Raaijmakers et al., 2013).
Understanding how dynein is regulated by Lis1, Nudel/NudE, and dynactin requires an assay system in which the function of the different protein components can be perturbed using different means and the consequences analyzed under physiologically relevant conditions. To this end, we have used a spindle assembly assay in Xenopus egg extracts with great success (Ma et al., 2009; Goodman et al., 2010; Wang and Zheng, 2011). Using this system in conjunction with MT gliding assays, we have discovered an antagonistic relationship between dynactin and Lis1 in regulating spindle pole organization.
RESULTS
Nudel enhances the binding of Lis1 to dynein by promoting interactions among Nudel, DIC, and Lis1
Previous studies have shown that depletion and/or functional blocking of dynein, dynactin, Lis1, or Nudel/NudE completely disrupts the pole organization of spindles assembled in Xenopus egg extracts (Heald et al., 1996; Gaetz and Kapoor, 2004; Ma et al., 2009; Wang and Zheng, 2011; Zylkiewicz et al., 2011). The requirement for Lis1 and Nudel/NudE, which are not required for dynein-based movement of membrane vesicles in vitro (Schroer and Sheetz, 1991; Yamada et al., 2008), suggests that spindle pole organization requires dynein to function under high load. The precise role of dynactin, which is also required in this context, has, however, remained unknown. We reasoned that, by studying the interactions among dynein, dynactin, Lis1, and Nudel/NudE, we could gain insights regarding how dynein is controlled by these regulators during spindle assembly. Because Nudel/NudE can bind both Lis1 and DIC, it has been suggested that Nudel increases the binding of Lis1 to dynein by forming a ternary complex among Lis1, Nudel, and DIC (Wang and Zheng, 2011; Zylkiewicz et al., 2011). We therefore tested ternary complex formation, which has not been explored previously, using purified Nudel, Lis1, and DIC.
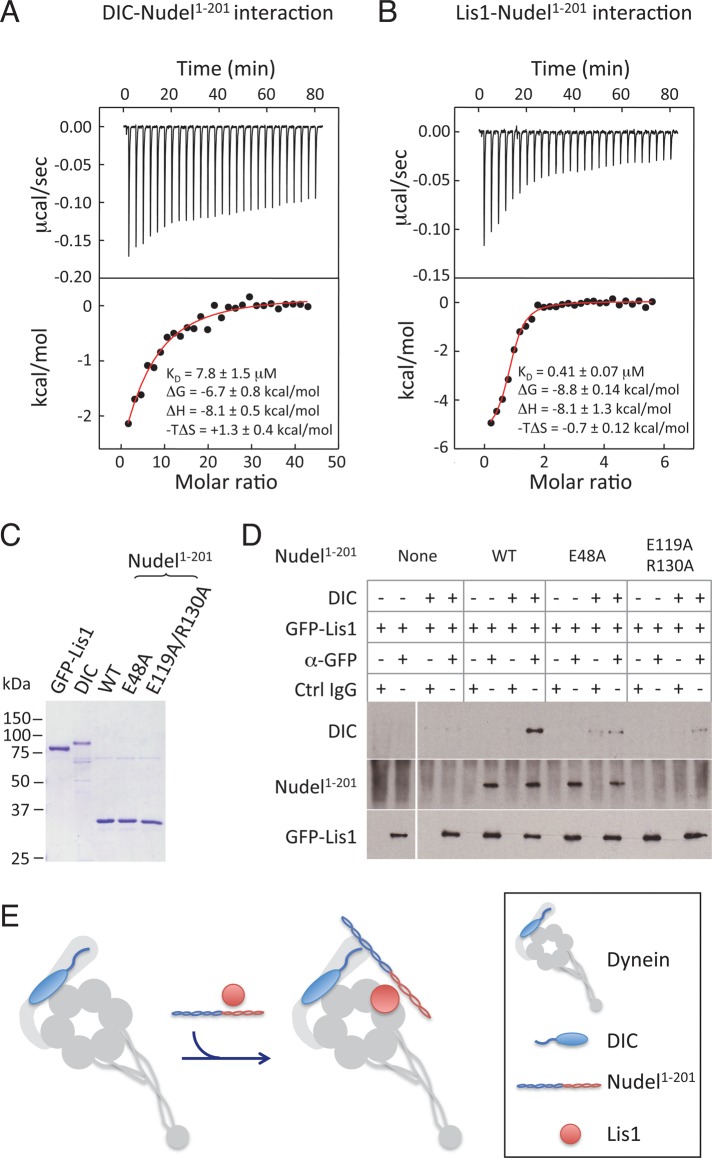
We first used isothermal titration calorimetry (ITC) to measure the binding affinities of Nudel to DIC and Nudel to Lis1. We showed previously that the N-terminal 201 amino acids of Nudel (Nudel1–201), which contains binding sites for both Lis1 (aa 103–153 of Nudel) and DIC (aa 1–80 of Nudel), was sufficient to replace endogenous Nudel/NudE in a spindle assembly assay in Xenopus egg extract when present at approximately twofold higher concentration than the endogenous Nudel/NudE (Wang and Zheng, 2011). We therefore used Nudel1–201 in the ITC measurements. We found that purified Nudel1–201 exhibited saturable binding to purified DIC with a _K_d of 7.8 ± 1.5 μM (Figure 1A). The affinity between Nudel1–201 and Lis1 was 0.41 ± 0.07 μM (Figure 1B), close to that measured previously (Tarricone et al., 2004).
FIGURE 1:
Nudel enhances binding of Lis1 to dynein by simultaneously interacting with Lis1 and DIC in vitro. (A and B) Affinity measurements of Nudel-DIC (A) or Nudel-Lis1 (B) interaction by ITC. Thermograph (top) and binding isotherms (bottom) show the titration of Nudel1–201 with DIC (A) or Lis1 (B). (C) Coomassie Blue–stained gel of the purified His-Nudel1–201 (WT); its mutants E48A and E119A/R130A; DIC; and GFP-Lis1. (D) Nudel1–201 facilitated the formation of Nudel1–201-Lis1-DIC ternary complex in vitro. GFP-Lis1 (1 μM) was mixed with 1 μM DIC in the absence and presence of 0.2 μM Nudel1–201, or two mutants (E48A and E119A/130A). Antibodies to GFP were used to pull down GFP-Lis1, and the presence of Nudel and DIC in immunoprecipitates was analyzed by Western blotting. Nonimmune IgG was used as a control. (E) A schematic illustration of the complex assembled by Nudel1–201, Lis1, and DIC. The squiggle on DIC represents the N-terminus that binds to Nudel and dynactin p150Glued.
To properly assess the interactions among Nudel, DIC, and Lis1, it is important to use concentrations found under physiological conditions. Although the concentrations that can be measured in cytosol do not necessarily reflect the amount of each protein that is engaged with spindle-associated dynein, the total amount of each protein present in the system will clearly impact its interaction with dynein. We therefore determined the concentrations of DIC, Lis1, and Nudel/NudE in Xenopus egg extracts known to support spindle assembly, using quantitative Western blotting with the purified recombinant proteins as standards. We found DIC and Lis1 both to be present at a concentration of ∼1 μM, whereas Nudel/NudE was present at 0.05–0.1 μM (Supplemental Figure S1). We also found p50/dynamitin, a subunit of dynactin, to be present at ∼0.7 μM, similar to DIC (Figure S1).
To perform binding assays, we expressed and purified green fluorescent protein (GFP)-Lis1 and DIC, using baculovirus in Sf9 cells. Nudel1–201 and the two point mutants E48A and E119A/R130A were purified from bacteria (Figure 1C). We showed previously that the single point mutant E48A failed to bind to DIC and that the double point mutant E119A/R130A failed to bind to Lis1 (see Figure 4B in Wang and Zheng, 2011). We verified that GFP-Lis1 was fully functional, because it could rescue spindle assembly in egg extracts depleted of Lis1 (unpublished data). Purified GFP-Lis1 was mixed with purified DIC and Nudel1–201 (or its point mutants) at the concentrations present in egg extracts, and GFP-Lis1 was immunoprecipitated with GFP antibody or control immunoglobulin G (IgG) beads. We found that Lis1 pulled down DIC much more efficiently in the presence of Nudel1–201 than in its absence (Figure 1D). The E48A mutant, which retained Lis1 binding but not DIC binding, failed to promote coprecipitation of DIC with Lis1. Similarly, the E119A/R130A mutant, which retained DIC binding but not Lis1 binding (Wang and Zheng, 2011), also failed to promote coprecipitation of DIC with Lis1. The above findings support the idea that Nudel enhances the binding of Lis1 to dynein by forming a Lis1-DIC-Nudel complex (Figure 1E).
Nudel promotes the binding of Lis1 and dynactin to dynein in Xenopus egg extracts
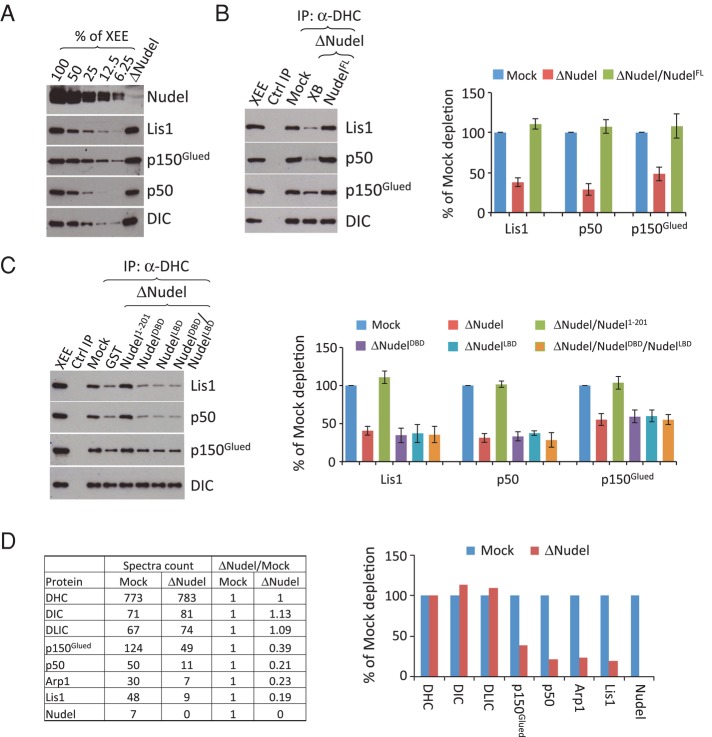
To further confirm that Nudel promotes Lis1 binding to dynein, we turned to Xenopus egg extracts. We immunodepleted Nudel/NudE from the egg extract using antibodies that we showed previously to efficiently remove >90% of both Nudel and NudE from Xenopus egg extract (Ma et al., 2009; Wang and Zheng, 2011) without obvious codepletion of other dynein pathway components (Figure 2A). The lack of codepletion is consistent with the idea that the interactions among dynein, dynactin, Lis1, and Nudel are weak. Despite the weak interaction, antibodies to one protein can immunoprecipitate a small amount of the other proteins. For example, the antibody produced using the last 325 amino acids of DHC coimmunoprecipitated 1–2% of Lis1, p150glued, and p50/dynamitin present in the input egg extracts (Figure S2). Using this antibody to assess dynein-Lis1 binding, we found that depletion of Nudel/NudE from the egg extracts reduced dynein-Lis1 association by ∼60% (Figure 2B).
FIGURE 2:
Nudel promotes the binding of Lis1 and dynactin to dynein in Xenopus egg extracts. (A) Western blot analyses of the efficiency of depleting Nudel/NudE. Decreasing amounts of mock-depleted egg extracts (% of XEE) were loaded along with the Nudel/NudE-depleted egg extracts, which were loaded equivalent to 100% of the mock-depleted egg extracts. Depletion of Nudel/NudE did not affect the amount of dynein or its other regulators present in the egg extracts. (B) Dynein and its associated proteins were immunoprecipitated using antibodies to DHC from mock- or Nudel/NudE-depleted egg extracts (XEE = Xenopus egg extract). The reduction of Lis1 and dynactin (as judged by p150Glued and p50/dynamitin levels) upon Nudel/NudE depletion was rescued by adding 0.1 μM of purified full-length Nudel but not GST. The amounts of Lis1 and dynactin in the immunoprecipitates were estimated by quantitative Western blotting and normalized against DIC. The graph on the right plots protein amounts as a percentage of what was immunoprecipitated from a mock-depleted extract. Error bars: SEM from three independent experiments. The XEE lane shows the relevant protein migration in egg extracts. (C) Effects of Nudel1–201, NudelDBD, and NudelLBD on dynactin and Lis1 association with dynein. The Nudel fragments were added at 0.2 μM to the Nudel/NudE-depleted egg extracts. The DHC antibody or control IgG was used to immunoprecipitate DHC and its associated proteins. Whereas Nudel1–201 was able to restore binding of dynein to Lis1 and dynactin in Nudel/NudE-depleted egg extracts, NudelDBD and NudelLBD, either added alone or together, failed to do so. Western blots were quantified in the plot to the right. Error bars: SEM from three independent experiments. The XEE lane shows the relevant protein migration in egg extracts. (D) Quantitative mass spectrometry analyses of DHC immunoprecipitates from the mock- or Nudel/NudE-depleted egg extracts. The table lists spectral counts of DHC and its associated proteins immunoprecipitated from the indicated conditions. The spectral counts for each protein were normalized against the spectral counts of DHC. The ratio of normalized spectral counts between mock- and Nudel/NudE-depleted samples are determined and plotted to the right.
Interestingly, we found that depletion of Nudel/NudE also reduced the binding of dynactin to dynein, as judged by Western blotting using antibodies to p50/dynamitin and p150Glued (Figure 2B). The reduced binding of dynein to Lis1 and dynactin could be fully rescued by supplementing the egg extract with 0.1 μM full-length Nudel but not by Xenopus buffer (XB) alone (Figure 2B). Supplementing the egg extracts with 0.2 μM glutathione _S_-transferase (GST)-Nudel1–201 also restored the binding of Lis1 and dynactin to dynein (Figure 2C), consistent with our previous finding that a twofold excess of Nudel1–201 could compensate for endogenous Nudel/NudE functions in spindle assembly (Wang and Zheng, 2011). On the other hand, addition of 0.2 μM Nudel fragment containing only the DIC-binding domain (GST-NudelDBD, aa 1–103) or the Lis1-binding domain (GST-NudelLBD, aa 100–201) either alone or in combination could not restore the binding of Lis1 and dynactin to dynein (Figure 2C). This suggested that a complex containing all three components—dynein, Lis1, and Nudel—is necessary for dynactin recruitment in this assay.
To verify that Nudel promotes the binding of Lis1 and dynactin to dynein, we subjected the dynein immunoprecipitates from mock- or Nudel/NudE-depleted egg extracts to comparative mass spectrometry analyses. By calculating the normalized spectral counts (see Materials and Methods), we found that depletion of Nudel/NudE significantly reduced the abundance of Lis1 and dynactin subunits (p150glued, p50/dynamitin, and Arp1) without affecting the abundance of DHC, DIC, and DLIC (Figure 2D). We noticed that reduction of p150glued was less than that of p50/dynamitin upon Nudel/NudE depletion from the above two independent assays. It is currently difficult to envision the cause of such difference. Nevertheless, we conclude that Nudel not only promotes the interaction between dynein and Lis1 but also between dynein and dynactin in egg extracts.
Nudel promotes the binding of dynactin to dynein through Lis1 in Xenopus egg extracts
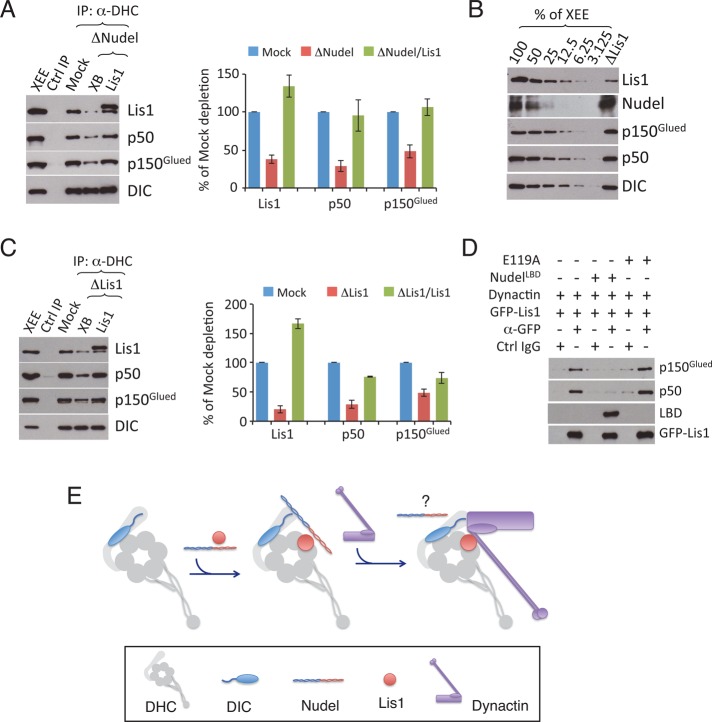
Nudel promotes Lis1 binding to dynein by creating a ternary complex of Nudel, DIC, and Lis1 (see Figure 1E). It is, however, surprising to observe that Nudel also promotes the dynein-dynactin interaction, given that Nudel/NudE and dynactin have been reported to bind a similar site on DIC (McKenney et al., 2011; Nyarko et al., 2012). We showed previously that 1 μM purified Lis1 could restore spindle assembly, a process that depends on the complex of dynein and dynactin, to egg extracts depleted of Nudel/NudE (Wang and Zheng, 2011). We reasoned that Nudel/NudE might be acting through Lis1 to promote dynactin binding to dynein by favoring Lis1 recruitment to dynein. To test this idea, we supplemented Nudel/NudE-depleted egg extracts with 1 μM purified Lis1 and examined the dynein-dynactin interaction by immunoprecipitation using the DHC antibody, as above. We found that addition of 1 μM purified Lis1, but not a buffer (XB) control, allowed normal amounts of dynactin to be coprecipitated with dynein (Figure 3A). To directly test whether Lis1 is required for full dynactin-dynein binding, we depleted Lis1. Western blot analyses revealed that Lis1 depletion did not reduce the amount of Nudel, dynactin (judged by p150glued and p50/dynamitin), or dynein (judged by DIC) in egg extracts (Figure 3B), but it did result in a marked reduction of dynactin in the DHC immunoprecipitates (Figure 3C). The interaction could be restored by adding purified Lis1 (Figure 3C). These findings suggest that Nudel/NudE promotes the dynein-dynactin interaction in a Lis1-dependent manner.
FIGURE 3:
Lis1 promotes the binding of dynactin to dynein in Xenopus egg extracts. (A) 2 μM purified Lis1 restored the association of dynactin with dynein in the Nudel/NudE-depleted extracts (XB = Xenopus buffer; a control for Lis1). Western blots were quantified in the plot to the right. Error bars: SEM from three independent experiments. The XEE lane shows the relevant protein migration in egg extracts. (B) Western blot analyses of the efficiency of depleting Lis1. Decreasing amounts of mock-depleted egg extracts (% of XEE) were loaded along with the Lis1-depleted egg extracts, which were loaded equivalent to 100% of the mock-depleted egg extracts. Depletion of Lis1 did not affect the amount of dynein or its other regulators present in the egg extracts. (C) Lis1 depletion from the egg extracts resulted in the reduction of dynactin and dynein association, which was rescued by 2 μM of the purified Lis1. Western blots were quantified in the plot to the right. Error bars: SEM from three independent experiments. The XEE lane shows the relevant protein migration in egg extracts. (D) Purified Lis1 directly binds to purified dynactin. Purified GFP-Lis1 (2 μM) was mixed with purified dynactin in the presence or absence of 4 μM NudelLBD or NudelLBDE119A. A GFP antibody or control IgG was used for immunoprecipitation. Western blots were quantified in the plot to the right. Error bars: SEM from three independent experiments. (E) A cartoon illustrating the interactions between dynein and its regulators. The binding of Nudel/NudE and Lis1 to dynein promotes the binding of dynactin to the complex. The cartoon reflects the known positions of Lis1 on the motor domain and DIC on the tail. The DIC N-terminus (squiggle) interacts with amino acids 415–530 of the dynactin p150Glued component, which is associated with the Arp1 filament (purple rectangle) at the base of the p150Glued projecting arm (aa 200–350). The precise contact site between Lis1 and dynactin is unknown. On dynactin binding, the fate of Nudel/NudE is unknown (question mark). Because Nudel/NudE has been suggested to bind to another site besides DIC on dynein, upon dynactin binding, Nudel/NudE could either shift from DIC to this other binding site or dissociate from dynein.
The p150Glued subunit of dynactin has been shown to bind the DIC N-terminus directly (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; King et al., 2003) with micromolar affinity (Morgan et al., 2011). It has been reported that p50/dynamitin and Lis1 can be coimmunoprecipitated in cells overexpressing the two proteins (Tai et al., 2002), suggesting an interaction between Lis1 and dynactin as well. To test whether Lis1 directly binds to dynactin, we mixed purified bovine dynactin with purified GFP-Lis1 and then used an antibody to GFP to pull down Lis1. We found that purified GFP-Lis1 indeed interacted with purified dynactin (Figure 3D). Because Lis1 also interacts directly with Nudel, we tested whether the interactions between Lis1 and Nudel or dynactin were mutually exclusive by adding just the Lis1-binding domain of Nudel (NudelLBD) to the binding reaction. NudelLBD blocked the binding of Lis1 to dynactin (Figure 3D), but a mutant version of the LBD (NudelLBDE119A) that exhibits only weak binding to Lis1 (Wang and Zheng, 2011) did not (Figure 3D). This suggests that Nudel and dynactin bind to the same site on Lis1.
Together these findings suggest the following testable hypothesis to explain the way in which dynein interacts with its essential cofactors. We propose that Nudel/NudE facilitates the loading of Lis1 onto dynein, forming a complex that allows dynein to function under high load. Dynactin may join the complex by displacing Nudel/NudE from DIC completely, or Nudel/NudE may remain associated via its other dynein-binding site(s) (Figure 3E).
NudelDBD inhibits spindle assembly by binding to DIC and inhibiting the interaction between dynactin and dynein
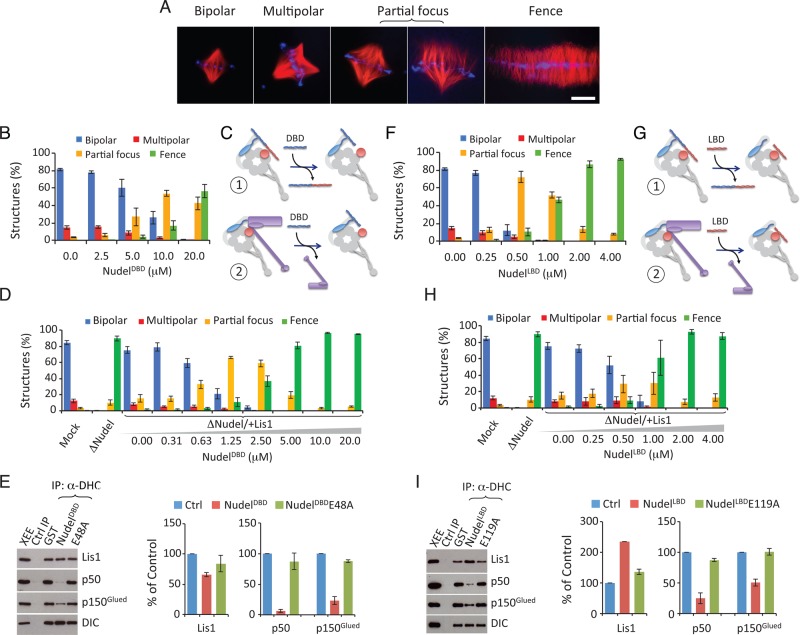
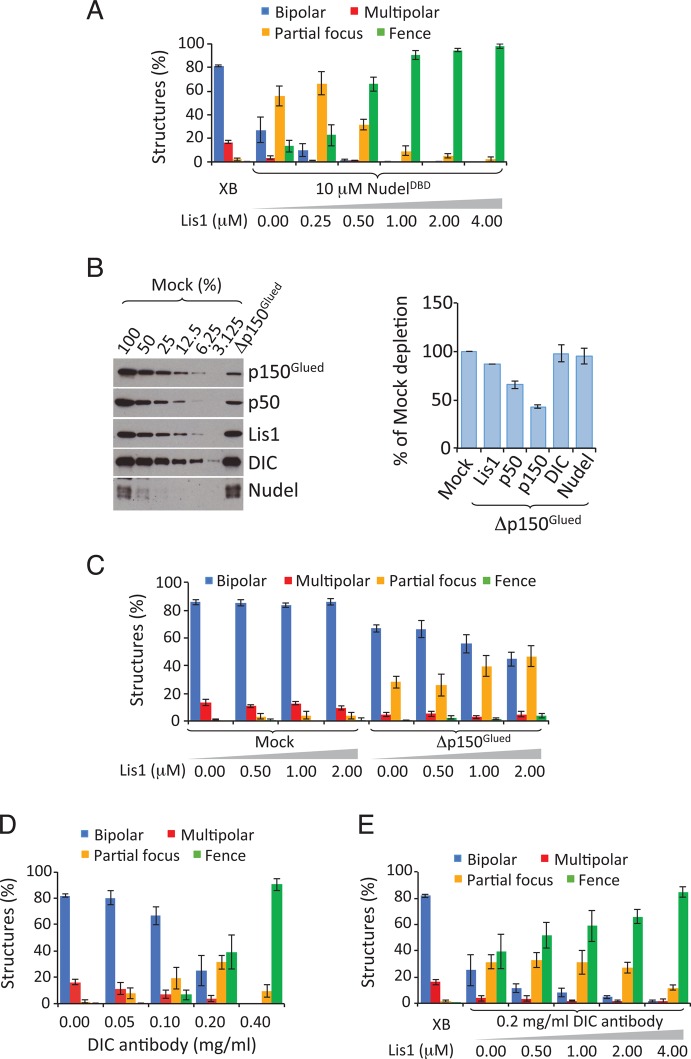
This interaction model (Figure 3E) makes several predictions that can be tested using the Xenopus spindle assembly assay supplemented with reagents designed to disrupt specific interactions. We showed previously that perturbing dynein activity in egg extracts results in defects of increasing severity (Figure 4A): spindles showing multiple poles, partially focused poles, or completely unfocused poles (“fence-like” spindles; Wang and Zheng, 2011). These defects can be reliably quantified and thus provide a measure of how different disrupting reagents affect interactions among dynein and its regulators.
FIGURE 4:
The interaction between dynein and dynactin promoted by Lis1 is necessary for spindle morphogenesis. (A) Images of representative spindles defined as bipolar, multipolar, partial focus, and fence. Scale bar: 20 μm. (B) Analysis of the effect of NudelDBD on spindle pole organization in egg extracts. Increasing concentrations of NudelDBD caused increasing spindle pole defects. Error bars: SEM of three independent experiments. (C) Cartoon illustrating that NudelDBD may inhibit dynein function by either competing with endogenous Nudel/NudE (scenario 1) or dynactin (scenario 2) to bind to dynein. See caption to Figure 3D for the keys for the graphic objects. (D) Analysis of the inhibitory effect of NudelDBD in egg extracts depleted of Nudel and supplemented with Lis1. NudelDBD inhibited spindle pole organization in a dominant-negative manner in the Nudel/NudE-depleted and Lis1-supplemented egg extract. (E) Addition of 10 μM of NudelDBD (but not NudelDBDE48A) into egg extracts reduced the interaction between dynactin and dynein as determined by DHC immunoprecipitation followed by Western blotting. The plot to the right shows the quantification of proteins immunoprecipitated by the DHC antibody. Error bars: SEM of three independent experiments. The XEE lane shows the relevant protein migration in egg extracts. (F) Analysis of the effect of NudelLBD on spindle pole organization in egg extracts. Increasing concentrations of NudelLBD caused increasing spindle pole defects. Error bars: SEM of three independent experiments. (G) Cartoon illustrating that NudelLBD may inhibit dynein function by competing with endogenous Nudel/NudE to bind to endogenous Lis1 (scenario 1) or by competing with dynactin to bind to dynein (scenario 2). See caption to Figure 3D for the key for the graphic objects. (H) Analysis of the inhibitory effect of NudelLBD in extracts depleted of Nudel and supplemented with Lis1. NudelLBD inhibited spindle pole organization in a dominant-negative manner in the Nudel/NudE-depleted and Lis1-supplemented egg extract. (I) Addition of 4 μM of NudelLBD (but not NudelLBDE119A) into egg extracts reduced the interaction between dynactin and dynein as determined by DHC immunoprecipitation followed by Western blot analysis. The plot to the right shows the quantification of proteins immunoprecipitated by the DHC antibody. Error bars: SEM of three independent experiments. The XEE lane shows the relevant protein migration in egg extracts.
Our interaction model predicts that disrupting the interaction between Nudel/NudE and DIC would inhibit spindle assembly. We used NudelDBD, which is known to bind DIC but not Lis1, for this purpose. NudelDBDE48A, which does not bind DIC, was used as a control. By adding increasing concentrations of NudelDBD or NudelDBDE48A to egg extracts and measuring the effect on spindle pole organization, we found that NudelDBD inhibited spindle pole assembly (Figure 4B), whereas NudelDBDE48A had no effect (Figure S3A). NudelDBD inhibited spindle assembly with an estimated effective concentration (EC50, wherein 50% of spindles assembled had abnormal morphology) of 5–10 μM (Figure 4B).
Although NudelDBD is likely to inhibit spindle assembly by preventing the binding of endogenous Nudel/NudE to DIC and blocking Lis1 recruitment, it is equally likely to interfere with the binding of dynactin to DIC, which would also inhibit spindle assembly (Figure 4C). To differentiate between these two possibilities, we simplified the system by removing endogenous Nudel/NudE and then assaying spindle assembly after supplementing the depleted extract with various components. As expected (Wang and Zheng, 2011), 1 μM purified Lis1 fully restored spindle formation in Nudel/NudE-depleted egg extracts (Figure 4D). We found that, as in mock-depleted control extracts (Figure 4B), NudelDBD inhibited spindle assembly in the Nudel/NudE-depleted and Lis1-supplemented egg extracts (Figure 4D). Interestingly, NudelDBD appeared to be a more potent inhibitor in this context, with an EC50 of 0.63–1.25 μM that was eightfold more potent than what was seen in control extracts (compare Figure 4D with 4B). This is consistent with our observation that full-length Nudel/NudE facilitates dynactin binding to dynein (Figure 2). In the absence of endogenous Nudel/NudE, NudelDBD would be expected to be highly effective at preventing the binding of dynactin to DIC. Together the above findings suggest that NudelDBD disrupts spindle assembly by interfering with the binding of dynactin to dynein (as illustrated in Figure 4C, scenario 2).
To verify that NudelDBD indeed inhibits dynactin-dynein binding, we immunoprecipitated dynein from egg extracts containing NudelDBD or NudelDBDE48A. Excess wild-type NudelDBD, but not NudelDBDE48A, not only slightly reduced Lis1 binding to dynein but also severely reduced dynein-dynactin interaction (Figure 4E). These findings suggest that NudelDBD could block the binding of dynactin to dynein by occupying the dynactin-binding site on DIC, thereby inhibiting spindle assembly.
NudelLBD inhibits spindle assembly by binding to Lis1 and inhibiting the interaction between dynactin and dynein
According to the interaction model (Figure 3E), excess NudelLBD should bind Lis1 and interfere with Lis1’s ability to promote dynactin and dynein interaction. To explore this possibility, we added increasing concentrations of NudelLBD to egg extracts, using NudelLBDE119A, which binds Lis1 very weakly (Wang and Zheng, 2011), as a control. We found NudelLBD, but not NudelLBDE119A, inhibited spindle assembly (Figures 4F and S3B). NudelLBD inhibited bipolar spindle assembly with an estimated EC50 of 0.25–0.5 μM (Figure 4F).
To determine whether NudelLBD inhibited spindle assembly by preventing the binding of endogenous Nudel/NudE to Lis1 or by interfering with the recruitment of dynactin to Lis1 in the dynein-Lis1 complex (Figure 4G), we again used egg extracts depleted of Nudel/NudE and supplemented with 1 μM Lis1. Once again, NudelLBD inhibited spindle assembly (Figure 4H). This suggests that it interferes with the binding of dynactin to dynein, inhibiting spindle assembly (Figure 4G, scenario 2).
Because the Lis1-dynein complex should still form under these circumstances, this result suggests that the inhibitory effect of NudelLBD was due to disruption of the dynein-dynactin interaction. To confirm this, we immunoprecipitated dynein from egg extracts in the presence of excess NudelLBD. NudelLBD, but not NudelLBDE119A, reduced the interaction between dynactin and dynein (Figure 4I), as predicted. Curiously, NudelLBD appeared to enhance the binding of Lis1 to dynein (Figure 4I). NudelLBD may induce a conformational change on Lis1 or dynein that increases Lis1-dynein affinity. If so, NudelLBD may not act as a simple competitive inhibitor, unlike NudelDBD. Alternatively, NudelLBD might affect other dynein regulators present in the egg extract that indirectly enhance Lis1-dynein binding. Although further work will be needed to understand the effect of NudelLBD on the association between dynein and Lis1, these results suggest that the increased binding of dynactin to dynein involves both DIC and Lis1.
Dynactin and Lis1 antagonistically regulate dynein activity during spindle assembly
Recent work suggests that Lis1 enhances the binding of dynein to MTs by locking dynein in a persistent force-producing state on MTs, which would allow dynein to exert force under high load (Yamada et al., 2008; McKenney et al., 2010; Torisawa et al., 2011; Yi et al., 2011; Huang et al., 2012). Although Nudel/NudE was shown to promote dynein motility by reducing Lis1-induced dynein stall on MTs in vitro (Yamada et al., 2008; McKenney et al., 2010; Torisawa et al., 2011), the molar ratio of Nudel/NudE to Lis1 used in these experiments was ∼10–20-fold higher than that found in egg extracts (Ma et al., 2009; Wang and Zheng, 2011), raising the possibility that Nudel/NudE does not facilitate detachment of dynein from MTs in vivo. To complicate matters further, dynactin has been shown to allow dynein to move more processively along MTs (King and Schroer, 2000; Ross et al., 2006; Kardon et al., 2009). The interaction model (Figure 3E) predicts that binding of dynactin to the dynein-Lis1 complex can relieve Lis1-induced dynein stall on MTs, allowing processive movement under high load conditions (e.g., maintenance of a focused spindle pole). In other words, Lis1 and dynactin may function antagonistically. If so, conditions that favor Lis1-dynein binding or disfavor dynactin-dynein binding would favor dynein stall and yield spindle assembly defects.
To explore this possibility, we started by reducing dynein-dynactin interactions in egg extracts by adding NudelDBD (10 μM) to partially suppress bipolar spindle assembly (Figure 5A). Further addition of purified Lis1 (up to 4 μM) resulted in increased disruption of bipolar spindle assembly (Figure 5A), whereas increasing the Lis1 concentration in a control egg extract had no effect (Figure S4A; see also Figure 5D). We then partially depleted dynactin using an antibody raised against p150Glued (Figure 5B). Quantitative blotting revealed ∼50% depletion of dynactin subunits with minimal impact on dynein, Lis1, or Nudel/NudE levels (Figure 5C). This treatment resulted in reduced formation of normal bipolar spindles and a corresponding increase in partially focused spindles (Figure 5D). Importantly, adding excess Lis1 to these egg extracts resulted in increased spindle defects (Figure 5D).
FIGURE 5:
Dynactin and Lis1 regulate dynein antagonistically during spindle assembly. (A) The spindle pole defects caused by NudelDBD were increased by addition of excess Lis1. NudelDBD added at 10 μM induced partial inhibition of spindle pole organization, which was further exacerbated by addition of increasing concentrations of Lis1. (B) Antibodies to p150Glued partially depleted dynactin as judged by Western blot analyses of p150glued and p50/dynamitin. Such depletion did not detectably reduce the amount of Nudel/NudE and DIC in the egg extract. The plot to the right shows the quantification of the degree of depletion. (C) Analysis of spindle pole organization in extracts with partial depletion of dynactin (or mock-depleted) in the presence of excessive Lis1. Addition of increasing concentrations of Lis1 increased the spindle pole organization without affecting spindle assembly in the mock-depleted egg extracts. (D) Analysis of spindle pole organization in extracts treated with increasing amounts of DIC antibody. Addition of increasing concentrations of the DIC antibody caused a gradual increase of spindle pole disorganization with an estimated EC50 between 0.1 and 0.2 mg/ml. (E) Analysis of spindle pole organization in extracts treated with DIC antibody to which increasing concentrations of purified Lis1 was added. The partial spindle pole disorganization caused by 0.2 mg/ml DIC antibody was increased by addition of increasing concentrations of purified Lis1. Error bars: SEM of three independent experiments.
Finally, we used an antibody raised against the N-terminal 65 amino acids of DIC to block the dynein-dynactin and dynein-Nudel/NudE interactions. Addition of increasing amounts of the DIC antibody, but not a control, resulted in an increasing number of defective spindles with an EC50 of 0.1–0.2 mg/ml (Figures 5E and S4B). Importantly, the partial inhibition seen at 0.2 mg/ml DIC antibody was exacerbated by addition of increasing amounts of Lis1, which is expected to further increase dynein stall on MTs (Figure 5E). Addition of Lis1 to a parallel egg extract containing control IgG had no effect (Figure S4C), as expected. Together these experiments show that weakening dynactin loading onto dynein exacerbates dynein stall on MTs caused by excess Lis1, even though addition of comparable levels of Lis1 to unperturbed egg extracts is not sufficient to cause spindle morphology defects. These findings indicate that Lis1 and dynactin have opposite effects on dynein activity during spindle assembly.
Dynactin overcomes Lis1-induced dynein stall on MTs in vitro
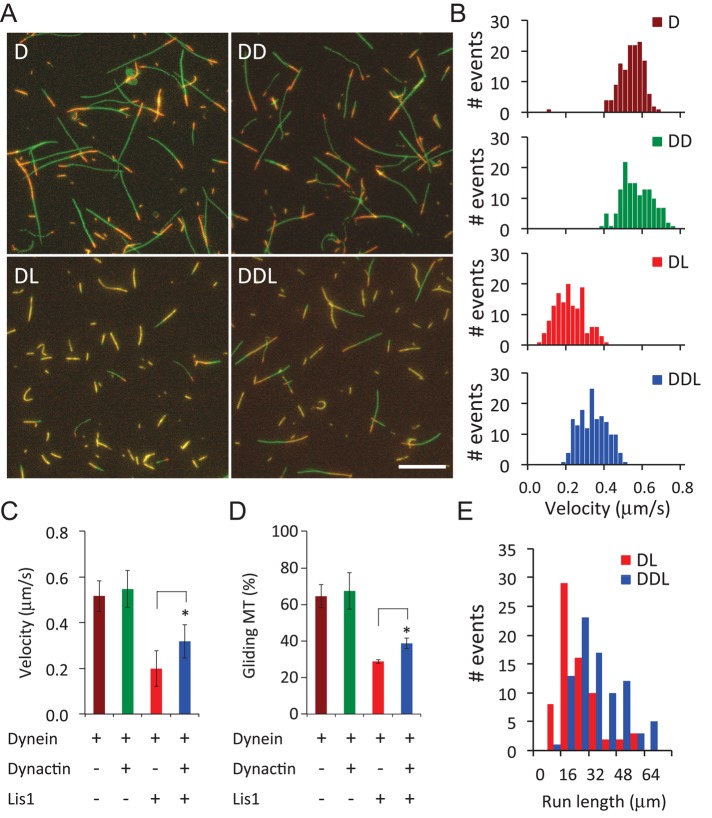
To directly test whether dynactin can relieve Lis1-induced dynein stall on MTs in the absence of Nudel/NudE, we used a dynein-mediated MT gliding assay. As previously reported (King and Schroer, 2000), we found that purified dynein could translocate MTs with plus ends leading at an average velocity of 0.52 ± 0.07 μm/s (n = 141). Addition of purified dynactin did not increase gliding velocity significantly (mean = 0.55 ± 0.08 μm/s, n = 141; Figure 6, A–C). Consistent with previous findings (Yamada et al., 2008; Torisawa et al., 2011), addition of Lis1 (2 μM) resulted in a reduction of the percentage of gliding MTs from 64.5 to 28.8% (Figure 6, A and D), and those that did move did so at a reduced velocity (0.20 ± 0.08 μm/s, n = 141; Figure 6C). Next we added Lis1 into flow chambers coated with a mixture of dynein and dynactin. Dynactin was found to partially reverse the Lis1-induced MT stall by increasing both the percentage of gliding MTs (from 28.8 to 38.7%) and gliding velocity (0.20 ± 0.08 μm/s to 0.32 ± 0.07 μm/s, n = 158) (Figure 6, B and D). Analysis of MT run lengths between pauses under the different conditions revealed that addition of dynactin to reactions containing dynein and Lis1 also yielded a significant increase in overall displacement (Figure 6E). These data suggest that dynactin can relieve Lis1-induced dynein stall on MTs and restore its ability to support long-range movement.
FIGURE 6:
Dynactin relieves Lis1-induced dynein stall on MTs in vitro. (A) Compiled images (one field per condition) of MT gliding on glass surfaces coated with purified dynein (D), dynein plus dynactin (DD), dynein plus Lis1 (DL), or dynein plus dynactin and Lis1 (DDL). Individual MTs are shown in orange. Their trajectories, which were constructed from 30 consecutive images taken at 1 frame/s, are shown with green lines. The longer the lines, the farther the MTs have moved. Scale bar: 10 μm. (B) Velocity histograms of gliding MTs driven by dynein (D), dynein plus dynactin (DD), dynein plus Lis1 (DL), or dynein plus Lis1 and dynactin (DDL). (C) Average velocities of MT gliding under the indicated conditions. (D) Percentages of gliding MTs under the indicated conditions. (E) MT run lengths between pauses under the indicated conditions. Each event is one gliding event between pauses. Error bars: SD. *, Student's t test (p < 0.001).
DISCUSSION
Dynactin, Lis1, and Nudel/NudE are among the most-studied dynein regulators. Although recent studies have shown that Lis1 promotes a stable MT-bound state for dynein, which may allow dynein to translocate heavy cargoes (Yamada et al., 2008; McKenney et al., 2010; Torisawa et al., 2011; Yi et al., 2011; Huang et al., 2012), it remains unclear whether and how the “clutch” function of Lis1 is integrated with the effects of dynactin to yield motility. Dynactin was originally identified as an activity that allowed purified dynein to move vesicles (a low load cargo) processively along MTs (Schroer and Sheetz, 1991). How dynactin contributes to movement of heavy cargoes, however, remains unclear.
Our findings suggest that dynactin and Lis1 function antagonistically to ensure spindle pole focusing in Xenopus egg extracts. They are consistent with a model in which Nudel/NudE promotes the binding of Lis1 to dynein, which in turn enhances the binding of dynactin to dynein. Dynactin-dynein binding may disengage the Lis1 clutch, thereby allowing dynein to move processively under high load. Of course, the egg extract contains other dynein regulators, whose activities may also have been impacted by our manipulations. Therefore, although we favor the idea that Lis1 and dynactin function together to support dynein-based movement under high load, it should be noted that other possible modes of dynein regulation cannot be ruled out.
Much previous work suggests that Nudel and NudE promote the binding of Lis1 to dynein (Sasaki et al., 2000; Liang et al., 2004; McKenney et al., 2010; Wang and Zheng, 2011; Zylkiewicz et al., 2011; Huang et al., 2012). Using purified proteins, we provide a molecular explanation for this activity. The close proximity of the DIC- and Lis1-binding sites in amino acids 1–201 of Nudel or NudE facilitates Lis1 recruitment to dynein by providing a multivalent interaction (Figure 1E). Strikingly, we find that both Nudel/NudE and Lis1 promote dynein-dynactin interactions using immunoprecipitation assays (Figures 2 and 3). Because excess Lis1 can promote dynactin binding to dynein in the absence of Nudel/NudE, we suggest that association of Lis1 with dynein creates an optimal “landing pad” for dynactin. Consistent with this, we find that purified Lis1 can bind purified dynactin directly (Figure 3C). It is well established that the dynactin p150glued subunit binds DIC directly (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; King et al., 2003), and our work suggests that Lis1 provides an additional contact site on the dynein-Lis1 complex for dynactin.
Although the difficulty of obtaining large amounts of dynactin has so far made it impossible to use only purified proteins to directly test whether Lis1 promotes the dynein-dynactin interaction, our findings encourage further studies aimed at elucidating how Lis1, Nudel/NudE, and dynactin could coordinate with one another to regulate dynein. For example, the finding that either NudelDBD or NudelLBD can inhibit the binding of dynactin to dynein in the absence of endogenous Nudel/NudE in egg extracts suggests that dynactin and Nudel/NudE share similar binding sites not only on DIC but possibly also on Lis1 (Figure 4). Additionally, the observation that NudelLBD enhances Lis1 binding to dynein (Figure 4I) suggests that dynactin may weaken Lis1-dynein binding by attenuating the interaction of Nudel/NudE with the motor complex, leading to disengagement of the Lis1 clutch. Our ongoing efforts to purify high concentrations of Lis1, Nudel/NudE, dynein, and dynactin will make it possible to test these interesting ideas in vitro.
We note that supplementing Lis1 up to fourfold over the endogenous level does not cause spindle defects in egg extracts that are mock-depleted or supplemented with control antibodies (Figures 5D and S4, A and C), whereas overexpression of Lis1 in mice results in brain defects (Bi et al., 2009). This discrepancy could be due to either a relatively higher concentration of dynactin in egg extracts than in mouse brain or a lack of sensitivity of the in vitro spindle assembly assay to excess Lis1. Although our findings suggest that dynactin and Lis1 function antagonistically to regulate dynein motility for spindle assembly, it is important to point out that our findings on spindle formation in Xenopus egg extracts may not apply to dynein-based transport of other cargoes. Moreover, additional study will be required to address whether and how Lis1 contributes to the motility of small vesicles and other low-load cargoes in animal cells in vivo.
MATERIALS AND METHODS
Cloning, protein expression, and purification
The cDNAs encoding the full-length Xenopus dynactin p150Glued (clone ID 4959212) and the partial human DHC (clone ID 3546415) were purchased from Open Biosystems (Huntsville, AL). The full-length DIC and its truncated forms were subcloned into either pGEX6P2 (GE Healthcare Life Sciences, Piscataway, NJ) or pET30a vectors (Novagen, Darmstadt, Germany). Dynactin p150Glued-CC1 (aa 174–505) and the C-terminal fragment of DHC (aa 4322–4646) were also subcloned into these two vectors. The full-length DIC and p150Glued were also subcloned into pFASTBacHT vector (Invitrogen, Carlsbad, CA) for expression in Sf9 cells. Expression and purification of recombinant proteins in Escherichia coli and in Sf9 cells were described previously (Wang and Zheng, 2011). GFP-tagged Lis1 or His-tagged Lis1 were produced in Sf9 cells and purified as described (Wang and Zheng, 2011). While GFP-Lis1 was used in the in vitro binding assays described in Figure 3C, the His-tagged Lis1 was used in all the other experiments. All proteins were dialyzed into XB buffer (10 mM K-HEPES, pH 7.7, 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose) and concentrated to appropriate concentrations before being frozen in liquid nitrogen and stored at −80°C.
Antibodies
The polyclonal antibody to human DHC used in dynein immunoprecipitation experiments was raised in rabbits using the purified GST-DHC C-terminal region (aa 4322–4646) and affinity purified using the same region of DHC but tagged with 6xHis. The rabbit dynactin p150Glued antibody was raised against GST-CC1 (aa 174–505) of Xenopus p150Glued and affinity purified using 6xHis-CC1. The rabbit DIC antibody was raised against GST-DIC N-terminal region (aa 1–65) of Xenopus DIC and affinity purified using GST-DIC1–65 after preabsorption of the anti-GST antibody in the serum. The antibodies mentioned above were affinity purified using the corresponding antigens coupled to Affi-Gel 10/15 (153-6098; Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The concentration of the purified antibody was determined by UV absorbance at 280 nm divided by the immunoglobulin extinct coefficient of 1.35. Additional antibodies to dynactin p150Glued and p50/dynamitin were purchased from BD Biosciences (611002 and 610473, respectively; Franklin Lakes, NJ). The anti-GFP antibody was purchased from Genscript (A01388; Piscataway, NJ). Other antibodies used in this study (DIC [74.1], Lis1, His tag, and Nudel/NudE) were described in an earlier study (Wang and Zheng, 2011).
ITC
ITC experiments were performed using a high-precision ITC200 titration calorimetric system from MicroCal/GE Healthcare (Northampton, MA). His-tagged DIC used in this study was expressed in Sf9 cells and purified using Ni-NTA agarose (Qiagen, Valencia, CA), whereas His-tagged Nudel1–201 was expressed and purified from E. coli. The protein concentration was determined by comparing the purified proteins with bovine serum albumin (BSA) standard on the same SDS–PAGE gel, and was converted into molar concentrations with the calculated molecular mass. For studies of the interaction between Nudel and DIC, Nudel1–201 at a concentration of 6 μM was titrated with 200 μM DIC. The titrant was added in steps of 1.4 μl, with preset intervals of 180 s. All solutions were degassed to avoid any formation of bubbles in the calorimeter during stirring. The reagents were dissolved in XB buffer, and all measurements were performed at 25°C. The heat released upon injection of the titrant was obtained from the integral of the calorimetric signal. The heat associated with the binding reaction was obtained by subtracting the heat of dilution from the heat of reaction. The individual heats were plotted against the molar ratio, and the values for the enthalpy change (Δ_H_) and association constant (_K_a = 1/_K_d) were obtained by nonlinear regression of the data.
Xenopus egg extract and sperm spindle assembly assay
Xenopus egg extracts and spindle assembly stimulated by Xenopus sperm DNA were carried out as previously described (Wang and Zheng, 2011). Immunodepletion was performed at 4°C using Affi-protein A beads (Bio-Rad, Hercules, CA) coupled with corresponding antibodies. One hundred micrograms of antibody was generally used to deplete 100 μl of Xenopus egg extract. Two rounds of immunodepletion using p150Glued antibody were required to remove 40–50% of dynactin. For mock depletion, purchased nonimmunized rabbit IgG (Sigma-Aldrich, St. Louis, MO) was used. For rescue experiments, the purified proteins were added into the immunodepleted extracts at a 1:20 dilution. For antibody inhibition experiments, nonimmunized rabbit IgG or purified anti-DIC antibody were dialyzed into XB and were added at a 1:20 dilution.
Immunoprecipitation, quantitative Western blotting, and mass spectrometry analyses
For studying the interactions among purified Nudel, Lis1, and DIC, 1 μM GFP-Lis1 and 1 μM His-DIC were incubated with either 0.2 μM His-Nudel1–201 or 0.2 μM His-Nudel1–201E48A or 0.2 μM His-Nudel1–201E119A/R130A in XB buffer in the presence of 4% BSA as a carrier. After 1 h of incubation at 4°C, the reaction mixture was centrifuged at 14,000 × g for 10 min in the cold room to clear any protein aggregates formed during incubation. The mixture was then incubated with the Affi-Prep protein A beads coupled with 1.5 μg GFP antibodies for 1 h at 4°C. The beads were washed five times with XB and were resuspended in 50 μl of 2× SDS sample buffer. Samples were diluted (1:10 for Lis1 and DIC; no dilution for Nudel1–201) and loaded 5 μl onto 10% SDS–PAGE gel and transferred onto nitrocellulose membranes for Western blot analyses. The dilution factors of antibodies used for immunoblotting were as follows: anti-Lis1, 1:2500; anti-DIC (74.1), 1:500; anti-His tag, 1:500 (H1029; Sigma-Aldrich).
For immunoprecipitation of DHC from Xenopus egg extract, 5 μg anti-DHC antibody was coupled to 10 μl of Affi-Prep protein A beads (slurry volume) by incubating overnight at 4°C on a rotating shaker. The extracts capable of assembling spindles were chosen for immunoprecipitation. The immunodepletion of Nudel/NudE and Lis1 was carried out as described earlier (Wang and Zheng, 2011). In general, 100 μl of extracts was depleted by 100 μg antibody. The immunoprecipitation was performed by incubating the DHC antibody beads with 100 μl of extracts for 1 h at 4°C. For dominant-negative perturbation experiments, 50 μl of extract was incubated with NudelDBD or NudelLBD at the indicated concentrations for 1 h at room temperature. The extracts were then immunoprecipitated by DHC antibody beads for 1 h at 4°C. The immunoprecipitates were washed for five times with XB in the cold room and were resuspended in 50 μl of 2× SDS loading buffer.
For quantitative Western blotting, samples were loaded in a series of dilutions and resolved by 7.5 or 10% SDS–PAGE gels. The amount of samples loaded onto gels is critical to get linear relationships of bands on x-ray films. The dilution factors of loading were as follows: p150Glued/p50/DHC, 1×; Lis1, 5×; DIC, 15×. The dilution factors of antibodies used for immunoblotting were as follows: anti-p150Glued, 1:2500; anti-dynamitin p50, 1:125 (overnight incubation); anti-Lis1, 1:2500; anti-DIC polyclonal, 1:5000. To reduce background, the secondary antibody used for polyclonal primary antibody in immunoblotting was Rabbit TrueBlot (18-1688-33; Rockland, Gilbertsville, PA). The relative protein level was determined as percentage of the control (i.e., the mock-depleted extract) and normalized against the protein level of DIC in immunoprecipitates. Quantification of band intensity was carried out in Photoshop Elements (Adobe, San Jose, CA).
For quantitative mass spectrometry analyses, the immunoprecipitated proteins were first denatured while still on the beads and reduced with 8 M urea, 5 mM Tris(2-carboxyethyl)phosphine in 100 mM Tris-HCl buffer (pH 8.5) at room temperature for 30 min and then alkylated by 10 mM iodoacetamide at room temperature for 15 min in the dark. The urea was then diluted to <2 M with 100 mM Tris-HCl buffer, and the proteins were subjected to tryptic digestion at enzyme:substrate ratio of 1:100 at 37ºC overnight. The digestion was stopped by adding formic acid to a final concentration of 1%. Beads were removed by centrifugation at 14,000 rpm for 10 min. The protein digest was first pressure-loaded onto a 250-μm i.d. trap column containing 2.5 cm of 5 μm Aqua C18 material (Phenomenex, Torrance, CA) and 2.5 cm 5 μm Partisphere strong cation exchanger (Whatman, Piscataway, NJ) with a Kasil frit at one end. The trap column was then washed with buffer containing 95% water, 5% acetonitrile, and 0.1% formic acid and further connected using a zero-dead volume union (IDEX Health & Science, Lake Forest, IL) to a 100-μm-i.d. analytical column with a 5-μm pulled tip and packed with 15 cm of 5 μm Aqua C18 material. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis was conducted on an Agilent 1100 quaternary high-performance liquid chromatography pump (Palo Alto, CA) and LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific, San Jose, CA). The tip flow rate was reduced to 300 nl/min using a 1:1000 split flow from the HPLC pump. The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A), 80% acetonitrile/0.1% formic acid (buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). A seven-step MudPIT analysis was performed by running 5 min with 0, 20, 40, 60, 80, and 100% buffer C and 10% bufferB–90% buffer C for each step, followed by a 2-h reversed-phase gradient. The repetitive 2-h gradients were from 100% buffer A to 10% buffer B over 3 min, 10% buffer B to 45% buffer B over 80 min, up to 100% B over 12 min, held at 100% B for 10 min, then back to 100% A for a 10-min column reequilibration. MS/MS spectra on the LTQ-Orbitrap were acquired in a data-dependent mode, with one full MS scan followed by eight MS/MS scans at ESI voltage of 2.5 kV. For MS data analysis, tandem mass spectra were extracted from raw files using RawExtract 1.9.9 McDonald et al., 2004) and searched using ProLuCID (Xu, 2006) against a combined database with their sequences reversed. The combined database includes the protein entries of Xenopus laevis and Xenopus tropicalis from Xenbase (www.xenbase.org) and Xenopus tropicalis predicted proteins (version 4.1) from the Joint Genome Institute. The search space included all full and semi-peptide candidates. Carbamidomethylation of cysteine was considered as a static modification. Peptide precursor and fragment mass tolerance were set at 50 and 600 ppm, respectively. DTASelect was used to filter protein candidates to a 1% false discovery rate based on the number of reverse sequences identified with 10 ppm peptide mass cutoff applied. Normalized spectral counting (Liu et al., 2004) was used to quantify the relative abundance of the identified proteins from Nudel-NudE depleted and mock-depleted conditions. Briefly, the total number of tandem MS spectra for each identified protein was first extracted; the normalized spectral count was then calculated by dividing the total number of the spectral count for a specific protein by that of the bait protein (DHC) for each sample; the relative protein abundance ratio from the two immunoprecipitation conditions was further calculated and compared for each protein.
MT gliding assay
Cytoplasmic dynein and dynactin were prepared from bovine brain as previously described (Bingham et al., 1998). The MT gliding assay was performed using the kinesin motility assay kit (BK-027; Cytoskeleton, Denver, CO). Before the experiment, 10 μl of 0.5 mg/ml purified cytoplasmic dynein was mixed with the equal volume of the dynactin storage buffer or purified dynactin (∼0.08 mg/ml) on ice for 15–20 min. The dynein or dynein/dynactin mixture was first perfused into the flow chamber and incubated for 5 min at room temperature. After being blocked with 5 mg/ml BSA at room temperature for 5 min, 2 μM Lis1 was perfused into the flow chamber and incubated for 5 min. Next the fluorescent, Taxol-stabilized MTs were perfused into each chamber and incubated for 5 min. Unbound MTs were washed away. Microtubule gliding was initiated by perfusion with 1.5 mM ATP in the motility buffer according to the user's manual for the kit (BK-027). Images were acquired at the speed of 1 or 5 s per frame for 20 min using a 60× oil lens on an inverted microscope (Nikon, Melville, NY). Microtubule trajectories were constructed in the MetaMorph software (Molecular Devices, Sunnyvale, CA) from 30 consecutive images in movies taken at the speed of 1 s per frame. The MT gliding velocity was analyzed by tracking individual MTs throughout the time-lapse series of images with MetaMorph. Microtubules usually ran continuously throughout the recording field. However, in the presence of Lis1, individual MTs typically exhibited motile events with intermittent pauses. So the run distance could be determined by measuring the distance between pauses for the conditions of dynein with Lis1 or dynein with Lis1 plus dynactin.
Supplementary Material
Supplemental Materials
Acknowledgments
We thank Bo Liu (University of California, Davis) and the members of the Zheng and Schroer labs for helpful suggestions. This work was supported by National Institutes of Health grants R01GM56312 (Y.Z.), R01GM44589 (T.A.S.), P41 RR011823 (J.Y.), and R01HL079442-08 (J.Y.).
Abbreviations used:
BSA
bovine serum albumin
DHC
dynein heavy chain
DIC
dynein IC
DLC
light chain
DLIC
light IC
GFP
green fluorescent protein
GST
glutathione _S_-transferase
IgG
immunoglobulin G
ITC
isothermal titration calorimetry
LC-MS/MS
liquid chromatography–tandem mass spectrometry
MT
microtubule
RNAi
RNA interference
Footnotes
REFERENCES
- Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham JB, King SJ, Schroer TA. Purification of dynactin and dynein from brain tissue. Methods Enzymol. 1998;298:171–184. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda U, et al. The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure. 2007;15:1467–1481. doi: 10.1016/j.str.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Efimov VP. Roles of NUDE and NUDF proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol Biol Cell. 2003;14:871–888. doi: 10.1091/mbc.E02-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Olson EC, Stukenberg PT, Flanagan LA, Kirschner MW, Walsh CA. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B, Channels W, Qiu M, Iglesias P, Yang G, Zheng Y. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. J Biol Chem. 2010;285:35238–35244. doi: 10.1074/jbc.M110.140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “Clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci USA. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nature Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, Gibbons IR, Burgess SA, Sutoh K. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J Cell Sci. 2010;123:202–212. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Yu W, Li Y, Yang Z, Yan X, Huang Q, Zhu X. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Ma L, Tsai MY, Wang S, Lu B, Chen R, Iii JR, Zhu X, Zheng Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat Cell Biol. 2009;11:247–256. doi: 10.1038/ncb1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR 3rd. MS1, MS2, and SQT—three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesngon MT, Tarricone C, Hebbar S, Guillotte AM, Schmitt EW, Lanier L, Musacchio A, King SJ, Smith DS. Regulation of cytoplasmic dynein ATPase by Lis1. J Neurosci. 2006;26:2132–2139. doi: 10.1523/JNEUROSCI.5095-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Song Y, Barbar E. Structural dynamics and multiregion interactions in dynein-dynactin recognition. J Biol Chem. 2011;286:39349–39359. doi: 10.1074/jbc.M111.296277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Nyarko A, Song Y, Barbar E. Intrinsic disorder in dynein intermediate chain modulates its interactions with NudE and dynactin. J Biol Chem. 2012;287:24884–24893. doi: 10.1074/jbc.M112.376038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J Neurosci. 2011;31:17207–17219. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. J Cell Biol. 2013;201:201–215. doi: 10.1083/jcb.201208098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, et al. ATP-driven remodeling of the linker domain in the dynein motor. Structure. 2012;20:1670–1680. doi: 10.1016/j.str.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C, Perrina F, Monzani S, Massimiliano L, Kim MH, Derewenda ZS, Knapp S, Tsai LH, Musacchio A. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–821. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Torisawa T, Nakayama A, Furuta K, Yamada M, Hirotsune S, Toyoshima YY. Functional dissection of LIS1 and NDEL1 towards understanding the molecular mechanisms of cytoplasmic dynein regulation. J Biol Chem. 2011;286:1959–1965. doi: 10.1074/jbc.M110.169847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y. Identification of a novel dynein binding domain in Nudel essential for spindle pole organization in Xenopus egg extract. J Biol Chem. 2011;286:587–593. doi: 10.1074/jbc.M110.181578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR 3rd. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- Yamada M, et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Penalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhuang L, Lee Y, Abenza JF, Penalva MA, Xiang X. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J Cell Sci. 2010;123:3596–3604. doi: 10.1242/jcs.075259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylkiewicz E, Kijanska M, Choi WC, Derewenda U, Derewenda ZS, Stukenberg PT. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J Cell Biol. 2011;192:433–445. doi: 10.1083/jcb.201011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials