Gastric and Colonic Zinc Transporter ZIP11 (Slc39a11) in Mice Responds to Dietary Zinc and Exhibits Nuclear Localization (original) (raw)
Abstract
Zinc transporters have been characterized to further understand the absorption and metabolism of dietary zinc. Our goal was to characterize zinc transporter Slc39a11 (ZIP11) expression and its subcellular localization within cells of the murine gastrointestinal tract of mice and to determine if dietary zinc regulates ZIP11. The greatest ZIP11 expression was in the stomach, cecum, and colon. Both Zip11 mRNA and ZIP11 protein were shown to be downregulated during dietary zinc restriction (<1 mg Zn/kg) in the murine stomach tissue but were unaffected in the colon. Acute repletion with zinc did not restore Zip11 mRNA levels in the stomach. Immunohistochemistry (IHC) revealed high ZIP11 levels in the lower regions of gastric glands and parietal cells of the stomach. IHC analysis of the colon showed a marked ZIP11 abundance within the cytoplasm of the colonic epithelial cells. IHC also showed an increase in ZIP11 expression in the colon during zinc restriction. There is a robust abundance of ZIP11 in the nuclei of cells of both stomach and colon. Our experiments suggest that when dietary zinc intake is compromised, the colon may increase zinc transporter expression to improve the efficiency for absorption via increased expression of specific zinc transporters, including ZIP11 and also zinc transporter Slc39a4. In conclusion, ZIP11 is highly expressed within the murine stomach and colon and appears to be partially regulated by dietary zinc intake within these tissues. ZIP11 may play a specialized role in zinc homeostasis within these tissues, helping to maintain mucosal integrity and function.
Introduction
Cellular channels and transporters are critical components that maintain metal metabolism and homeostasis. Two protein families, Zinc transporter (ZnT)6 and Zrt, Irt-like protein (ZIP), are particularly important for regulation of zinc metabolism (1). ZnT transport activity lowers the cytosolic ionic zinc concentration. In contrast, ZIP activity increases. Expression of these transporters is tissue specific. Within the gastrointestinal tract, where considerable zinc transport activity is directed at absorption of dietary zinc, zinc transporter Slc39a4 (ZIP4) appears to have a major role. Mutations of ZIP4 in humans produce the zinc malabsorption syndrome, Acrodermatitis enteropathica (2). ZIP4 is located at the apical membrane of enterocytes (3, 4). The zinc efflux transporter zinc transporter Slc30a1 (ZnT1) is located at the basolateral membrane of enterocytes and is thought to be the principal transporter responsible for zinc translocation from enterocytes into the systemic circulation (5).
Zinc transporter Slc39a5 (ZIP5) and zinc transporter Slc39a14 (ZIP14) are also highly expressed in the gastrointestinal tract but have not received extensive attention at the physiological level. ZIP5 may exhibit basolateral membrane localization when dietary zinc intake is adequate but degrades during zinc deficiency (3). ZIP14 does not exhibit zinc regulation but is influenced by proinflammatory mediators (6).
We have investigated the expression of zinc transporter Slc39a11 (ZIP11) throughout the gastrointestinal tract. Limited structure, function, or regulatory information is available for the zinc transporter ZIP11. Zip11 is the sole member of the gufA subfamily of ZIP transporters (1). Zip11 is well conserved across several species. Mouse Zip11 is found on the antisense strand of chromosome 11, whereas human ZIP11 is found on the antisense strand of chromosome 17 (7). Very recently, ZIP11 was shown to act as a zinc transporter in transfected HEK293T cells and the modest increase in Zip11 expression, due to acute oral zinc loading in mice, is related to multiple metal response elements (MREs) found in the Zip11 promoter (8).
In this report, we investigated the topology of murine ZIP11 and demonstrated that ZIP11 is highly expressed in the stomach and colon of mice, responsive to dietary zinc restriction in the stomach, and localized to the gastric parietal cells and lower regions of the gastric glands and in the colonic epithelial cells, where the protein partially colocalizes with the nucleus.
Materials and Methods
Mice and dietary treatments.
Mice of the C57BL/6 strain were given free access to tap water and a commercial rodent diet (Harlan Teklad-7912) and maintained with a 12-h-light/-dark cycle. Both male and female mice (8–12 wk) were used. To examine responses to zinc intake, the AIN76 diet (Research Diets) formulated with egg white protein and the appropriate amount of zinc carbonate to provide low zinc [zinc depletion (ZnD); <1 mg Zn/kg diet], adequate zinc (ZnA; 30 mg Zn/kg diet), or high zinc (ZnR; 180 mg Zn/kg diet)] was used (9, 10). When mice were fed the purified zinc diets, they were placed in hanging stainless steel cages and had free access to Milli-Q water and food for 1–2 wk (10). At the end of the comparison period, the mice were anesthetized using isoflurane and killed by cardiac puncture and exsanguination. Protocols were approved by the University of Florida Institutional Animal Care and Use Committee.
qPCR analysis.
After blood was collected, sections of the gastrointestinal tract, pancreas, liver, and kidney were excised, flushed with ice-cold PBS containing 1× Halt Protease Inhibitor (Pierce) solution, and then placed in RNAlater (Ambion), flash frozen with liquid nitrogen, or fixed in 10% formalin. Serum zinc was measured by flame atomic absorption spectrophotometry as previously described (10). Tissue samples were removed from RNAlater and homogenized (Polytron) in TRI Reagent (Molecular Research Center) (4, 10). Residual DNA contamination was removed from RNA by using TurboDNase (Ambion) and qPCR was conducted as previously described (10). The primer and probe sequences for Zip11 mRNA were: forward 5′-CACTGAGTGGAAGGCATCTTTCT-3′ reverse 5′-TGAGGTGTTGAAGTTGAGTCTAGTGA-3′ probe 5′-FAM-TCGAGGCTAACCCCTACTTGTCCCACC-BHQ1–3′.
Relative quantitation for assays used 18S rRNA or TATA binding protein (TBP) mRNA as normalizers and an RNA standard curve as previously described in detail (11).
Western analysis.
The peptide used to generate the in-house ZIP11 polyclonal rabbit antibody was CPALMKKSDPRDPTS. IgG fractions were affinity purified (Pierce). For Western-blot analysis, tissues were placed in ice-cold Tris-Triton buffer (1 mol/L Tris-HCl, pH 7.4, 1% Triton X-100, 1× Halt Protease Inhibitor) homogenized using a Polytron as previously reported (11, 12). Cytoplasmic, membrane, and nuclear fractions were isolated from colonic mucosa scraped from the serosal layers using the NE-PER nuclear and cytoplasmic extraction kit from Thermo Scientific. The primary polyclonal rabbit antibodies used were tubulin (1:5000, Abcam), TBP monoclonal mouse antibody (1:2000, Abcam), ZIP4 (2 _μ_g/mL, Thermo Scientific), and our in-house antibody ZIP11 (2 _μ_g/mL). The ZIP11 antibody bound to a protein at 35 kDa (the predicted size of the ZIP11). To test the specificity of the antibody, the primary antibody was preincubated with a 200-fold molar excess of the Zip11 peptide that was used as the antigen. Immunoreactivity was visualized by enhanced chemiluminescence (Thermo Scientific) and a FluorChem E Imager (ProteinSimple).
Immunohistochemistry.
Stomach, cecum, and colon tissue were collected and placed in 10% formalin for 24 h at room temperature and then in cold PBS and kept at 4°C. Two different immunohistochemistry (IHC) methods were used. For immunoperoxidase staining, the Vectastain Elite ABC (Vector Laboratories) protocol was followed, using the ZIP11 primary antibody (10 _μ_g/mL) diluted in the blocking serum. To test the specificity of the antibody for IHC, the primary antibody solution was preincubated as mentioned above. Chromophor development used a biotinylated secondary antibody and a peroxidase substrate (Vector Laboratories). The slides were also stained with hematoxylin (Santa Cruz) to visualize the nuclei. Staining was viewed with a light microscope (Zeiss, Axiovert S100). For immunofluorescence, after deparaffinization, stomach tissues were incubated with antibodies for ZIP11 (10 _μ_g/mL) and H,K-ATPase (1:2000, Novus). Colon tissues were incubated with the ZIP11 (10 _μ_g/mL) and β-catenin (1:200, Cell Signaling) antibodies. The secondary antibodies were donkey, anti-rabbit IgG labeled with AlexaFluor 594 (1:300) and goat anti-mouse IgG labeled with AlexFluor 488 (1:400), both from Invitrogen. Nuclei were detected with 4′,6-diamidino-2-phenylindole (DAPI; 1:200, Molecular Probes). Samples were visualized using either a Laser Scanning Confocal Fluorescence Microscope (Leica TCS-SP2) or a Spinning Disk Confocal Fluorescence Microscope (Olympus IX2-DSU).
Statistical analysis.
Data are presented as the means ± SDs. Significant changes between groups were analyzed by ANOVA with the Tukey Kramer multiple comparison test. Significance was set at P < 0.05.
Results
Predicted ZIP11 topology.
The murine Zip11 gene consists of 10 exons, 9 of which are translated from the coding sequence. Zip11 mRNA has 2 known isoforms with 21 bp removed during alternative slicing from the end of exon 5, forming isoform 2 (13). The predicted protein structure of ZIP11 has 6 transmembrane domains with the N terminus having an extracellular location (Fig. 1). A histidine-rich loop is usually found in Zip transporter proteins (1), but this region is not found in the murine ZIP11 protein. The topology of murine ZIP11 was analyzed using several Web sites that use specific transmembrane prediction software (14–16). The variable region of the ZIP11 isoforms (light gray box) and location of the sequence used to make the ZIP11 antibody (dark gray box) are depicted. Phylogenetic analyses show that ZIP11 proteins are present in metazoans, with only limited occurrence of orthologous proteins in lower organisms.
FIGURE 1.
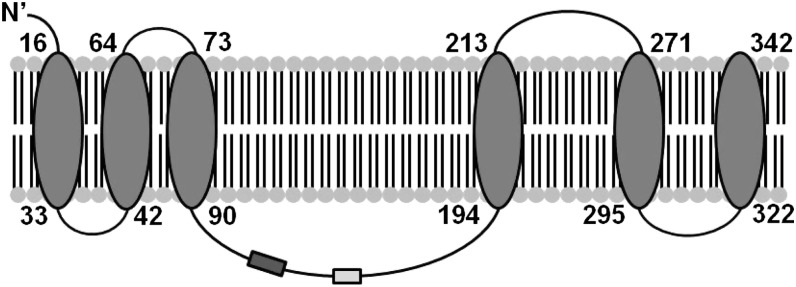
Predicted topology for murine ZIP11 showing 6 transmembrane-spanning regions. The light gray box indicates the variable region of the isoforms. The dark gray box indicates the region used to produce our ZIP11 antibody. ZIP11, zinc transporter SLC39A11.
Zip11 RNA and protein expression in mice fed a commercial diet.
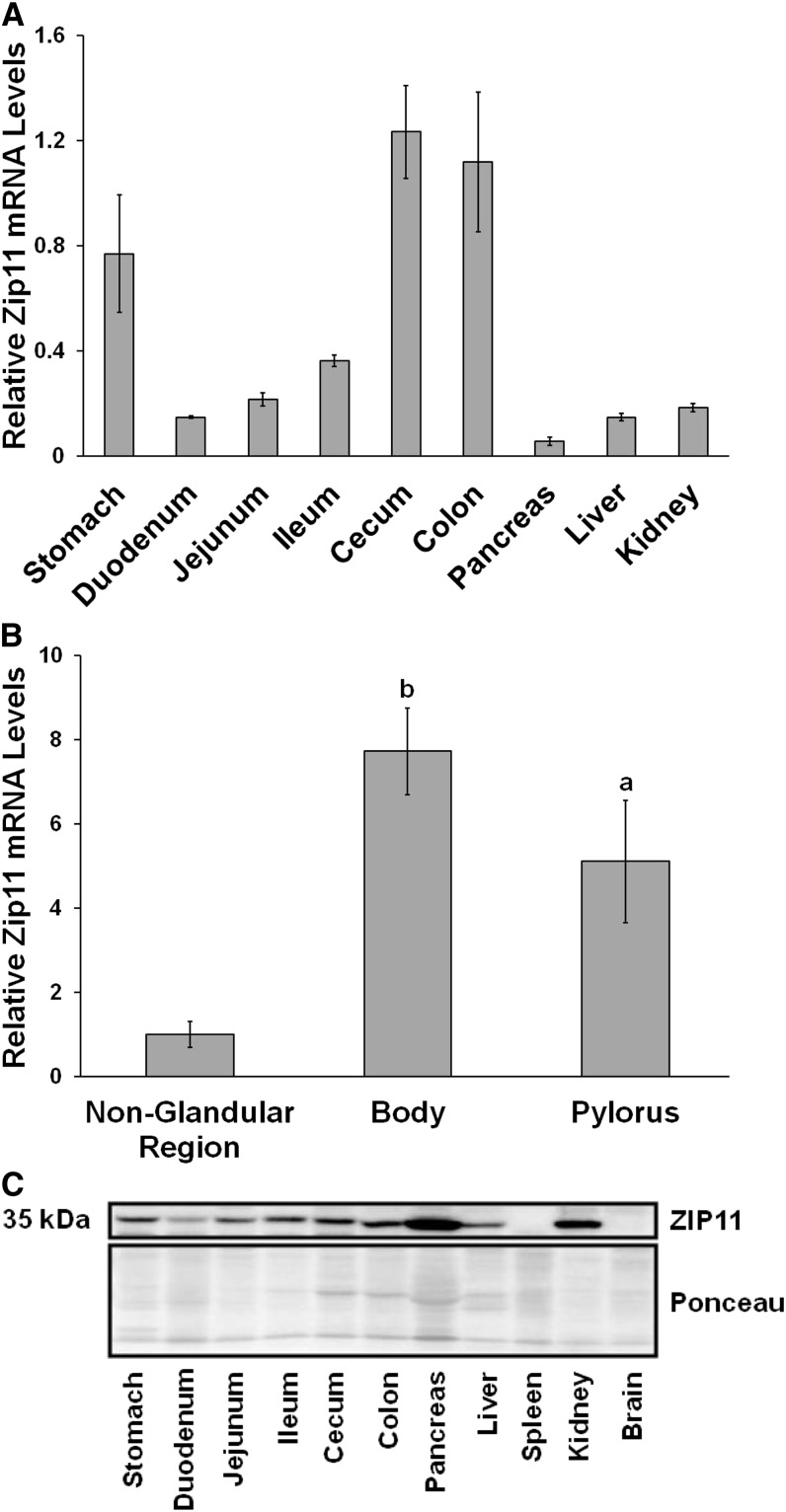
Zip11 expression in the gastrointestinal tract of C57BL/6 mice under normal husbandry conditions fed a commercial rodent diet (Harlan Teklad-7912) comprised of feed-grade ingredients was examined. The distribution of Zip11 transcripts was very abundant in the stomach, cecum, and colon (Fig. 2A). The stomach was separated into 3 regions (nonglandular, body, and pylorus) and Zip11 mRNA was greatest in the body and pyloric regions (Fig. 2B). The in-house ZIP11 antibody was used for the Western analysis, with ZIP11 visualized at 35 kDa (Fig. 2C). Ponceau staining of the membrane was used as the loading control. The protein data revealed similar results to the transcript data, with the stomach and colon having the highest expression of ZIP11 protein within the gastrointestinal tract. Zip11 mRNA levels in the pancreas and kidney were low relative to other tissues; however, the ZIP11 protein content of both tissues was greater on a relative basis.
FIGURE 2.
Tissue distribution of the Zip11 gene transcripts and ZIP11 protein in C57BL/6 mice. RNA was extracted from tissues and used for qPCR analysis of Zip11 mRNA. (A) Zip11 mRNA tissue distribution with pancreatic expression set to 1. (B) Zip11 mRNA expression distribution in the 3 regions of the mouse stomach. The nonglandular region expression was set to 1. Values were normalized to 18S. Values are means ± SDs, n = 4–5 for each tissue sample. Significant differences compared to the nonglandular region are indicated (a, P < 0.05; b, P < 0.01). (C) Western blots of total tissue lysates. ZIP11 is shown as a 35-kDa band. Ponceau staining from the blot was used as the loading control. ZIP11, zinc transporter SLC39A11.
ZIP11 mRNA expression during zinc depletion and repletion.
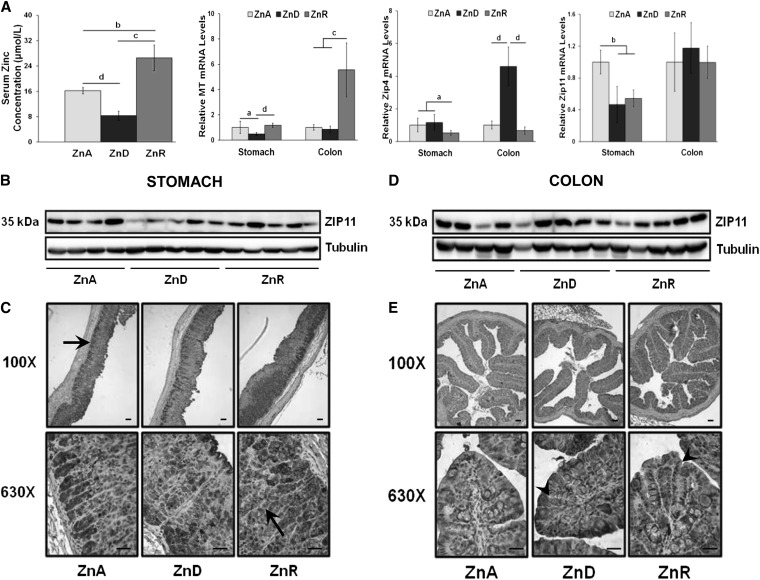
Mice were fed the ZnA or ZnD diet for 2 wk. Because our focus was on the stomach and colon due to their relatively high ZIP11 expression, we used a design where both dietary zinc restriction (ZnD) and an acute, 1-d zinc repletion (ZnR) were used to test the responsiveness of Zip11 expression to dietary zinc. The acute repletion was with the ZnR diet provided for 1 d. The effectiveness of the ZnD diet to decrease serum zinc and the acute ZnR to significantly elevate the concentration above the ZnA group is shown in Figure 3A. Metallothionein (MT) _m_RNA levels were used as a positive control as a classical zinc-regulated gene to evaluate the effectiveness of the diets at the tissue level (Fig. 3A). This mRNA was reduced in the stomach when the ZnD diet was fed but was unchanged in the colon. Stomach and colon MT mRNA of the ZnD mice significantly increased in response to the ZnR diet. By comparison, Zip4 mRNA, another positive control, markedly increased in response to the ZnD diet in the colon but not in the stomach. A significant decrease in Zip11 transcripts was seen in response to the ZnD diet in the stomach but not in the colon (Fig. 3A). No change in response to ZnR in Zip11 mRNA was observed in the stomach or colon, suggesting that Zip11 does not respond rapidly to dietary zinc repletion as observed with either MT or Zip4.
FIGURE 3.
Dietary ZnD and ZnR produce changes in ZIP11 expression in the stomach and colon compared with mice fed a ZnA diet. (A) Serum zinc concentration and relative expression of MT mRNA, Zip4 mRNA, and Zip11 mRNA in stomach and colon. mRNA levels for ZnA were set to 1. Data are expressed as means ± SDs, n = 3–5. Significant differences are indicated (a, P < 0.05; b, P < 0.01; c, P < 0.001; d, P < 0.0001). (B) Representative Western blot of stomach tissue lysate showing an abundance of the ZIP11 protein at 35 kDa, normalized to tubulin. (C) Immunoperoxidase staining of ZIP11 in the mouse stomach, visualized with a light microscope using 100× and 630× magnification. The stomach shows strong ZIP11 staining (dark gray) in the ZnA samples with a decrease in the ZnD samples. Black arrows indicate strong ZIP11 staining in the chief cell region and parietal cells of the gastric glands. ZIP11 staining returned to comparable levels seen in ZnA samples in the stomach after ZnR. (D) Representative Western blot of colon lysate showing abundance of the ZIP11 protein with tubulin as the loading control. (E) Immunoperoxidase staining of ZIP11 in the mouse colon is visualized at 100× and 630× magnification. The colon samples show an increase in staining in the ZnD samples compared with ZnA samples, and the staining remained the same after 24 h repletion. Black arrowheads indicate colonic crypts. Black lines in the 630× images represent 25 _μ_m and those in the 100× images represent 50 _μ_m. MT, Metallothionein; ZIP4, zinc transporter Slc39a4; ZIP11, zinc transporter SLC39A11; ZnA, zinc adequate; ZnD, zinc depletion; ZnR, acute zinc repletion.
ZIP11 protein expression is variable in stomach and colon during zinc depletion.
Using our in-house ZIP11 antibody, we investigated ZIP11 protein expression in the stomach and colon of individual mice. In the stomach, zinc restriction with the ZnD diet decreased ZIP11 and expression returned to ZnA levels after zinc repletion with the ZnR diet (Fig. 3B). A peptide competition assay provided evidence of the specificity of the ZIP11 antibody. ZIP11 was detected at 35 kDa, but when the Zip11 peptide was first incubated with the antibody, the 35-kDa protein was no longer detected (Supplemental Fig. 1A). IHC showed strong ZIP11 staining in the ZnA stomach tissue (Fig. 3C). There was less staining in the ZnD sample and a return to ZnA staining patterns in the ZnR group. Darker staining for ZIP11 was also visualized in the lower regions of the gastric glands, where chief cells reside, and in the parietal cells (black arrows) (Fig. 3C). Western analysis of ZIP11 protein levels in the colon showed they were constant during ZnD or ZnR (Fig. 3D). IHC of the colon revealed distinct staining of ZIP11 throughout the crypts (black arrowheads) of the colonic mucosa (Fig. 3E). Unlike the Western blot, the IHC showed a high intensity of ZIP11 staining during zinc restriction but also during acute repletion.
ZIP11 localization in stomach and colon.
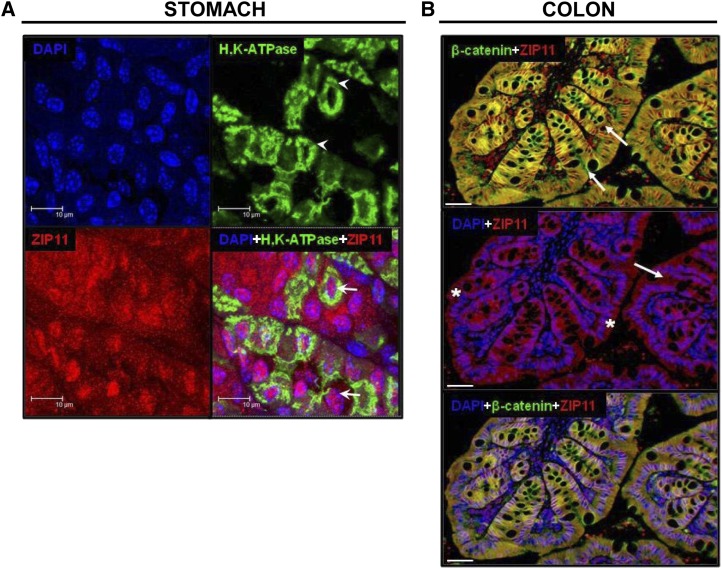
Because ZIP11 is highly expressed in the gastric mucosa, we hypothesized that ZIP11 might be localized at the plasma membrane of the parietal cell, the acid-secreting cell of the stomach. Using H,K-ATPase as a membrane marker for parietal cells (Fig. 4A, white arrowheads), we found that ZIP11 was not colocalized with the parietal cell marker; rather, in most cells, ZIP11 colocalized with DAPI, the nuclei marker. The white arrows indicate the colocalization of DAPI and ZIP11, which appears in pink (Fig. 4A). Extensive red fluorescent ZIP11 staining also appears to be in the cytoplasm of cells found in the lower portions of the gastric glands (data not shown). Specificity of the ZIP11 antibody used in this application was shown by peptide competition (Supplemental Fig. 1_B_). Expression of this specific zinc transporter in the stomach could relate to gastric mucosal damage caused by zinc deficiency and the increase of gastric acidity seen with zinc supplementation. After detecting high expression of ZIP11 within the colon with Western blots and IHC, we examined ZIP11 localization in colonic epithelial cells. ZIP11 (red) was found to colocalize with β-catenin (green), a plasma membrane marker. This colocalization created a yellow fluorescence (Fig. 4B, top). Colonic crypts are indicated by the white arrows. Abundant fluorescence from β-catenin is seen at the basolateral membrane of the epithelial cells (Fig. 4B). In contrast, ZIP11 (red) was localized to specific compartments within the epithelial cells (white asterisks) (Fig. 4B, middle). The bottom image of Figure 4B is a merged image of all 3 markers (ZIP11, β-catenin, and DAPI) showing the localization of ZIP11 at the plasma membrane and nuclei of the cells. Colonic samples from ZnD mice showed an increase in ZIP11 compartmentalization in the colon during zinc restriction (ZnD) as was seen by immunoperoxidase staining (data not shown). Specificity of the ZIP11 antibody in colonic tissue was shown by peptide competition (Supplemental Fig. 1_B_).
FIGURE 4.
IF imaging of ZIP11 in the mouse stomach and colon. (A) IF images of stomach are shown at 63× with 4× zoom. The tiled figure shows individual and merged fluorescence images. Images show the nuclei marker, DAPI (blue), the parietal cell marker, H,K-ATPase (green) (white arrowheads), and ZIP11, indicated by the red fluorescence. A merged image of all 3 antibodies indicated a localization of ZIP11 to the cytoplasm (red) and nuclei (pink) (white arrows) of gastric cells. The white bars represent 10 _μ_m. (B) IF images of ZIP11 in the mouse colon are shown at 20×. The colonic folds are presented at high magnification to show the invaginations (colonic crypts) indicated by white arrows. The top merged image of ZIP11 (red) and β-catenin (green) indicates colocalization (yellow) of ZIP11 at the colonic epithelial cell plasma membrane. The middle merged image of ZIP11 (red) and DAPI (blue) shows colocalization (purple) of ZIP11 to the colonic epithelial cell nuclei (white asterisks). The bottom panel shows a merged image of ZIP11, β-catenin, and DAPI. This image indicates the localization of ZIP11 both at the colonic epithelial plasma membrane (yellow) and in the nuclei (purple). The white bars represent 25 _μ_m. IF images were obtained using a laser spinning disc confocal microscope. DAPI, 4′,6-diamidino-2-phenylindole; IF, immunofluorescence; ZIP11, zinc transporter SLC39A11.
Localization of ZIP11 in the nucleus was further investigated with Western-blot analysis of isolated cell fractions (cytoplasmic, membrane, and nuclear) of mouse colon. Immunoblotting detected ZIP11 in all 3 fractions (Fig. 5). In contrast, the apical zinc transporter ZIP4 is strongly detected in the membrane fraction and serves as a negative control. As was expected, TBP is highly abundant in the nuclear fraction and tubulin in the cytoplasmic fraction, which constitutes respective positive controls. With these controls and the immunofluorescence (Fig. 4), we concluded from this evidence that ZIP11 is present in the cytoplasm, membrane, and nuclear fractions of the mouse colon.
FIGURE 5.
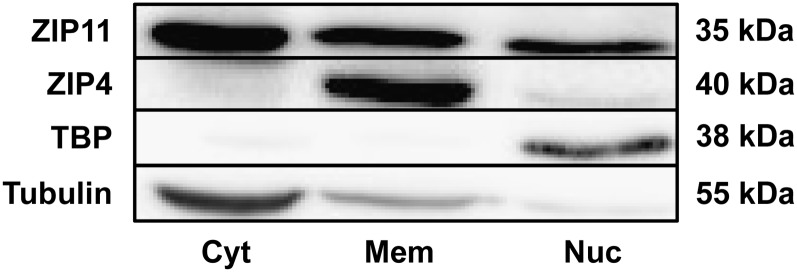
Western-blot analysis of ZIP11 from the subcellular fractionation of mouse colonic mucosal scrapings. Mucosal lysate was separated into a cytoplasmic fraction (Cyt), membrane fraction (Mem), and nuclear fraction (Nuc). ZIP11 was present in all three fractions. Controls for each fraction are shown: ZIP4 (mem), TBP (nuc), and tubulin (cyt). TBP, TATA binding protein; Zip4, zinc transporter Slc39a4; ZIP11, zinc transporter SLC39A11.
Discussion
Zinc is now a recognized factor in cell signaling (17). Zinc-regulated processes likely have an influence in gastrointestinal-related diseases, such as irritable bowel disease (18). Hence, a more robust understanding of zinc metabolism and signaling events that are influenced by zinc in specific tissues are important to define. Zinc transporters are key to this understanding. Bioinformatic analysis of murine Zip11 revealed that numerous MRE consensus sequences could be found in the 5′ region of the gene. When activated through zinc binding to the metal responsive transcription factor-1 (MTF-1) and subsequent nuclear translocation, MREs enable changes in transcription (19). The interaction usually produces an increase in transcription, as observed with MT (Fig. 3A) and ZnT1 genes. However, MRE placement can also lead to repression of transcription with nuclear translocation of MTF-1. This mode of zinc-responsive repression has been shown for the zinc transporter gene Zip10 (11). Consequently, the modest response of Zip11 mRNA to dietary zinc intake we have shown was surprising considering the number of MREs in the Zip11 promoter. Nevertheless, a very recent study with mice that was limited to the spleen also showed only a modest increase in Zip11 mRNA in response to administered acute, oral, massive amounts of zinc (8).
Data shown in this report demonstrate that within the murine gastrointestinal tract, dietary changes in zinc intake that produce major changes for the zinc-responsive gene, MT, produce only modest responsiveness of the Zip11 gene. Although ZIP11 is modestly downregulated by zinc deficiency in the stomach, nevertheless, this ZIP transporter may have important roles in zinc processing and/or functions, particularly in the stomach and colon. The colon has been shown to absorb zinc, particularly when the functional capacity of the small intestine has been compromised (20, 21). Hence, absorption from the colon may be enhanced in zinc deficiency, because ZIP4 is upregulated (Fig. 3A) and ZIP11 expression remains constant. Colonic zinc absorption could be extremely important in addressing supplementation therapies to treat diarrheal disease in zinc-deficient humans. Zinc-containing formulations, once hydrolyzed by colonic microorganisms, could provide available zinc for colonic absorption via ZIP4- and ZIP11-mediated transport processes. We found that relative Zip11 mRNA levels in the stomach and colon were comparable and much greater than those of the small intestine. This difference occurred when the diet consisted of less purified components, e.g., soybean meal (Fig. 2A), and when the AIN 76 purified diet was fed (Fig. 3A). In this regard, our results differ from those of Yu et al. (8), where Zip11 expression in the colon was much lower. This difference is possibly related to variations in microflora and/or colonic fermentation in the different murine colonies. In that regard, it was recently shown that microbiota in the gastrointestinal tract compete with the host for available zinc (22).
The localization of ZIP11 to the parietal and chief cell region of the stomach is of interest. The parietal cell is the site of acid secretion in the stomach. Zinc has been shown to be important in the maturation of tubulovesicles within the parietal cell and may play a role in maintaining the integrity of these cells (23). Zinc deficiency has also been shown to negatively affect the gastric mucosa and secretion of acid (24). ZIP11 decreases in the stomach during zinc deficiency, which could indicate a role for the transporter and zinc in a region of the parietal cell involved with tubulovesicles and acid secretion. The high ZIP11 expression in the polarized parietal cells shows that future research could focus on cultured cells isolated by collagenase digestion to study correlation with H,K-ATPase activity and localization at the apical membrane. With the strong staining of ZIP11 in the chief cell region of the gastric glands, this zinc transporter could also influence secretion of pepsinogen, the precursor for pepsin. The importance of zinc transporters and secretory function was previously shown with the transport of zinc into pancreatic zymogen granules by ZnT2 (25) as well as tight regulation of zinc metabolism within other secretory tissues, such as the prostate and mammary glands (26). Localization of ZIP11 to the colonic epithelial cells indicates that ZIP11 may be involved in maintaining mucosal integrity within the colon or may possibly play a role in zinc sequestration/conservation during zinc deficiency. Colocalization of ZIP11 and β-catenin shows that ZIP11 is at the plasma membrane surface of the colonic epithelial cell, suggestive of a role in absorption. Colonic pH is also tightly regulated as it is in the stomach. Hence, a unifying concept of gastric and colonic cells, showing high ZIP11 expression, could relate to the transporter supplying zinc for a process required to maintain pH balance.
ZIP11 is the sole member of the gufA subfamily of ZIP transporter proteins and our analysis of topology predicts 6 transmembrane domains (Fig. 1). This finding is unique among the ZIP family, suggesting that ZIP11 has a functional role in zinc biology that has not, to our knowledge, been elicited. Localization of ZIP11 to the nucleus is also novel for a ZIP protein. Some studies have shown zinc trafficking in and out of the nucleus. This is usually as a zinc-protein complex such as zinc-finger proteins, MTF-1- or MT-bound zinc (27). Hence, at this time, we can only speculate that ZIP11 transports nuclear Zn2+ back into the cytoplasm, as would be expected of a ZIP protein. Furthermore, protein phosphatase activities within the nucleus could provide sites for zinc signaling for control of cell cycle progression, RNA polymerase II phosphorylation, and phosphorylation of signaling molecules (28). That function may require a regulated supply of nuclear zinc. Also, very relevant is the recent finding that the iron efflux transporter Ferroportin 1 and Divalent Metal Transporter-1, an importer of iron, have been found associated with the nucleus (29). The finding that ZIP11 is in the nucleus opens the opportunity to investigate nuclear zinc fluxes during variations in the colonic environment. It also suggests unique foci for zinc-iron interactions within the colon.
In summary, the main findings of our study revealed that dietary zinc affected Zip11 expression differently in tissues of the mouse gastrointestinal tract. Zip11 expression was greatest in the stomach and colon. ZnD decreased expression of Zip11 in the stomach; however, expression was unaffected in the small and large intestine. IHC revealed the decrease in ZIP11 expression in the stomach but an increase in ZIP11 protein in the colon during ZnD. ZIP11 was found to be partially colocalized in the nuclei of parietal and colonic epithelial cells, and subcellular fractionation of colonic mucosal confirmed ZIP11 expression within the nuclear, cytoplasmic, and membrane fractions. ZIP11 could play an integral role in the zinc homeostasis required to maintain mucosal integrity, function, and pH within the mouse stomach and colon.
Supplementary Material
Online Supporting Material
Acknowledgments
A.B.M. and R.J.C. designed research; A.B.M., T.B.A., G.J.G., and S.-M.C. conducted research; A.B.M., D.A.S., and R.J.C. analyzed data; and A.B.M., T.B.A., and R.J.C. wrote the paper. All authors read and approved the final manuscript.
Footnotes
6
Abbreviations used: DAPI, 4′,6-diamidino-2-phenylindole; IHC, immunohistochemistry; MRE, metal response element; MT, Metallothionein; MTF-1, metal responsive transcription factor-1; TBP, TATA binding protein; ZIP, Zrt, Irt-like protein; Zip4, zinc transporter Slc39a4; Zip5, zinc transporter Slc39a5; Zip11, zinc transporter Slc39a11; Zip14, zinc transporter Slc39a14; ZnA, adequate zinc; ZnD, zinc depletion; ZnR, acute zinc repletion; ZnT1, zinc transporter Slc30a1.
Literature Cited
- 1.Lichten LA, Cousins R. Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr. 2009;29:153–76. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Zhou B, Kuo Y, Zemansky J, Gitschier J. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am J Hum Genet. 2002;71:66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Kim BE, Petris MJ, Eide DJ. The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J Biol Chem. 2004;279:51433–41. [DOI] [PubMed] [Google Scholar]
- 4.Liuzzi JP, Bobo JA, Lichten LA, Samuelson DA, Cousins RJ. Responsive transporter genes within the murine intestinal-pancreatic axis form a basis of zinc homeostasis. Proc Natl Acad Sci USA. 2004;101:14355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahon RJ, Cousins RJ. Regulation of the zinc transporter ZnT-1 by dietary zinc. Proc Natl Acad Sci USA. 1998;95:4841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, Ganz T, Cousins RJ. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc Natl Acad Sci USA. 2005;102:6843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.e!Ensembl. Joint project of EMBL-EBI and Wellcome Trust Sanger Institute. Version current September 2013 [cited 2013 Jan 13]. Available from: http://useast.ensembl.org/index.html.
- 8.Yu Y, Wu A, Zhang Z, Yan G, Zhang L, Shen X, Hu R, Zhang Y, Zhang K, Wang F. Characterization of the GufA subfamily member SLC39A11/Zip11 as a zinc transporter. J Nutr Biochem. 2013;24:1697–708. [DOI] [PubMed] [Google Scholar]
- 9.Huber KL, Cousins RJ. Maternal zinc deprivation and interleukin-1 influence metallothionein gene expression and zinc metabolism of rats. J Nutr. 1988;118:1570–6. [DOI] [PubMed] [Google Scholar]
- 10.Moore JB, Blanchard RK. McCormack WT, Cousins RJ. cDNA array analysis identifies thymic LCK as upregulated in moderate murine zinc deficiency before t-lymphocyte population changes. J Nutr. 2001;131:3189–96. [DOI] [PubMed] [Google Scholar]
- 11.Lichten LA, Ryu MS, Guo L, Embury J, Cousins RJ. MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS ONE. 2011;6:e21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beker Aydemir T, Chang SM, Guthrie GJ, Maki AB, Ryu MS, Karabiyik A, Cousins RJ. Zinc transporter ZIP14 functions in hepatic zinc, iron and glucose homeostasis during the innate immune response (endotoxemia). PLoS ONE. 2012;7:e48679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Center for Biotechnology Information. Gene database. Version current 31 July 2013 [cited 2013 Jan 13]. Available from: http://www.ncbi.nlm.nih.gov/gene/.
- 14.ExPASy Bioinformatics Resource Portal. TMpred program (TMbase). Version 1993 [cited 2013 April 15]. Available from: http://www.ch.embnet.org/software/TMPRED_form.html.
- 15.Center for Biological Sequence Analysis. TMHMM Server v. 2.0. Version current 12 June 2013 [cited 2013 Apr 15]. Available from: http://www.cbs.dtu.dk/services/TMHMM-2.0/.
- 16.Institute of Enzymology. HMMTOP. Version 2.0 2001 [cited 2013 Apr 15]. Available from: http://www.enzim.hu/hmmtop/index.php.
- 17.Haase H, Rink L. Zinc signaling. In: Rink L, editor. Zinc in human health. Berlin: IOS Press; 2011. p. 94–118. [Google Scholar]
- 18.Soybel DI, Kohler JE. Zinc and the gastrointestinal tract. In: Rink L, editor. Zinc in human health. Berlin: IOS Press; 2011. p. 448–72. [Google Scholar]
- 19.King JC. Cousins RJ. Zinc. In: AC Ross, B Caballero, RJ Cousins, KL Tucker, TR Ziegler, editors. Modern nutrition in health and disease. 11th ed. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2013. p. 189–205. [Google Scholar]
- 20.Naveh Y, Lee-Ambrose LM, Samuelson DA, Cousins RJ. Malabsorption of zinc in rats with acetic acid-induced enteritis and colitis. J Nutr. 1993;123:1389–95. [DOI] [PubMed] [Google Scholar]
- 21.Hara H, Konishi A, Kasai T. Contribution of the cecum and colon to zinc absorption in rats. J Nutr. 2000;130:83–9. [DOI] [PubMed] [Google Scholar]
- 22.Gielda LM, DiRita VJ. 2012. Zinc competition among the intestinal microbiota. MBio. 2012;3:e00171–12. [DOI] [PMC free article] [PubMed]
- 23.Gerbino A, Hofer A, McKay B, Lau B, Soybel D. Divalent cations regulate acidity within the lumen and tubulovesicle compartment of gastric parietal cells. Gastroenterology. 2004;126:182–95. [DOI] [PubMed] [Google Scholar]
- 24.Naik HB, Beshire M, Walsh B, Liu J, Soybel D. Secretory state regulates Zn2+ transport in gastric parietal cell of the rabbit. Am J Physiol Cell Physiol. 2009;297:C979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Lichten L, Ryu M, Liuzzi J, Wang F, Cousins R. STAT5-glucocorticoid receptor interaction and MTF-1 regulate the expression of ZnT2 (Slc30a2) in pancreatic acinar cells. Proc Natl Acad Sci USA. 2010;107:2818–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelleher SL, McCormick NH, Velasquez V, Lopez V. Zinc in specialized secretory tissues: roles in the pancreas, prostate, and mammary gland. Adv Nutr. 2011;2:101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zalewski P. Chapter 5. Zinc in mammalian cell cycle and cell death. In: Rink L, editor. Zinc in human health. Berlin: IOS Press; 1999. p. 63–93. [Google Scholar]
- 28.Moorhead BG. Trinkle-Mulcahy L, Ulke-Lemée A. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234–44. [DOI] [PubMed] [Google Scholar]
- 29.Naz N, Malik IA, Sheikh N, Ahmad S, Khan S, Blaschke M, Schultze F, Ramadori G. Ferroportin-1 is a ‘nuclear’-negative acute-phase protein in rat liver: a comparison with other iron-transport proteins. Lab Invest. 2012;92:842–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Supporting Material