Characterization of Ferroplasma Isolates and Ferroplasma acidarmanus sp. nov., Extreme Acidophiles from Acid Mine Drainage and Industrial Bioleaching Environments (original) (raw)
Abstract
Three recently isolated extremely acidophilic archaeal strains have been shown to be phylogenetically similar to Ferroplasma acidiphilum YT by 16S rRNA gene sequencing. All four Ferroplasma isolates were capable of growing chemoorganotrophically on yeast extract or a range of sugars and chemomixotrophically on ferrous iron and yeast extract or sugars, and isolate “_Ferroplasma acidarmanus_” Fer1T required much higher levels of organic carbon. All four isolates were facultative anaerobes, coupling chemoorganotrophic growth on yeast extract to the reduction of ferric iron. The temperature optima for the four isolates were between 35 and 42°C and the pH optima were 1.0 to 1.7, and “_F. acidarmanus_” Fer1T was capable of growing at pH 0. The optimum yeast extract concentration for “_F. acidarmanus_” Fer1T was higher than that for the other three isolates. Phenotypic results suggested that isolate “_F. acidarmanus_” Fer1T is of a different species than the other three strains, and 16S rRNA sequence data, DNA-DNA similarity values, and two-dimensional polyacrylamide gel electrophoresis protein profiles clearly showed that strains DR1, MT17, and YT group as a single species. “_F. acidarmanus_” Fer1T groups separately, and we propose the new species “_F. acidarmanus_” Fer1T sp. nov.
Acidophilic ferrous iron-oxidizing microorganisms have been implicated in the production of acid mine drainage (AMD), whereby metal sulfides are solubilized by oxidative dissolution, releasing metals and acid (32; for a review, see reference 30). The discharge of AMD causes considerable environmental damage via the release of metal-rich acidic effluents into groundwater. It was initially thought that the most important microorganism implicated in AMD was Acidithiobacillus ferrooxidans (21). However, recent advances in molecular phylogenetic techniques have shown that other species are numerically dominant at certain acid-generating sites. One example of this is Iron Mountain, Calif., where the pyrite concentration in the ore is 95%, resulting in the generation of extremely acidic solutions (20). The microorganism population at the Iron Mountain acid-generating site is dominated by an archaeon of the genus Ferroplasma. Organisms from this genus were shown to constitute 85% ± 7% of the microorganism population by fluorescent in situ hybridization. An isolate obtained from the acid-leaching biofilm has been provisionally named “_Ferroplasma acidarmanus_” Fer1 (10). The “_F. acidarmanus_” Fer1T genome has been sequenced (97% complete), and draft results are available at the U.S. Department of Energy web site (http://genome.jgi-psf.org/draft_microbes/ferac/ferac.home.html). Details of the chemical and microbial aspects of sulfide dissolution at Iron Mountain (9) as well as an evaluation of the use of fluorescent in situ hybridization at the site (2) have been published.
Acidophilic microorganisms have been extensively exploited in biooxidation and bioleaching operations to remove metals from ore flotation concentrates, and three other Ferroplasma strains have been isolated from these environments. The type strain for the genus, Ferroplasma acidiphilum strain Y, was isolated from a pilot plant bioreactor for biooxidation of a gold-bearing arsenopyrite-pyrite concentrate (11). F. acidiphilum YT is described as being obligatory aerobic and autotrophic, as it is only capable of growth on Fe(II) or Mn(II) with the addition of a small concentration of organic carbon as a growth factor. F. acidiphilum YT is reported to be incapable of growth on organic carbon alone and is not capable of aerobic growth on reduced inorganic sulfur compounds.
Another member of the genus Ferroplasma is strain MT17, isolated from a South African pilot scale bioleaching reactor oxidizing a polymetallic sulfide concentrate at 45°C (22). This isolate was described as unable to grow in the absence of organic carbon but capable of chemomixotrophic growth on yeast extract either with Fe(II) or the reduced inorganic sulfur compound tetrathionate. MT17 was also reported to grow on yeast extract alone and was described as heterotrophic. It was also capable of anaerobically oxidizing yeast extract coupled to the reduction of Fe(III), but it was not ascertained if this was coupled to growth.
The final isolate, strain DR1, was cultured from a separate pilot scale biooxidation plant in South Africa and was shown to be similar to the genus Ferroplasma (by 16S ribosomal DNA [rDNA] sequence analysis). No phenotypic characterization of this isolate has been published.
For this study, we aimed to elucidate the genotypic and phenotypic characteristics of the genus Ferroplasma. Sequence comparisons of 16S rRNA gene sequences suggest that the isolates studied are closely related. However, key questions and inconsistencies regarding this genus need to be verified. These include chemoautotrophic versus chemoorganotrophic growth, oxidation of reduced inorganic sulfur compounds, and anaerobic growth. This study includes the first use of two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) protein profile similarities to estimate the phylogenetic relationships between microorganisms.
MATERIALS AND METHODS
Isolates.
The Ferroplasma isolates used in this study were the type strain for the genus, F. acidiphilum Y (11), acquired from Deutsche Sammlung von Mikroorganismen (DSM12658T); “_F. acidarmanus_” Fer1T (10), kindly provided by J. F. Banfield and K. J. Edwards; isolate MT17 (22), supplied by D. B. Johnson; and isolate DR1, generously provided by D. E. Rawlings.
Growth conditions.
Unless otherwise stated, all growth experiments were carried out in mineral salts medium (MSM) containing trace elements (7), 20 g of FeSO4 · 7H2O liter−1, and 0.02% (wt/vol) yeast extract. The MSM containing yeast extract was adjusted to pH 1.2 with H2SO4 and autoclaved before filter-sterilized trace elements and ferrous iron were added. To ensure that cells were placed under standard conditions for phenotypic characterization and comparison, all growth conditions and substrate utilization experiments were inoculated with steady-state cells from continuous culture vessels. The cultures were inoculated with a viable cell mass equivalent to 10 μg of cell protein (measured by using a Bio-Rad protein assay reagent, with bovine serum albumin as the standard) from continuous culture vessels grown with MSM, trace elements, ferrous iron, and 0.02% (wt/vol) yeast extract (6a). The batch cultures were incubated on a rotary shaker at 150 rpm and 37°C for 63 h (unless otherwise stated). Growth curves were monitored by direct cell counts with a hemocytometer (Hawksley) on an Olympus BX50 phase-contrast microscope or by measurements of the optical density at 600 nm (in a Philips PU8730 spectrometer). The cells for determining growth rates on yeast extract alone were previously subcultured in MSM and yeast extract. Unless otherwise stated, all experiments were performed in triplicate, with results reported as means ± standard deviations (SD) (calculated as the square roots of the sums of the squares for the test and control).
Cellular response to organic and inorganic substrates.
“_F. acidarmanus_” Fer1T was either grown chemomixotrophically on ferrous iron and yeast extract or chemoorganotrophically on yeast extract alone. After growth, cells were harvested by centrifugation at 10,000 × g for 10 min (Heraeus 400R Labofuge), washed, resuspended in MSM, pH 1.2, and then stored on ice for no more than 2 h before use. Oxygen consumption was measured by using a Clark-type oxygen electrode (Rank) at 37°C. Cells were equilibrated for 3 min prior to the addition of substrate. Each substrate was added in the presence and absence of cells to determine biological and chemical oxygen consumption. The substrate-stimulated consumption of oxygen was recorded as a plus or minus response in (at least) duplicate (n = 2 to 5). The following substrates were tested (0.1% [wt/vol] final concentration, unless specified otherwise) on Fe(II)- and yeast extract-grown cells: 70 mM ferrous iron, 0.02% yeast extract, sucrose, d-glucose, starch, d-galactose, d-fructose, l-maltose, l-arabinose, d-sorbitol, d-xylose, d-mannitol, lactose, Casamino Acids, l-alanine, l-arginine, l-asparagine, l-aspartic acid, l-cysteine, l-glutamine, l-glutamic acid, l-glycine, l-histidine, l-isoleucine, l-leucine, l-lysine, l-methionine, l-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, l-tyrosine, and l-valine. The vitamin mix was prepared according to the method of Golyshina et al. (11). Ferrous iron and yeast extract were also tested with chemoorganotrophic yeast extract-grown cells.
Investigation of growth substrates.
Those substrates that were found to stimulate oxygen consumption were tested to see if they could support the growth of Ferroplasma isolates. One-hundred-milliliter batch cultures of MSM amended with various substrates were incubated with Ferroplasma isolates for 63 h, and the protein concentrations were measured (as described above). Each of the substrates was added to a final concentration of 1 mM unless otherwise specified. Autotrophic growth was also tested in continuous culture vessels containing MSM, trace elements, and 20 g of FeSO4 · 7H2O liter−1 (6a). The protein concentration was monitored (as described above), and after three medium changes, the presence of viable cells was confirmed by the inoculation of 100-ml shake flasks containing MSM, Fe(II), and yeast extract and the detection of growth (protein analysis was done as described above). It is possible that the strains could grow on organic carbon provided by impurities in the water used to prepare the MSM. To lower this probability, we prepared the MSM with ultrapure water (Millipore). The theoretical washout rates of protein and yeast extract were calculated by utilizing the equation Xt = X_0e−_Dt, where Xt is the protein or yeast extract concentration at a certain time point after measurement was started, _X_0 is the initial protein or yeast extract concentration, D is the dilution rate, and t is the time after measurement was commenced (24).
Growth in the presence of the reduced inorganic sulfur compound tetrathionate was monitored by measuring the absorbance at 600 nm and the utilization of tetrathionate by cyanolysis (28).
Anaerobic growth.
Anaerobic growth was tested in Hungate tubes containing MSM supplemented with growth substrates as indicated (total volume, 10 ml). The medium was sparged with N2 for 30 min to remove oxygen before it was inoculated with a cell mass equivalent to 10 μg of protein from the continuous culture vessel. Prior to inoculation, cells were washed in MSM to remove any Fe(III) carried over from the continuous culture vessel. To ensure the exclusion of oxygen, the Hungate tubes were placed in anaerobic gas jars (CO2 atmosphere) and then incubated stationary at 37°C for 22 days. Cell growth was determined by measuring the protein concentration (as described above).
Growth on solid media.
Solid media were prepared by using Sigma Type I and BRL Ultrapure agarose as gelling agents. Both types of agarose were autoclaved as 1% (wt/vol) stock solutions in ultrapure water (Millipore), to which double-strength sterile MSM (pH 1.2), trace elements, and the desired growth substrates, as indicated, were added to make a final concentration of 0.5% agarose. Plates were incubated at 37°C for 21 days.
Antibiotic sensitivity and heavy metal tolerance.
The growth of Ferroplasma isolates in the presence of antibiotics and heavy metals was measured in batch cultures containing MSM, trace elements, Fe(II), and yeast extract (as described above). Cell growth was determined by measuring the protein concentration after 63 h (as described above). Initially, the isolates “_F. acidarmanus_” Fer1T, MT17, and DR1 were grown in the presence and absence of heavy metals. A standard amount of cell mass equivalent to 10 μg of protein of the metal-exposed and nonexposed cells was then used to inoculate fresh medium containing a range of heavy metal concentrations (0.1 to 1 g of metal liter−1). After an incubation of 63 h, the growth was measured as the change in protein concentration.
CO2 fixation.
Fixation of 14CO2 was carried out with all four Ferroplasma isolates based on the method of Golyshina et al. (11), with the following alterations. Experiments were carried out at 37°C in MSM containing the following (final concentrations): cell mass corresponding to 50 μg of protein with and without both 50 mM Fe(II) and 0.02% yeast extract. Conical glass inserts (300 μl) containing 5 μl of NaH14CO3 (total activity, 29.6 kBq per assay) were utilized to generate 14CO2. Controls were cells grown at 4°C as well as no-cell controls. Samples (100 μl) were removed at time frames ranging from 10 min to 17 h, filtered (0.2-μm-pore-size cellulose acetate filter; Whatman), and washed with 1 ml of MSM. The filters were immersed in 10 ml of liquid scintillation cocktail (Optiphase Supermix; Perkin Elmer) and counted on a Wallac 1409 liquid scintillation counter.
Electron microscopy.
Transmission electron micrographs were taken of “_F. acidarmanus_” Fer1T whole cells grown on Fe(II) and yeast extract and on yeast extract alone. For the measurement of cell diameter, 5-μl samples of the cell suspensions were pipetted onto 200-mesh copper grids and 5 μl of 2% (vol/vol) aqueous uranyl acetate was added. The mix was fixed for 1 min and blotted dry, and the cells were viewed at 80 kV on a JEOL 2000X STEM electron microscope.
Cells were fixed in a 50% (vol/vol) mix of 5% glutaraldehyde in sodium cacodylate for 30 min. Postfixing was done with 1% OsO4 (wt/vol) in sodium cacodylate for 1 h, followed by washing twice in distilled water for 5 min. The cells were dehydrated through an ethanol gradient series (10 min each in 30, 50, 70, 90, 98, and 100% ethanol) followed by 10 min in a solution containing 50% London resin (Agar Scientific) and 50% ethanol (vol/vol) for 1 h. The ethanol-London resin mix was replaced by 100% London resin, left to polymerize for 20 to 24 h at 60°C, and sectioned on an LKB ultramicrotome set at 70 to 100 nm. The sections were mounted on 100-mesh copper grids, double stained with salts of uranium and lead (14), and viewed at 80 kV on an LKB STEM electron microscope.
DNA extraction, 16S rDNA sequencing, and phylogenetic analysis.
DNA was extracted via freeze-thaw, lysis, and phenol-chloroform purification (4). The 16S rRNA gene was amplified separately by using primer pair 21Fa (6) and 1492R (18) and primer pair 515F (15) and 1492R. The PCR incubation sequence was 94°C for 5 min followed by 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s, and then finally 72°C for 5 min on a Touchgene thermocycler (Techne). The PCR fragments were cleaned by using Chroma Spin + TE-1000 columns (Clontech Laboratories), and sequencing reactions were done with a Big Dye kit (Perkin-Elmer Biosystems), with incubations of 94°C for 30 s followed by 30 cycles of 95°C for 30 s, 45°C for 15 s, and 60°C for 4 min. The primers used for sequencing were 21Fa, 515F, 906F (18), 519R (18), 907R (18), and 1492R. PCR fragments for sequencing were cleaned by using SigmaSpin postreaction purification columns (Sigma) and were analyzed on an ABI Prism 377 DNA sequencer. Evolutionary analyses of sequence data were performed by distance methods, using ARB (29) and PAUP (31), and by parsimony and maximum likelihood algorithms in PAUP. Distances were calculated in ARB according to the substitution algorithm of Jukes and Cantor (17), and phylogenetic trees were assembled by neighbor joining. Maximum likelihood analysis was done by using the GTR model (25), and heuristic searching was performed for the parsimony and maximum likelihood analyses.
DNA-DNA hybridization and G+C content.
G+C contents were calculated (in triplicate), with the type strain of Methanibrevibacter ruminantium serving as a reference. For establishment of the DNA-DNA reassociation values between Ferroplasma genomic DNAs, the spectrophotometric renaturation rate kinetic procedure adapted by Bowman et al. (5) from Huss et al. (16) was used. The optimal temperature for renaturation (25°C below melting temperature) was calculated, assuming a G+C content of 37 mol%, to be 64°C.
2D-PAGE.
Cells for 2D-PAGE were harvested, washed, and prepared in urea-thiourea cell lysis buffer (13). The cell extract was focused in the first dimension by loading 200 μg of protein on pH 4 to 7 strips (Amersham) and then was separated in the second dimension in PAGE gels (6a). PAGE gels were stained with silver nitrate (1) and scanned by the Proteomic imaging system (Perkin-Elmer Life Sciences). Gels were analyzed with ProteomWeaver, version 1.3 (Definiens), and spot profiles were used to produce similarity values utilizing the Dice-Sorensen index (23), from which an unrooted neighbor-joining tree was constructed in PAUP (31).
Nucleotide sequence accession number.
The DR1 16S rDNA sequence was elucidated and deposited into GenBank under accession number AY222042.
RESULTS
Morphology.
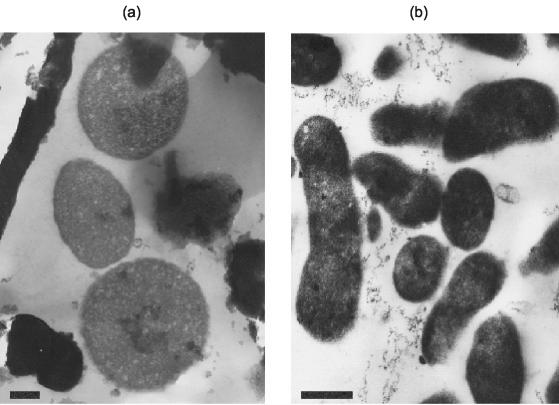
The Ferroplasma isolates were either pleomorphic or irregular cocci, and the “_F. acidarmanus_” Fer1T diameter was found to be 0.66 ± 0.18 and 0.57 ± 0.20 μm (n = 52) for cells grown on Fe(II) and yeast extract (Fig. 1a) and for those grown on yeast extract alone (Fig. 1b), respectively. The size difference between the two sets of cells was analyzed by Student's t test and was found to be significant, with a confidence of 98.4%.
FIG. 1.
Transmission electron micrographs of “_F. acidarmanus_” Fer1T cells grown chemomixotrophically on ferrous iron and yeast extract (a) and chemoorganotrophically on yeast extract (b). Bars, 250 nm.
Growth rates and temperature and pH optima.
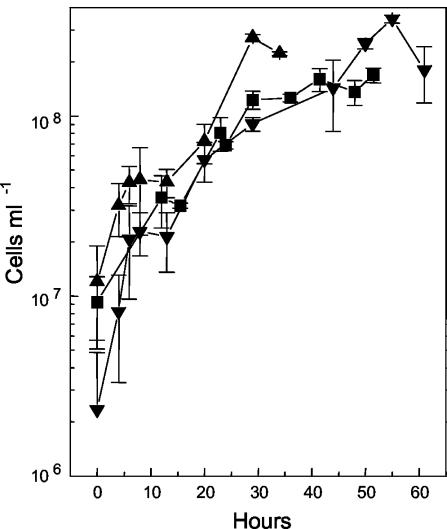
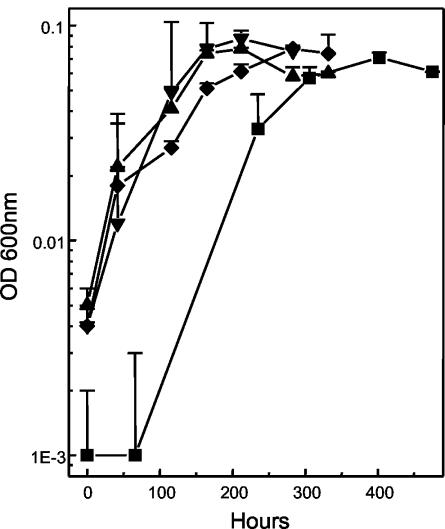
“_F. acidarmanus_” Fer1T was capable of growing chemomixotrophically on Fe(II) plus yeast extract (Fig. 2) and chemoorganotrophically on yeast extract alone (Fig. 3). The generation times for the chemomixotrophic and chemoorganotrophic cultures were 4.22 and 16.13 h, respectively. “_F. acidarmanus_” Fer1T grew to a higher peak cell density on Fe(II) plus yeast extract (1.4 × 108 ± 2.9 × 107 cells ml−1; n = 9) than on yeast extract alone (8.9 × 107 ± 3.7 × 107 cells ml−1; n = 12) (data not shown). Compared to “_F. acidarmanus_” Fer1T, strains MT17 and DR1 grew to higher cell densities, reaching 2.7 × 108 ± 1.2 × 107 cells ml−1 (n = 2) and 3.5 × 108 ± 1.7 × 107 cells ml−1 (n = 2), respectively (Fig. 2). The four Ferroplasma isolates were also grown chemoorganotrophically on yeast extract, and growth was measured by optical density readings (Fig. 3). Compared to the other isolates, chemoorganotrophically grown “_F. acidarmanus_” Fer1T had an extended lag phase and again achieved a lower cell density. With two further subcultures in MSM plus yeast extract, the generation time of the chemoorganotrophically grown culture decreased to 9.62 h and the lag phase decreased from 56 to 14 h (data not shown). We also tested whether the oxidation of manganese (72 mM) could support the growth of “_F. acidarmanus_” Fer1T and F. acidiphilum YT in the presence of 0.02% yeast extract. A statistically significant amount of growth was measured for both isolates, but it was less than their respective growth rates on yeast extract alone (data not shown).
FIG. 2.
Growth of Ferroplasma isolates in batch culture. “_F. acidarmanus_” Fer1T (▪), MT17 (▴), and DR1 (▾) were grown chemomixotrophically on ferrous iron and yeast extract. The data points are means ± SD (n ≥ 2).
FIG. 3.
Chemoorganotrophic growth of “_F. acidarmanus_” Fer1T (▪), MT17 (▴), DR1 (▾), and F. acidiphilum YT (♦) in batch cultures as measured by the optical density at 600 nm. The data points are means ± SD (n ≥ 2).
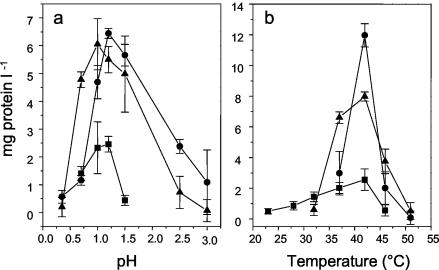
“_F. acidarmanus_” Fer1T grew at pHs 0.2 to 2.5, with an optimum at 1.2 (Fig. 4a), and had a temperature optimum of 42°C (Fig. 4b). Although “_F. acidarmanus_” Fer1T did not appear to grow at pH 0.35 (Fig. 4a) after 63 h, at 115.5 h it grew to 0.27 ± 0.15 and 2.60 ± 1.35 μg of protein ml−1 (n = 3) at pHs 0.2 and 0.5, respectively. Within the time frame tested (63 h), isolates MT17 and DR1 grew at pHs 0.35 to 3 and 0.35 to 2.5, respectively (Fig. 4a), and at 37 to 51°C and 32 to 51°C, respectively (Fig. 4b).
FIG. 4.
Temperature and pH dependency of Ferroplasma strains. “_F. acidarmanus_” Fer1T (▪), MT17 (•), and DR1 (▴) were grown on ferrous iron and yeast extract for 63 h at a range of pHs (a) and temperatures (b), and growth was measured as an increase in protein concentration. The results are averages of three experiments ± SD.
Oxygen consumption and growth by Ferroplasma isolates.
An increase in the oxygen consumption rate above that of the cell-free controls was observed for all of the substrates listed in Materials and Methods, except d-galactose, d-fructose, and sorbitol (data not shown). The increased O2 consumption suggests that these substrates are at least taken up, if not utilized, by “_F. acidarmanus_” Fer1T. It is possible that other Ferroplasma strains can grow by utilizing these organic substrates, but this was not tested. The four Ferroplasma isolates were tested to see if they could grow on each of the possible substrates. Those substrates that supported a statistically higher level of growth than the no-substrate control are listed in Table 1. All growth values are means of the differences between the final protein concentration (n ≥ 3) and the concentration in the respective no-substrate controls for each of the isolates. The optimum concentrations of yeast extract for chemomixotrophic growth with Fe(II) were 0.05, 0.01, and 0.02% (wt/vol) for Fer1T, MT17, and DR1, respectively. We also attempted to grow all four isolates chemoautotrophically on tetrathionate alone and chemomixotrophically on yeast extract and tetrathionate, as was previously reported for strain MT17 (22). In contrast to the previously published results for MT17, no tetrathionate oxidation was observed for any of the isolates under either of the growth conditions.
TABLE 1.
Growth of Ferroplasma isolates on various substrates (1 mM final concentration, unless specified otherwise)
| Growth condition or substrate | Protein concn (μg ml−1) in straina | |||
|---|---|---|---|---|
| Fer1T | MT17 | DR1 | YTb | |
| Chemoorganotrophic | ||||
| Yeast extract (0.02%) | 1.34 ± 0.38a | 4.09 ± 1.09 | 4.18 ± 0.68 | 1.81 ± 0.48 |
| Casamino Acids (2%) | 1.56 ± 0.26 | 2.66 ± 0.37 | 3.12 ± 0.37 | 0.59 ± 0.48 |
| Sucrose | 3.02 ± 0.36 | 2.56 ± 0.36 | 1.95 ± 0.29 | 2.19 ± 0.34 |
| Sorbitol | 1.98 ± 0.16 | 0.33 ± 0.24 | 2.61 ± 0.42 | 1.63 ± 0.36 |
| Lactose | 2.81 ± 0.41 | 2.90 ± 0.25 | 2.82 ± 0.34 | 1.24 ± 0.36 |
| Chemomixotrophic [Fe(II) plus substrate] | ||||
| Yeast extract (0.02%) | 2.74 ± 0.18 | 6.83 ± 0.20 | 5.50 ± 0.47 | Growth |
| Sucrose | 1.86 ± 0.27 | 2.86 ± 0.17 | 1.30 ± 0.32 | 1.06 ± 0.64 |
| Glucose | 1.50 ± 0.45 | 1.84 ± 0.18 | 1.84 ± 0.28 | ND |
| Galactose | 2.22 ± 0.38 | 1.26 ± 0.17 | 1.46 ± 0.28 | ND |
| Fructose | 3.57 ± 0.20 | 1.26 ± 0.17 | 1.25 ± 0.18 | ND |
| Maltose | 3.23 ± 0.46 | 1.86 ± 0.17 | 1.60 ± 0.28 | ND |
| Xylose | 2.58 ± 0.23 | 1.85 ± 0.34 | 0.47 ± 0.28 | ND |
| Mannitol | 3.50 ± 0.33 | 1.10 ± 0.25 | 0.49 ± 0.30 | ND |
| Sorbitol | 2.04 ± 0.34 | 0.27 ± 0.23 | 2.17 ± 0.30 | 2.57 ± 0.41 |
| Lactose | 2.28 ± 0.21 | 2.52 ± 0.23 | 3.38 ± 0.29 | 1.05 ± 0.61 |
| Thiamine | 0.26 ± 0.14 | 2.01 ± 0.18 | 0.63 ± 0.35 | ND |
| Casamino Acids (2%) | 2.78 ± 0.17 | 4.40 ± 0.25 | 4.90 ± 0.34 | Growth |
| Asparagine | 0.48 ± 0.15 | 0.66 ± 0.23 | 0.65 ± 0.32 | ND |
| Aspartic acid | 0.37 ± 0.17 | 0.60 ± 0.22 | 0.72 ± 0.29 | ND |
| Glutamic acid | 0.28 ± 0.18 | 1.21 ± 0.21 | 0.00 ± 0.28 | ND |
| Methionine | 0.25 ± 0.17 | 0.08 ± 0.34 | 1.37 ± 0.31 | ND |
| Phenylalanine | 2.76 ± 0.41 | 0.67 ± 0.20 | 1.01 ± 0.38 | ND |
| Serine | 0.44 ± 0.15 | 0.00 ± 0.17 | 0.00 ± 0.32 | ND |
| Tyrosine | 0.30 ± 0.18 | 0.00 ± 0.18 | 0.68 ± 0.31 | ND |
| Valine | 0.26 ± 0.15 | 0.13 ± 0.23 | 0.68 ± 0.31 | ND |
| Absence of added organic carbon | ||||
| Fe(II) | 0.00 ± 0.21 | 2.72 ± 0.25 | 3.08 ± 0.39 | 2.42 ± 0.32 |
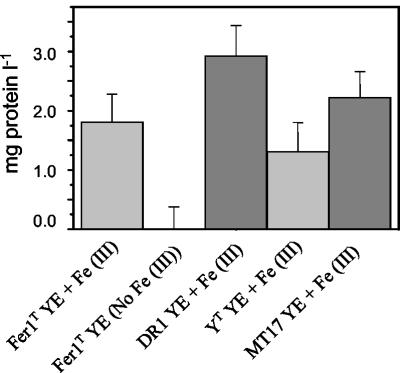
After 22 days, growth was observed in Hungate tubes under anaerobic conditions for all four Ferroplasma isolates. Growth occurred on 0.02% (wt/vol) yeast extract plus 10 mM Fe(III) (Fig. 5), and Fe(III) reduction was detected (data not shown). Anaerobic growth of all isolates in the absence of Fe(III) was very low (Fig. 5) and was similar to that of the no-substrate (absence of Fe(III) and yeast extract) controls.
FIG. 5.
Anaerobic growth of Ferroplasma strains on MSM supplemented with 0.02% (wt/vol) yeast extract (YE) plus 10 mM Fe(III) or yeast extract alone.
After 3 weeks of incubation at 37°C, small, white, round colonies appeared on plates containing MSM plus trace elements and yeast extract, but no growth was evident on plates grown with Fe(II) and yeast extract or ferrous iron plus Casamino Acids. The plating efficiencies for BRL Ultrapure and Sigma Type I agarose were 10.3% ± 2.1% and 7.7% ± 1.4% (n = 3), respectively. Single colonies were reinoculated from both agarose types into shake flasks with Fe(II) plus yeast extract or yeast extract alone. After 8 days, all of the cultures had grown (data not shown).
Analysis of autotrophy.
F. acidiphilum YT, DR1, and MT17 grew in MSM plus Fe(II) in shake flasks (Table 1). This suggested that strains YT, MT17, and DR1 could fix CO2 for autotrophic growth, as had previously been reported for F. acidiphilum YT (11). Attempts to stimulate chemolithoautotrophic growth of “_F. acidarmanus_” Fer1T by the use of increased CO2 (2% [vol/vol] in air) in shake flasks and also after a slow adaptation to reduced levels of yeast extract and increased CO2 (2% [vol/vol]) in the continuous culture vessel were unsuccessful.
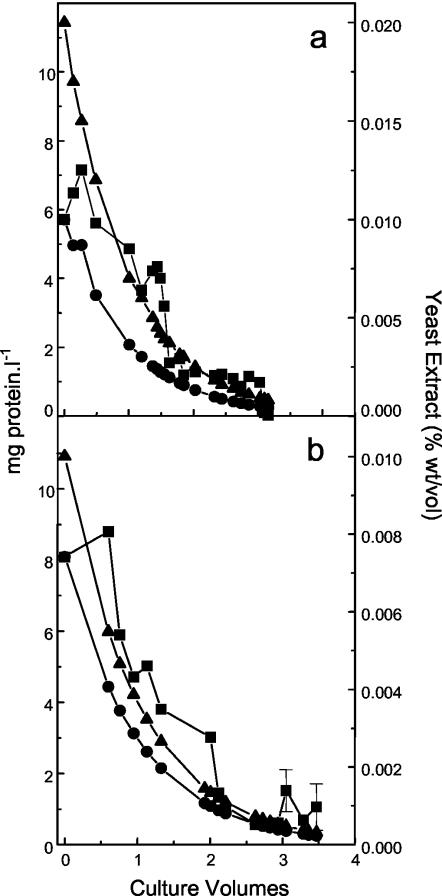
As a further test, the Ferroplasma isolates were maintained in continuous culture vessels in the absence of organic carbon. Neither DR1 nor “_F. acidarmanus_” Fer1T maintained a steady state after 3 volume changes in the absence of organic carbon (Fig. 6), although all isolates except “_F. acidarmanus_” Fer1T maintained a statistically valid protein concentration and could be revived in MSM plus Fe(II) and yeast extract (data not shown). The protein and yeast extract theoretical washout rates were also plotted, and the DR1 and “_F. acidarmanus_” Fer1T protein concentrations were maintained above the washout rate until the theoretical yeast extract concentration reached 0.0045 or 0.0003% (wt/vol), respectively. MT17 and F. acidiphilum YT also failed to maintain a steady state above the washout curve (data not shown).
FIG. 6.
Growth of “_F. acidarmanus_” Fer1T (a) and isolate DR1 (b) in continuous culture vessels in the absence of organic carbon. The culture volume started when the vessel was no longer fed organic carbon, and protein concentrations (▪) and theoretical protein (dashed line) and yeast extract (solid line) washout rates are presented. Protein concentrations for which SD are presented are means ± SD (n = 3).
We attempted to measure CO2 fixation on five separate occasions with various conditions and sampling times, but at no stage was a statistically significant concentration of 14CO2 incorporated into “_F. acidarmanus_” Fer1T compared to no-cell controls or no-growth controls (data not shown). We also tested whether F. acidiphilum YT, DR1, and MT17 incorporated more 14CO2 than the no-growth controls. No increase in 14CO2 incorporation was detected (data not shown).
Antibiotic sensitivity and heavy metal tolerance.
With the exception of gentamicin, “_F. acidarmanus_” Fer1T was at least partly susceptible to all of the antibiotics at the concentrations tested (Table 2). “_F. acidarmanus_” Fer1T was only slightly inhibited by ampicillin, chloramphenicol, and kanamycin and was strongly inhibited by rifampin and tetracycline (Table 2). In general, “_F. acidarmanus_” Fer1T was less sensitive to inhibition by antibiotics than F. acidiphilum YT (11).
TABLE 2.
Resistance of “_F. acidarmanus_” Fer1T to antibiotics
| Antibiotic | Concn (μg ml−1) | Growth of strain Fer1T (μg of protein ml−1)a |
|---|---|---|
| No antibiotic | 0 | 2.74 ± 0.18 |
| Ampicillin | 10 | 2.18 ± 0.60 |
| 100 | 1.43 ± 0.78 | |
| Chloramphenicol | 1 | 2.01 ± 0.39 |
| 50 | 1.93 ± 0.18 | |
| Kanamycin | 10 | 2.22 ± 0.37 |
| 100 | 1.15 ± 0.28 | |
| Rifampin | 2.5 | 0.98 ± 0.23 |
| 100 | 0.11 ± 0.23 | |
| Tetracycline | 0.2 | 1.68 ± 0.20 |
| 10 | 0.69 ± 0.42 | |
| Gentamicin | 0.2 | 2.10 ± 0.51 |
| 10 | 2.45 ± 0.46 |
All three of the Ferroplasma isolates tested exhibited a degree of resistance to cadmium, copper, arsenate, and arsenite (Table 3), although “_F. acidarmanus_” Fer1T appeared to be less sensitive to heavy metal inhibition. Increased levels of resistance to some of the metals could be induced by a previous exposure to subtoxic concentrations (for example, cadmium inhibition of DR1) (Table 3). We also found that in the presence of low concentrations of arsenate, “_F. acidarmanus_” Fer1T grew to a higher cell density than in the absence of As(V) (Table 3).
TABLE 3.
Metal resistance in strains “_F. acidarmanus_” Fer1T, MT17, and DR1 in uninduced and induced cells
| Metal | Concn (g liter−1) | Growth of strain (μg of protein ml−1)a | ||
|---|---|---|---|---|
| Fer1T | MT17 | DR1 | ||
| No metal | 0 | 2.74 ± 0.18 | 6.83 ± 0.20 | 5.50 ± 0.47 |
| Cd | 0.1 | 2.41 ± 0.24 | 1.86 ± 0.18 | 1.60 ± 0.49 |
| Reinoculated Cd | 0.4 | 2.53 ± 0.30 | 2.39 ± 0.19 | 2.79 ± 0.37 |
| Cu | 1 | 1.85 ± 0.45 | 0.77 ± 0.19 | 3.90 ± 0.38 |
| Reinoculated Cu | 1 | 1.97 ± 0.59 | 1.18 ± 0.25 | 1.06 ± 0.33 |
| As(III) | 0.05 | 1.81 ± 0.29 | 2.43 ± 0.19 | 1.57 ± 0.35 |
| Reinoculated As(III) | 0.1 | 2.73 ± 0.62 | 2.57 ± 0.31 | 1.94 ± 0.39 |
| As(V) | 0.15 | 7.85 ± 0.22 | 0.58 ± 0.40 | 3.00 ± 0.50 |
| Reinoculated As(V) | 0.15 | 2.37 ± 0.45 | 2.12 ± 0.17 | 2.32 ± 0.43 |
16S sequence data, DNA hybridization, and 2D-PAGE protein profiles.
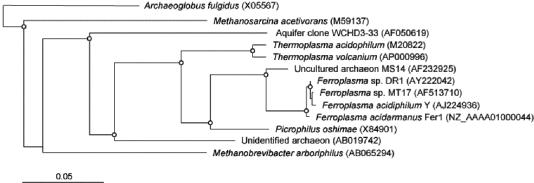
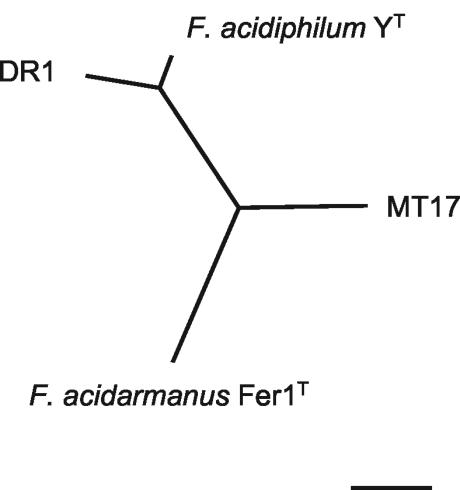
The DR1 16S rDNA sequence was elucidated, and a similarity matrix was calculated for the four isolates (Table 4). A phylogenetic tree was constructed for the Ferroplasma isolates and related microorganisms (Fig. 7). It can be seen that although the 16S rDNA similarities are very close, “_F. acidarmanus_” Fer1T forms a separate group from the other three strains, and this relationship is supported by all phylogenetic estimations (Fig. 7). The DNA-DNA similarities between the four isolates are also listed in Table 4, and an unrooted tree was constructed (Fig. 8). The results clearly show that strains DR1 and YT are from the same species and that MT17 is more closely related to DR1 and YT than to “_F. acidarmanus_” Fer1T. 2D-PAGE protein profiles were obtained with whole-cell extracts from all four Ferroplasma isolates grown chemomixotrophically on Fe(II) and 0.02% yeast extract (6a). The unrooted neighbor-joining tree constructed from the protein profile data had the same topology as that produced from DNA-DNA similarity data (Dopson et al., submitted), with the longest branch separating “_F. acidarmanus_” Fer1T from the other isolates.
TABLE 4.
Degrees of 16S rDNA (upper sector) and DNA-DNA (lower-sector) similarity among the four Ferroplasma strains
| Strain (for DNA-DNA similarity) | % 16S rDNA or DNA-DNA similarity with indicated straina | ||
|---|---|---|---|
| Fer1T | DR1 | MT17 | YT |
| Fer1T | 99.7 | 99.6 | 98.9 |
| DR1 | 67.6 ± 6.0 | 99.7 | 99.7 |
| MT17 | 75.6 ± 10.6 | 72.8 ± 5.5 | 99.7 |
| YT | 71.7 ± 7.0 | 91.3 ± 3.1 | 74.5 ± 5.2 |
FIG. 7.
Phylogenetic relationship of 16S rRNA gene sequences with representative sequences from databases. The dendrogram was generated by distance matrix and neighbor-joining methods and was rooted thereafter at the branch with Archaeoglobus fulgidus as the outgroup. Accession numbers for sequences are given in parentheses. The scale bar represents 0.05 substitutions per site. Branch points supported by distance, maximum likelihood, and parsimony estimations are indicated by open circles.
FIG. 8.
Unrooted neighbor-joining tree of DNA-DNA similarity. The scale bar represents 10% changes.
DISCUSSION
This paper is a detailed study of the phenotypic and genotypic characteristics of four isolates from the genus Ferroplasma which were isolated from different acid bioleaching and AMD environments. A summary of all of the phenotypic and genotypic characteristics of the four isolates is given in Table 5. The results from this study make possible an assessment of the biogeochemical role of these Ferroplasma isolates in acid-leaching environments.
TABLE 5.
Phenotypic characteristics of Ferroplasma isolatesa
| Characteristic | Result for strainb | |||
|---|---|---|---|---|
| Fer1T | MT17 | DR1 | YT | |
| Morphology | Pleomorphic | Irregular cocci | Irregular cocci | Pleomorphic |
| Chemoautotrophic growth | − | ± | ± | ± |
| Chemomixotrophic growth | + | + | + | + |
| Chemoorganotrophic growth | + | + | + | + |
| Organic carbon requirement | High | low | Low | Low |
| Anaerobic growth | + | + | + | + |
| Growth temperature | ||||
| Range (°C) | 23-46 | 32-51 | 32-51 | 15-45 |
| Optimum (°C) | 42 | 42 | 42 | 35 |
| pH | ||||
| Range | <0-1.5 | 0.35-3.0 | 0.35-3.0 | 1.3-2.2 |
| Optimum | 1.2 | 1.2 | 1.0 | 1.7 |
| Yeast extract optimum (%) | 0.05 | 0.02 | 0.01 | 0.02 |
| DNA G+C content (%) | 36.8 | 36.5 | 37.0 | 36.5 |
The question of autotrophy is central to the classification of the genus Ferroplasma, and at least two definitions are presented in the literature. The first, used by Golyshina et al. (11), is that an autotroph fixes CO2 as its carbon source “except perhaps for some vitamins that may be required for growth” (34), whereas the second definition (used in this study) is that an autotroph fixes CO2 as its sole source of organic carbon (19). Although F. acidiphilum YT has been described as strictly chemoautotrophic, the culture medium used in that study included 0.02% (wt/vol) yeast extract (11). It was argued that the amount of yeast extract in the medium was insufficient to be a carbon and energy source, and therefore by the definition utilized by Golyshina et al., F. acidiphilum YT grew autotrophically. In this study, we have shown that the four Ferroplasma isolates could grow chemoorganotrophically on the same concentration of yeast extract as that used in the F. acidiphilum YT study. Therefore, under the conditions described, F. acidiphilum YT cannot be described as chemoautotrophic and must be referred to as chemomixotrophic.
The protein concentration in continuous culture vessels in the absence of organic carbon was initially maintained above the theoretical protein washout rate. After 3 volume changes, DR1 (Fig. 6b), F. acidiphilum YT, and MT17 (data not shown) maintained a statistically valid level of protein above zero, but “_F. acidarmanus_” Fer1T did not (Fig. 6a). This reflects the relative requirement for organic carbon for the four isolates. We also attempted to measure 14CO2 uptake with all four Ferroplasma isolates, but we were unsuccessful. As in a previous study (11), low levels of 14CO2 uptake were detected. However, these levels were not statistically higher than those for no-cell controls or cells incubated in the presence of 14CO2 at 4°C. Therefore, our results clearly show that F. acidiphilum YT, DR1, and MT17 have a much lower requirement for organic carbon than “_F. acidarmanus_” Fer1T and that they possibly fix organic carbon. This pattern of carbon requirement among the isolates was supported by the revival of all isolates except “_F. acidarmanus_” Fer1T after three culture volume changes in continuous culture vessels in the absence of added organic carbon. Also, protein production was detected in batch cultures in the absence of organic carbon for all isolates except “_F. acidarmanus_” Fer1T (Table 1). The annotation of the 97% complete genome sequence of “_F. acidarmanus_” Fer1T does not indicate the presence of genes for a complete CO2 fixation pathway. The presence of genes for CO2 fixation cannot be completely ruled out given the number of hypothetical proteins with unknown functions and given that the genome sequence is incomplete. However, in compliance with the organic carbon requirements detected in this study, it is possible that the other Ferroplasma isolates (not “_F. acidarmanus_” Fer1T) have genes encoding autotrophy.
Our results provide some insights into the possible activities of these organisms in their habitats. Mixotrophic growth oxidizing Fe(II) is a key phenotypic trait which is important for growth and survival of these microorganisms in acid-leaching environments. This trait would also be key to contributing to the production of acid-leaching solutions affected by the biological generation of the strong oxidant Fe(III) and protons. The lack of chemoautotrophic growth by “_F. acidarmanus_” Fer1T was surprising considering its high level of biofilm growth in association with fungi in a low-organic-carbon-level environment (3). It is possible that “_F. acidarmanus_” Fer1T is autotrophic, but not under the conditions tested, or that it may require a growth factor that was not supplied in the laboratory but that may be provided in its natural environment, possibly by a fungus. Another alternative is that the natural population contains both chemolithoautotrophic and nonchemolithoautotrophic strains that are indistinguishable by the 16S rDNA probe used. “_F. acidarmanus_” Fer1T grows at both lower temperatures and over a wider temperature range (23 to 46°C) than the other three strains. This possibly reflects the fact that it was isolated from a natural mine site, where it would be expected to experience a wider range of temperatures than the controlled environment in a biooxidation vessel. Also, “_F. acidarmanus_” Fer1T grew at a lower and narrower pH range and has previously been reported to grow at pH 0 (10). It is likely that this reflects the extremely low pH of the Iron Mountain AMD site (20). One other phenotypic difference is that “_F. acidarmanus_” Fer1T grows to an approximately 10-fold-lower cell density (based on protein concentration [11]) than F. acidiphilum YT and has significantly lower cell counts than strains DR1 and MT17. This could be due to the culture conditions for “_F. acidarmanus_” Fer1T being below optimum. For example, “_F. acidarmanus_” Fer1T has a higher requirement for organic carbon than the other strains do.
Also, in contrast to a previous report (11), we found that manganese could not support the growth of “_F. acidarmanus_” Fer1T or F. acidiphilum YT, as the amount of chemomixotrophic growth was less than that of chemoorganotrophic growth on yeast extract alone. This suggests that the manganese added (72 mM) was not only unable to be used as an energy source but was also slightly inhibitory for both strains. Finally, in contrast to the F. acidiphilum YT study, all four Ferroplasma isolates have been demonstrated to grow anaerobically via the oxidation of yeast extract coupled to the reduction of Fe(III). In natural AMD and bioleaching environments, Ferroplasma species grow in the form of biofilms (4), which is an advantageous form of growth for avoiding washout. Anaerobic growth is an important phenotype for Ferroplasma, as the dissolved oxygen concentrations in biofilms vary greatly, and thus within the interior Fe(III) reduction may be important for activity and cell maintenance.
As “_F. acidarmanus_” Fer1T does not possess a cell wall, it was unsurprising that ampicillin was ineffective. Similar results were found for F. acidiphilum YT (11) and for members of another acidophilic archaeal genus, Picrophilus (27). A difference between the antibiotic sensitivities of “_F. acidarmanus_” Fer1T and Picrophilus was that “_F. acidarmanus_” Fer1T was resistant to >50 μg of chloramphenicol ml−1 (a concentration expected to be inhibitory), while Picrophilus was susceptible. “_F. acidarmanus_” Fer1T, DR1, and MT17 all exhibited a degree of resistance to cadmium, copper, arsenite, and arsenate. This is not surprising given the metal-rich environments from which they were isolated. In some instances, either induction of the Ferroplasma isolates to metal resistance or adaptation to higher levels of metals occurred. It was also observed that small quantities of arsenate stimulated the growth of “_F. acidarmanus_” Fer1T. An increase in the oxidation rate of reduced inorganic sulfur compounds in the presence of low levels of arsenite has also been observed for Acidithiobacillus caldus (12), possibly due to the increased energy demand for efflux via the ArsB protein (8). Also, in its natural environment “_F. acidarmanus_” Fer1T grows in very high metal concentrations, including arsenopyrite at the Iron Mountain site (20), and it is possible that it may require small quantities of metals for optimal growth.
The four Ferroplasma isolates have 16S rRNA gene similarities between 98.9 and 99.7%, although phylogenetic trees constructed using distance, maximum likelihood, and parsimony estimations all supported branch points that separated “_F. acidarmanus_” Fer1T from the other three Ferroplasma strains (Fig. 7). DNA-DNA similarity data are currently viewed as the most reliable method of defining taxonomic relationships, and a similarity of 70% or higher has been defined as the species border (33). Even though 16S rDNA sequence similarities of >97% are accepted as a single species, it has been found that isolates with >97% similarity have <70% DNA-DNA similarity (26). The DNA-DNA similarity values for the Ferroplasma isolates suggest that DR1 and F. acidiphilum YT are definitely of the same species, and strain MT17 is just above the 70% cutoff. However, similarity values comparing “_F. acidarmanus_” Fer1T with YT and DR1 are at the species cutoff borderline.
Phenotypic studies that are useful for comparisons include assays of substrates that support growth, assays to measure optima of temperature and pH, and chemotaxonomic assays such as those measuring the cellular production of proteins. This study presents the first use of 2D-PAGE profiles for phylogenetic analysis. This analysis gave the same tree topology for the relationship between the four Ferroplasma strains as the DNA-DNA similarity analysis did (Fig. 8) (6a). Consequently, this method may be very useful for classifying microorganisms, particularly for distinguishing between closely related isolates and when other phylogenetic methods are inconclusive.
The relationship of “_F. acidarmanus_” Fer1T to the other Ferroplasma isolates is summarized in Table 5. Based on 16S rDNA sequence data and 2D-PAGE profiles, “_F. acidarmanus_” Fer1T groups as a separate phylotype, and it is phenotypically and genotypically different from the other Ferroplasma strains. While DNA-DNA similarity values are borderline, we propose that it is a separate species and retain it as “_F. acidarmanus_” Fer1T.
Updated description of Ferroplasma genus based on the work of Golyshina et al. (11).
Grows chemoorganotrophically, chemomixotrophically, and possibly chemoautotrophically. Facultative anaerobes. Colonies formed on yeast extract solid medium are small, white, and round. G+C content is 36.5 to 37 mol%. The type strain is Ferroplasma acidiphilum YT and there are three other isolates, namely DR1, MT17, and “_Ferroplasma acidarmanus_” Fer1T.
Description of F. acidarmanus sp. nov. Dopson, Baker-Austin, Hind, Bowman, and Bond 2004.
F. acidarmanus (a.cid.ar′man.us. L. n. acidus, acid; L. adj. armanus, pertaining to Arman, the owner of the mine from which the species was isolated; N.L. adj. acidarmanus, an acidophilic archaeon isolated from a mine belonging to Arman).
Isolated from an AMD site in California. Morphology as described for the genus. G+C content of 36.8 mol%. Growth occurs between 23 and 46°C (mesophilic) and pHs 0 and 1.5 (acidophilic). Grows chemoorganotrophically on yeast extract, Casamino Acids, and sugars and chemomixotrophically on Fe(II) and the above organic carbon sources. Facultative anaerobe, coupling oxidation of yeast extract to Fe(III) reduction. “_F. acidarmanus_” Fer1T has been deposited in the American Type Culture Collection in the patent pending collection.
Acknowledgments
We acknowledge Jillian F. Banfield and Katrina J. Edwards for the provision of “_F. acidarmanus_” Fer1T and for fruitful discussions. Doug Rawlings and D. Barrie Johnson are gratefully acknowledged for providing strains DR1 and MT17, respectively. Barrie Johnson is also thanked for kindly sharing data on MT17 prior to publication. The technical assistance of Linda Flegg was greatly appreciated and Richard Evans-Gowing is acknowledged for taking the electron micrographs.
This work was funded by a BBSRC research grant.
REFERENCES
- 1.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant-proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8**:**93-99. [Google Scholar]
- 2.Bond, P. L., and J. F. Banfield. 2001. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41**:**149-161. [DOI] [PubMed] [Google Scholar]
- 3.Bond, P. L., G. K. Druschel, and J. F. Banfield. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66**:**4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66**:**3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, J. P., S. A. McCammon, J. L. Brown, and T. A. McMeekin. 1998. Glaciecola punicea gen. nov., sp. nov.: psychrophilic bacteria from Antarctic sea-ice habitats. Int. J. Syst. Bacteriol. 48**:**1213-1222. [DOI] [PubMed] [Google Scholar]
- 6.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89**:**5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Dopson, M., C. Baker-Austin, and P. L. Bond. First use of 2-dimensional polyacrylamide gel electrophoresis to determine phylogenetic relationships. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 7.Dopson, M., and E. B. Lindstrom. 1999. Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl. Environ. Microbiol. 65**:**36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dopson, M., E. B. Lindstrom, and K. B. Hallberg. 2001. Chromosomally encoded arsenical resistance of the moderately thermophilic acidophile Acidithiobacillus caldus. Extremophiles 5**:**247-255. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, K. J., P. L. Bond, G. K. Druschel, M. M. McGuire, R. J. Hamers, and J. F. Banfield. 2000. Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem. Geol. 169**:**383-397. [Google Scholar]
- 10.Edwards, K. J., P. L. Bond, T. M. Gihring, and J. F. Banfield. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287**:**1796-1799. [DOI] [PubMed] [Google Scholar]
- 11.Golyshina, O. V., T. A. Pivovarova, G. I. Karavaiko, T. F. Kondrat'eva, E. R. B. Moore, W. R. Abraham, H. Lunsdorf, K. N. Timmis, M. M. Yakimov, and P. N. Golyshin. 2000. Ferroplasma acidiphilum gen. nov., sp nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int. J. Syst. Evol. Microbiol. 50**:**997-1006. [DOI] [PubMed] [Google Scholar]
- 12.Hallberg, K. B., M. Dopson, and E. B. Lindstrom. 1996. Arsenic toxicity is not due to a direct effect on the oxidation of reduced inorganic sulfur compounds by Thiobacillus caldus. FEMS Microbiol. Lett. 145**:**409-414. [Google Scholar]
- 13.Hesketh, A., D. Fink, B. Gust, H. U. Rexer, B. Scheel, K. Chater, W. Wohlleben, and A. Engels. 2002. The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 46**:**319-330. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, J. D., P. R. Smith, R. Evans-Gowing, and D. J. Richardson. 1995. Bacterial photoprotection through extracellular cadmium sulfide crystallites. Photochem. Photobiol. 62**:**1022-1026. [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180**:**4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huss, V. A. R., H. Festl, and K. H. Schleifer. 1983. Studies on spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4**:**184-192. [DOI] [PubMed] [Google Scholar]
- 17.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 18.Lane, D. J. 1985. 16S/23S sequencing, p. 115-176. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 19.Madigan, M. T., J. M. Martinko, and J. Parker. 1997. Brock Biology of microorganisms, 8th ed. Prentice-Hall International, London, United Kingdom.
- 20.Nordstrom, D. K., and C. N. Alpers. 1999. Negative pH, efflorescent mineralogy, and consequences for environmental restoration at the iron mountain superfund site, California. Proc. Natl. Acad. Sci. USA 96**:**3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norris, P. R. 1990. Acidophilic bacteria and their activity in mineral sulfide oxidation, p. 3-27. In H. L. Ehrlich and C. L. Brierley (ed.), Microbial mineral recovery. McGraw Hill, New York, N.Y.
- 22.Okibe, N., M. Gericke, K. B. Hallberg, and D. B. Johnson. 2003. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred-tank bioleaching operation. Appl. Environ. Microbiol. 69**:**1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pielou, E. C. 1972. Niche width and niche overlap: a method for measuring them. Ecology 53**:**687-692. [Google Scholar]
- 24.Pritchard, R. H., and D. W. Tempest. 1982. Growth: cells and populations, p. 99-123. In J. Mandelstam, K. McQuillen, and I. Dawes (ed.), Biochemistry of bacterial growth, 3rd ed. Blackwell Scientific Publications, Oxford, United Kingdom.
- 25.Rodríguez, F., J. F. Oliver, A. Marín, and J. R. Medina. 1990. The general stochastic model of nucleotide substitutions. J. Theoret. Biol. 142**:**485-501. [DOI] [PubMed] [Google Scholar]
- 26.Rossello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25**:**39-67. [DOI] [PubMed] [Google Scholar]
- 27.Schleper, C., G. Puehler, I. Holz, A. Gambacorta, D. Janekovic, U. Santarius, H. P. Klenk, and W. Zillig. 1995. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J. Bacteriol. 177**:**7050-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorbö, B. 1957. A colorimetric method for the determination of thiosulfate. Biochim. Biophys. Acta 23**:**412-416. [DOI] [PubMed] [Google Scholar]
- 29.Strunk, O., and W. Ludwig. 2002. ARB-a software environment for sequence data. Department of Microbiology, Technical University of Munich, Munich, Germany.
- 30.Suzuki, I. 2001. Microbial leaching of metals from sulfide minerals. Biotechnol. Adv. 19**:**119-132. [DOI] [PubMed] [Google Scholar]
- 31.Swofford, D. L. 2002. PAUP. Phylogenetic analysis using parsimony, 4.0b5 ed. Sinauer Associates, Sunderland, Mass.
- 32.Torma, A. E. 1977. The role of Thiobacillus ferrooxidans in hydrometallurgical processes. Adv. Biochem. Eng. 6**:**1-37. [Google Scholar]
- 33.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad-hoc-committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37**:**463-464. [Google Scholar]
- 34.White, D. 1995. The physiology and biochemistry of prokaryotes. Oxford University Press, New York, N.Y.