A type I IFN–Flt3 ligand axis augments plasmacytoid dendritic cell development from common lymphoid progenitors (original) (raw)
Type I interferon promotes the differentiation of plasmacytoid dendritic cells in part by up-regulating expression of Flt3 on common lymphoid progenitors.
Abstract
During infections and inflammation, plasmacytoid dendritic cells (pDCs) are the most potent type I interferon (IFN-I)–producing cells. However, the developmental origin of pDCs and the signals dictating pDC generation remain incompletely understood. Here, we report a synergistic role for IFN-I and Flt3 ligand (FL) in pDC development from common lymphoid progenitors (CLPs). Both conventional DCs (cDCs) and pDCs were generated from CLPs in response to FL, whereas pDC generation required higher concentrations of FL and concurrent IFN-I signaling. An absence of IFN-I receptor, impairment of IFN-I signaling, or neutralization of IFN-I significantly impeded pDC development from CLPs. Furthermore, FL induced IFN-I expression in CLPs, which in turn induced Flt3 up-regulation that facilitated survival and proliferation of CLPs, as well as their differentiation into pDCs. Collectively, these results define a critical role for the FL/IFN-I/Flt3 axis in pDC differentiation from CLPs.
Plasmacytoid DCs (pDCs) are a subset of DCs that are capable of producing large quantities of type I IFN (IFN-I) upon stimulation (Liu, 2005). pDCs are short-lived and require constant replenishment from their precursors in the BM. DCs, including pDCs, are generated from Flt3-expressing progenitors of myeloid lineages, including common myeloid progenitor (CMP), macrophage/DC progenitor, and common dendritic progenitor (CDP), and of lymphoid lineages such as common lymphoid progenitor (CLP; D’Amico and Wu, 2003; Karsunky et al., 2003; Shigematsu et al., 2004; Naik et al., 2007; Onai et al., 2007; Liu et al., 2009). Targeted deletion of Flt3 or FL in mice leads to reduced numbers of DC progenitors and impaired DC development (McKenna et al., 2000; Waskow et al., 2008). Ectopic expression of Flt3 on Flt3- megakaryocyte/erythrocyte progenitors permits them to generate DCs (Onai et al., 2006). These results suggest that the Flt3 ligand (FL) is one of the most physiological relevant cytokines for maintaining homeostasis of steady-state DCs.
IFN-I, a pluripotent cytokine, regulates different aspects of DC biology. Mouse pDCs depend on IFN-I for migration and clustering in the marginal zone of the spleen (Asselin-Paturel et al., 2005). pDCs developed in the presence of IFN-I fail to produce IFN-I in response to CpG (Li et al., 2011). IFN-I induced during systemic viral infections and inflammation trigger apoptosis of pDCs and cDCs, resulting in decline of the populations in the spleen (Swiecki et al., 2011). Therefore, IFN-I appears to govern migration, function, apoptosis, and homeostasis of DCs. Whereas cDCs and pDCs can be generated from myeloid or lymphoid lineages, the developmental origin and signals controlling pDC development remain elusive. Here, we showed that FL controlled the developmental program of cDCs and pDCs from CLPs in a dose-dependent manner. FL also induced IFN-I in CLPs, resulting in up-regulation of Flt3 that facilitated survival, proliferation, and differentiation of CLPs. These results establish a critical role for IFN-I in FL-dependent pDC development from CLPs.
RESULTS AND DISCUSSION
pDC development is impaired in the absence of IFN-I signaling
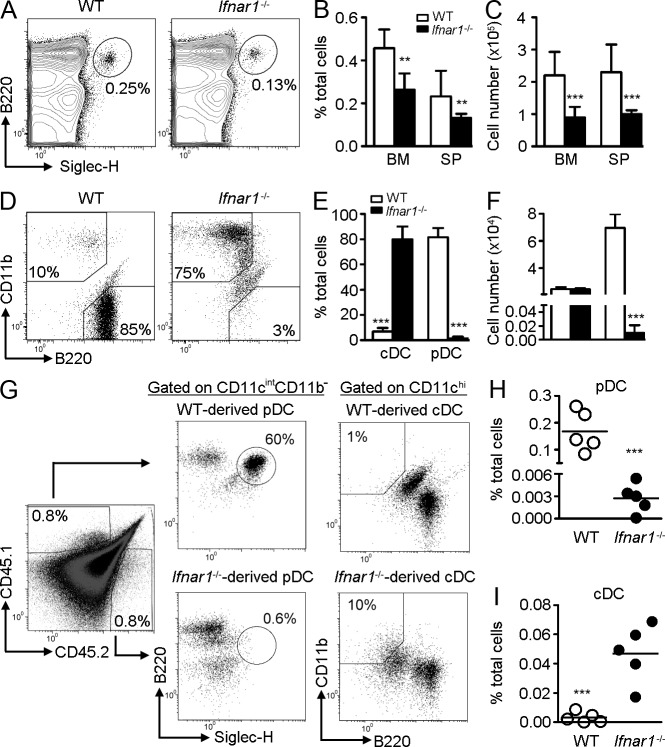
We have previously shown that Stat2 hypomorphic mutant mice (Stat2m/m) are hyporesponsive to IFN-I and have reduced numbers of DCs, particularly pDCs, in the spleen (Chen et al., 2009). Because STAT2 is predominantly activated by type I and III IFN, we used Ifnar1−/− mice to investigate whether the phenotype was due to impaired IFN-I signaling. Indeed, Ifnar1−/− mice showed significantly lower percentages and numbers of pDCs in the BM and spleen compared with WT mice (Fig. 1, A–C). A similar phenotype was observed in Stat1−/− and Stat2m/m mice (unpublished data), suggesting that IFN-I receptor and its downstream signals are involved in pDC homeostasis.
Figure 1.
Impaired pDC development from _Ifnar1_−/− CLPs is cell autonomous. (A) BM or splenocytes from WT or _Ifnar1_−/− mice were stained, gated on CD11cintCD11b−, and analyzed for pDCs (B220+Siglec-H+). Mean percentages (B) and numbers (C) of pDCs are shown (n = 4). Results represent four experiments. (D) CLPs from the BM of WT or _Ifnar1_−/− mice were co-cultured with AC-6.21, supplied with 100 ng/ml FL, and gated on CD11c+. cDC (CD11b+B220−) and pDCs (CD11b−B220+) populations were enumerated. Mean percentages (E) and numbers (F) of DCs are shown (n = 4–8). Results represent three experiments. (G) Competitive adoptive transfer was performed by co-transferring 104 WT (CD45.1) and 104 Ifnar1−/− (CD45.2) CLPs with 2 × 105 BM cells (CD45.1xCD45.2) into lethally irradiated donors (CD45.1xCD45.2). After 12 d, BM was analyzed for CLP-derived DCs. Mean percentages of pDCs (H) and cDCs (I) are shown (n = 5). Results represent two experiments.
Because pDCs can be generated from myeloid or lymphoid progenitors, we next examined whether the generation of pDC from CDPs (myeloid) or CLPs (lymphoid) is affected by IFN-I signaling. In an AC-6.21 feeder system, FL primarily supported cDC development from CDPs of both WT and Ifnar1−/− mice (unpublished data). In sharp contrast to the development from CDPs, FL showed poor ability to support cDC development from WT CLPs. Instead, pDCs were preferentially generated (Fig. 1, D and E). However, this process was reversed by the absence of IFN-I signaling, as Ifnar1−/− CLPs predominantly developed cDCs but not pDCs. The total numbers of DCs generated from Ifnar1−/− CLPs were also lower than those from WT CLPs. Therefore, the numbers of Ifnar1−/− CLP-derived cDCs were actually comparable to those from WT CLPs, whereas the numbers of Ifnar1−/− CLP-derived pDCs were markedly reduced (Fig. 1 F). A similar result was also observed using BM cells as feeder (unpublished data). We next performed competitive adoptive transfer to examine whether the developmental defect is cell intrinsic. As shown in Fig. 1 (G–I), the reconstitution rate of WT and Ifnar1−/− CLPs in the BM was comparable. However, the percentages of Ifnar1−/− CLP-derived pDCs remained significantly lower than those from WT CLPs. Conversely, the percentages of Ifnar1−/− CLP-derived cDCs were higher than those from WT CLPs. A similar phenotype was observed in the spleen and thymus (unpublished data). These results suggest that the requirement of IFN-I for CLP-driven pDC development is cell autonomous.
Ifnar1−/− CLPs express lower Flt3 and show impaired survival, proliferation, and differentiation in response to FL
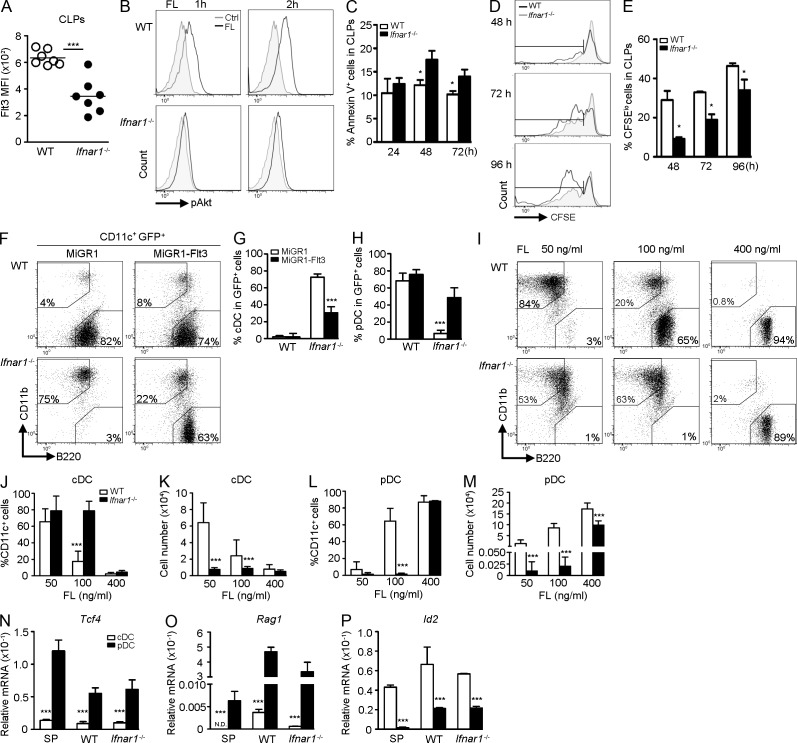
Because DC development is tightly regulated by the expression of Flt3 on progenitor cells, we next examined whether the impaired pDC generation from Ifnar1−/− CLPs is caused by aberrant Flt3 expression. Indeed, Ifnar1−/− CLPs expressed ∼50% less Flt3 than did WT CLPs, as judged by reduced mean fluorescent intensity (Fig. 2 A), even though the percentage of Flt3-expressing CLPs was comparable (not depicted). Concomitantly, FL-stimulated Akt phosphorylation was attenuated in lin−Flt3+ cells of Ifnar1−/− mice compared with those of WT mice (Fig. 2 B). In addition, Ifnar1−/− CLPs also showed increased apoptosis (Fig. 2 C), and decreased proliferation (Fig. 2, D and E) in response to FL. Interestingly, enforced expression of Flt3 in Ifnar1−/− CLPs significantly increased the ability to produce pDCs compared with control (Fig. 2, F–H), suggesting that the developmental defect in Ifnar1−/− CLPs is strongly linked to decreased Flt3 expression.
Figure 2.
_Ifnar1_−/− CLPs display reduced Flt3 expression and impaired survival, proliferation, and differentiation in response to FL. (A) CLPs were isolated from WT or _Ifnar1_−/− mice and Flt3 expression was analyzed by flow cytometry (n = 7). Results represent five experiments. (B) BM was stimulated with or without 100 ng/ml of FL for the indicated times, cells were gated on lin−Flt3+ cells, and expression of pAkt was analyzed by flow cytometry (n = 3). One experiment out of three is shown. (C) CLPs were stimulated with 100 ng/ml of FL for the indicated times and stained for Annexin V (n = 4). Results represent four experiments. (D) Same as in C, except CLPs were CFSE labeled before FL stimulation for the indicated times. (E) Mean percentages of CFSElo population are shown (n = 4). Results represent two experiments. (F) CLPs that had been transduced with MiGR1 or MiGR1-Flt3 were co-cultured with AC-6.21 supplied with 100 ng/ml of FL, gated on CD11c+GFP+, and analyzed for DC populations. Mean percentages of GFP+ cDCs (G) and pDCs (H) are shown (n = 4). Results represent four experiments. (I) CLPs were co-cultured with AC-6.21 supplied with the indicated doses of FL, gated on CD11c+, and analyzed for DC populations. Mean percentages (J and L) and numbers (K and M) of DCs are shown (n = 3–9). Results represent five experiments. Same as in I, except 400 ng/ml of FL was supplied. CLP-derived cDCs or pDCs were sorted, and expression of Tcf4 (N), Rag1 (O), or Id2 (P; n = 3) was evaluated by RT-qPCR. One experiment out of three is shown.
We reasoned that if reduced Flt3 expression was the cause of attenuated signaling and, thus, defective pDC development, increasing the concentration of FL might rescue the development. Interestingly, low-dose FL (50 ng/ml) predominantly induced cDC development in WT CLPs, whereas high-dose FL (100 ng/ml or 400 ng/ml) preferentially promoted pDC formation (Fig. 2, I–K), revealing a dose-dependent effect of FL on the differentiation potential of CLPs. Although 100 ng/ml of FL failed to rescue pDC development from Ifnar1−/− CLPs, increasing the dose to 400 ng/ml did recover the ability to differentiate into pDCs. To investigate whether pDCs derived from Ifnar1−/− CLPs remained physiologically intact, we examined the expression of pDC-specific genes, such as Tcf4 (E2-2) and Rag1. mRNA levels of these two genes in WT and Ifnar1−/− CLP-derived pDCs were comparable and, like splenic pDCs, were higher than those of cDCs (Fig. 2, N-O). However, the expressions of Id2, a cDC-specific gene, in WT and Ifnar1−/− pDCs were lower than those of cDCs (Fig. 2 P). Collectively, these results suggest that there is a dose-dependent effect of FL on pDC development from CLPs.
FL induces IFN-α in CLPs, which promotes FL-dependent pDC development
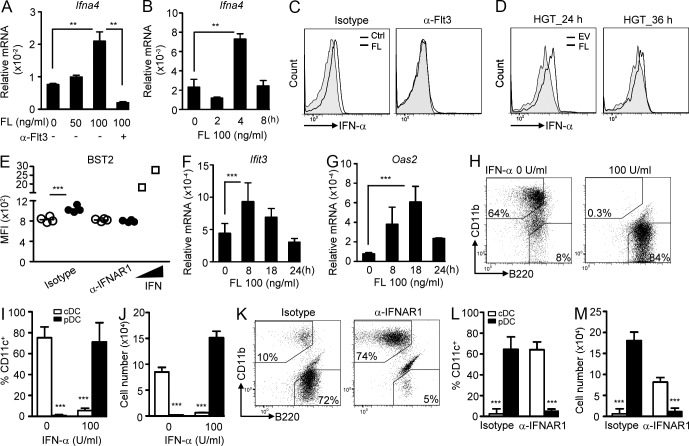
We have demonstrated that IFN-I activity was involved in FL-dependent DC development from CLPs. One possibility is that FL induces IFN-I in CLPs. Indeed, FL induced IFN-α expression and production in CLPs in vitro in a dose- and time-dependent manner, which was blocked by neutralization of Flt3 (Fig. 3, A–C). In vivo FL delivery also induced IFN-α in CLPs (Fig. 3 D). Moreover, IFN-I downstream genes, including BST2, Ifit3, and Oas2 were up-regulated in FL-treated CLPs (Fig. 3, E–G). Given that IFN-α is induced by FL in CLPs, we reasoned that exogenous IFN-I might facilitate pDC generation from CLPs. Indeed, the addition of exogenous IFN-α promoted pDC development even at low-dose FL (50 ng/ml) treatment compared with control (Fig. 3, H–J). Conversely, neutralization of IFNAR1 abrogated pDC formation upon stimulation with 100 ng/ml of FL (Fig. 3, K–M), mimicking the phenotype seen in Ifnar1−/− CLPs under the same culture conditions (Fig. 1 D). Together, these results suggest that FL induces IFN-I, which in turn facilitates pDC generation from CLPs.
Figure 3.
FL induces IFN-α production in CLPs, which promotes FL-dependent pDC development. (A) WT CLPs were stimulated with the indicated doses of FL plus anti-Flt3 or isotype antibody (1 µg/ml each) for 4 h, and expression of Ifna4 was evaluated by RT-qPCR. (B) Same as in A, except CLPs were stimulated with 100 ng/ml of FL for the indicated times (n = 3). One experiment out of three is shown. (C) WT CLPs were stimulated with or without 100 ng/ml of FL plus anti-Flt3 or isotype antibody (1 µg/ml each) for 24 h, and IFN-α expression was evaluated by intracellular staining (n = 3). One experiment out of three is shown. (D) EV or FL-expressing vector was delivered in vivo using the HGT method, followed by sorting out CLPs and intracellular staining for IFN-α (n = 3). One experiment out of three is shown. (E) Same as in C, except anti-IFNAR1 or isotype antibody (1 µg/ml each) was used. The treated cells were stained for BST2. CLPs stimulated with IFN-α4 (125 and 250 U/ml) served as positive controls (n = 4). Results represent three experiments. (F and G) WT CLPs were stimulated with 100 ng/ml of FL for the indicated times, and Ifit3 (F) and Oas2 (G) expression was examined by RT-qPCR (n = 3). One experiment out of three is shown. (H) WT CLPs were co-cultured with AC-6.21 cells and supplied with 50 ng/ml of FL in the presence or absence of IFN-α (100 U/ml). Cells were gated on CD11c+ and analyzed for DC populations. Mean percentages (I) and numbers (J) of DCs are shown (n = 3). Results represent two experiments. (K) Same as in H, except 100 ng/ml of FL and anti-IFNAR1 or isotype antibody were used (1 µg/ml each). Mean percentages (L) and numbers (M) of DCs are shown (n = 5). Results represent three experiments.
FL enhances Flt3 expression on CLPs in an IFN-I–dependent mechanism
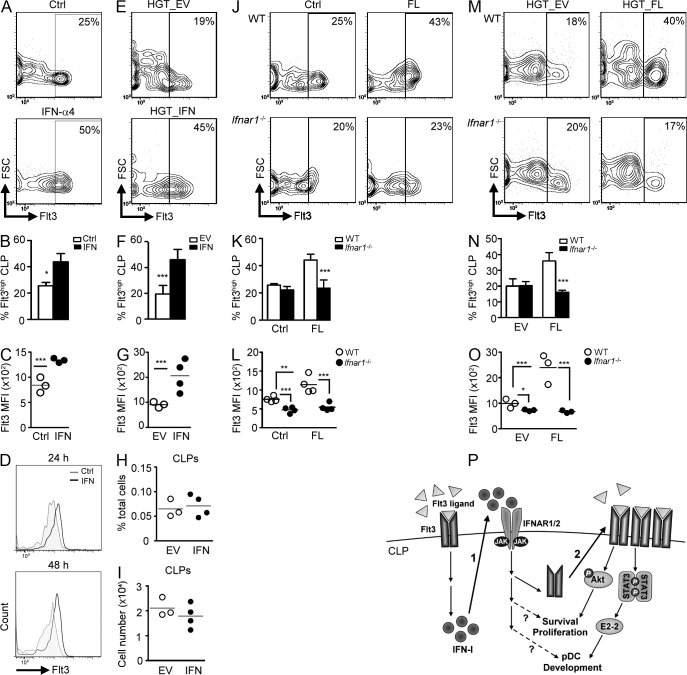
It was proposed that FL induces Flt3 expression on progenitors through up-regulation of PU.1, which in turn controls DC development (Merad, 2010). Because IFN-I accentuated FL-dependent pDC differentiation, we examined whether IFN-I could induce Flt3 expression. IFN-α4 treatment in BM cells significantly augmented Flt3 expression on CLPs (Fig. 4, A–C). The effect of IFN on CLPs was direct, as the same phenomenon was observed when treating CLPs with IFN-α4 (Fig. 4 D). In vivo administration of IFN-α4 also enhanced Flt3 levels on WT CLPs, as opposed to empty vector (EV) control (Fig. 4, E–G). It has been reported that IFN-α can activate dormant HSCs, inducing their proliferation and reestablishing homeostasis (Essers et al., 2009). Although it is still not clear if IFN-α will enhance the production of Flt3+CLPs, in vivo delivery of IFN-α4 did not alter the percentages or numbers of CLPs in the BM, at least, within 24 h (Fig. 4 H-I). The requirement of IFN-I for FL-dependent Flt3 induction was also investigated. In vitro FL treatment markedly enhanced Flt3 expressions on WT CLPs, whereas IFNAR1 deficiency or neutralization of IFNAR1 blocked this process (Fig. 4, J–L, and not depicted). In vivo FL delivery also enhanced Flt3 expression on WT but not on Ifnar1−/− CLPs (Fig. 4, M–O). We next examined the transcriptional levels of Flt3 after stimulation of CLPs with FL or IFN-α4. FL did not alter the mRNA levels of Flt3, whereas IFN-α4 stimulation, surprisingly, reduced the expression in CLPs (not depicted). These results suggest that FL- or IFN-I–stimulated surface Flt3 expression on CLPs is probably independent of transcriptional activity of Flt3. Together, these results suggest that FL induces Flt3 expression on CLPs through an IFN-I–dependent mechanism.
Figure 4.
IFN-I is required for FL-dependent up-regulation of Flt3 on CLPs. (A) BM cells stimulated with or without IFN-α4 (1,000 U/ml) for 24 h and Flt3+ CLPs were enumerated by flow cytometry. Mean percentages of Flt3hi CLPs (B) and MFI of Flt3 on CLPs (C) are shown (n = 3). Results represent three experiments. (D) Same as in A, except sorted CLPs were stimulated for the indicated times. (E) EV or IFN-α4–expressing vector (1 µg each) was delivered in vivo using the HGT method for 24 h, and Flt3 expression on CLPs was evaluated by flow cytometry. Mean percentages of Flt3hi CLPs (F) and MFI of Flt3 on CLPs (G) are shown (n = 3–4). Results represent two experiments. (H and I) Same as in E, except percentages (H) and numbers (I) of CLPs were evaluated by flow cytometry. (J) Same as in A, except BM was stimulated with or without 100 ng/ml of FL. Mean percentages of Flt3hi CLPs (K) and MFI of Flt3 on CLPs (L) are shown (n = 4). Results represent two experiments. (M) Same as in E, except EV or FL-expressing vector (5 µg each) was delivered into mice. Mean percentages of Flt3hi CLPs (N) and MFI of Flt3 on CLPs (O) are shown (n = 3). (Results represent three experiments.) (P) A model for FL/IFN–I/Flt3 axis in promoting pDC development from CLPs. High-dose FL triggers the production of IFN-I (1), which facilitates FL-stimulated up-regulation of Flt3 (2), resulting in increased survival, proliferation, and differentiation into of CLPs. It remains to be determined whether IFN-I is directly involved in these processes.
We have demonstrated here that IFN-I augments FL-dependent pDC development from CLPs through up-regulation of Flt3. Moreover, there is a dose-dependent effect of FL on DC lineage commitment from CLPs. This finding also supports the instructive model of DC differentiation (Schmid et al., 2010). However, this effect seems to be CLP-specific, as CMPs (Sathe et al., 2013) and CDPs (not depicted), two myeloid progenitors with DC potential, still predominantly develop cDCs at high doses of FL. Since STAT3, a downstream signal of Flt3, stimulates the expression of pDC-specific gene Tcf4 (Li et al., 2012), it is tempting to speculate that high-dose FL promotes pDC development from CLPs through increased STAT3 activation and elevated E2-2 expression. Together, we hypothesize a model for the FL–IFN-I–Flt3 axis in promoting pDC development from CLPs (Fig. 4 P).
The effect of IFN-I on DC development, maturation, and function has been controversial (Honda et al., 2003; Hahm et al., 2005; Li et al., 2011; Swiecki et al., 2011). Measles virus and lymphocytic choriomeningitis virus infection–induced IFN-I blocks FL-mediated DC development (Hahm et al., 2005). Moreover, IFN-I also contributes to systemic viral infection–induced decline of pDC numbers (Swiecki et al., 2011). In contrast, IFN-I promotes pDC development in the Peyer’s patches in the gut (Li et al., 2011). Our results also support the beneficial effect of IFN-I for pDC development. Several possibilities may account for these seemingly contradictory results. First, IFN-I may exert differential effects on different subsets of DCs or DC progenitors through altered intracellular signaling networks (Longman et al., 2007). Second, IFN-I may trigger opposite effects in a dose-dependent manner (Dalod et al., 2002; Gautier et al., 2005). FL and IFN-I are both induced by chemo- or radiotherapy, inflammation, and viral infections (Levy et al., 2011; Stirewalt and Radich, 2003). Therefore, the FL–IFN-I–Flt3 axis can function under physiological settings, triggering the amplification of DC compartments, particularly pDCs, thereby enhancing immunity. On the other hand, dysregulated pDC activation and IFN-α production is strongly linked to the development of systemic lupus erythematosus and psoriasis (Gilliet et al., 2008). Therefore, targeting the production or actions of IFN-I may provide a therapeutic approach to reduce pDC numbers and ameliorate the autoimmune diseases.
MATERIALS AND METHODS
Mice.
All mice used in this study were from the C57BL/6 background. Mice were bred and kept under specific pathogen–free conditions. C57BL/6 (CD45.2) wild-type mice were purchased from the National Laboratory Animal Center, Taiwan. Ifnar1−/− mice (provided by M. Karin, University of California, San Diego, CA, through G.-Y. Yu at National Health Research Institute, Taipei, Taiwan), which have been backcrossed to C57BL/6 for at least six generations. C57BL/6-Thy1.1 (CD45.1) congenic mice and CD45.1xCD45.2 mice were rederived and bred at the Laboratory Animal Center of National Taiwan University College of Medicine. Procedures and the use of the animals were reviewed and approved by the Institutional Animal Care and Use Committee.
Antibodies and flow cytometry.
Staining was performed at 4°C with Fc Block (hybridoma 2.4G2) in FACS staining buffer (0.5% FBS and 0.1% NaN3 in PBS). The following antibodies were purchased from eBioscience: biotin-anti-CD11c (N418), PE-anti-CD11b (M1/70), PE-anti-CD3 (17A2), PE-anti-CD8 (53–6.7), PE-anti–Gr-1 (RB6-8C5), PE-anti-CD19 (eBio1D3), PE-anti-TER119 (2E2), PE-anti-MHCII (NIMR-4), PE-anti-Thy1.1 (HIS51), PE-anti-NK1.1 (PK136), APC-anti-B220 (RA3-6B2), PerCP-eFluor710-anti-Siglec-H (eBio440c), PerCP-eFluor710-anti–c-Kit (2B8), APC-anti–M-CSFR (AFS98), PE/Cy7-anti–IL-7Rα (A7R34), biotin-anti-Flt3 (A2F10), FITC-anti–Sca-1 (D7), APC-Cy7-Streptavidin, FITC-anti-BrdU, and anti-IFNAR1 (MAR1-5A3). Purified anti-Flt3 (A2F10), anti-BST2 (927), and FITC-Annexin V were purchased from BioLegend. FITC-anti–IFN-α (RMMA-1) was purchased from PBL Biomedical Laboratories. Flow data were analyzed with FlowJo (Tree Star) or Kaluza (Beckman Coulter) software.
Progenitor analysis and cell sorting.
After lysis of RBCs, BM cells were stained with PE-conjugated lineage markers, including anti-CD3, anti-CD8, anti-B220, anti-CD19, anti-CD11b, anti–Gr-1, anti-TER119, anti-Thy1.1, anti-NK1.1, and anti-MHC II. CLPs defined as lin−c-kitintSca-1intM-CSFR−IL-7Rα+ (Kondo et al., 1997), were analyzed with the BD FACSCanto II or sorted with the BD FACSAria III.
In vitro DC development.
In vitro DC development was performed as previously described (Chicha et al., 2004; Naik et al., 2005) with minor modifications. For AC-6.21 (Whitlock et al., 1987) feeder system, AC-6.21 (kindly provided by I. Weissman, Stanford University, Stanford, CA) were seeded at 5.9 × 104 into 12-well plates 1 d before to reach a confluency of 80%, followed by γ-irradiation with 3,000 rad (30 Gy). Sorted CLPs at 500 cells/well were co-cultured with the AC-6.21 cells supplied with the indicated doses of huFL-Ig for 20 d. FL was produced in-house using the FL expression system provided by M. Manz (University Hospital Zürich, Zürich, Switzerland; Onai et al., 2006). Progeny cells from AC-6.21 feeder system were stained with antibodies to CD11c, CD11b, and B220. cDCs and pDCs are CD11c+CD11b+B220− and CD11c+CD11b−B220+, respectively. For IFN-α treatment, different doses of mouse IFN-α (Merck) or IFN-α4 (produced in-house) were added into the co-culture system supplied with 50 ng/ml of FL. For antibody-blocking experiments, the indicated doses of anti-IFNAR1 (MAR1-5A3; eBioscience), anti-Flt3 (A2F10, eBioscience), or their isotype antibodies was added.
Competitive adoptive transfer.
10,000 CLPs sorted from C57BL/6 (CD45.1) and C57BL/6 (CD45.2)-Ifnar1−/− mice were co-transferred intravenously with 2 × 105 C57BL/6 (CD45.1xCD45.2) BM cells into C57BL/6 (CD45.1xCD45.2) mice that had been lethally irradiated with 600 rad (6 Gy) twice at 4-h intervals. 12 d later, BM cells were stained and gated on the CLP-derived cells (CD45.1+ or CD45.2+) and analyzed for pDCs (CD11cintCD11b−B220+Siglec-H+) or cDC (CD11c+CD11b+B220−).
Apoptosis and proliferation assays.
For apoptosis assay, sorted CLPs co-cultured with AC-6.21 were treated with 100 ng/ml FL for the indicated times, followed by staining with FITC-Annexin V (BioLegend) according to the manufacturer’s instructions. For proliferation assay, WT BM cells (CD45.1) and Ifnar1−/− (CD45.2) were mixed first, and then stained with 5 µM CFSE (Invitrogen), followed by sorting out CFSEhi CLPs, co-culturing with AC-6.21, and stimulation with 100 ng/ml of FL. The cells were collected and analyzed with flow cytometry at the indicated times.
Hydrodynamic gene transfer (HGT).
Mice were injected intravenously via tail veins with plasmid DNA in 10% body-weight sterile PBS. DNA injection was completed within 5 s. For IFN-α4 treatment, 0.25–1 µg pcDNA3.1 EV or the vector expressing mIFN-α4 (provided by L.-H. Hwang, National Yang-Ming University, Taipei, Taiwan) was injected. For FL treatment, 2.5–5 µg of pcDNA3 EV or the vector expressing mFL (provided by T.-C. Wu, Johns Hopkins University, Baltimore, MD) was injected.
Intracellular staining.
For intracellular pAkt staining, BM cells treated with FL were fixed with 1% paraformaldehyde for 10 min, followed by permeabilization with 90% methanol for 30 min at room temperature. After the removal of methanol, the cells were resuspended and stained with antibodies to lineage markers, Flt3, and pAkt (pSer473; Cell Signaling Technology). F(ab’)2 anti–rabbit IgG-FITC (eBioscience) was used for secondary labeling. For intracellular IFN-α staining, 3–5 × 104 sorted CLPs were stimulated with FL in vitro for 20 h. Monensin (2 µM, eBioscience) was added into the culture for an additional 4 h before performing intracellular staining. For in vivo treatment, CLPs were sorted directly into 1% paraformaldehyde after HGT with mFL expressing plasmid for the indicated times. The treated cells were fixed in BD Cytofix/Cytoperm solution. Staining was performed in BD Perm/Wash buffer using FITC anti–mouse IFN-α antibody (RMMA-1; PBL Biomedical Laboratories).
RT-qPCR.
For gene expression analysis, sorted CLPs (5 × 104/treatment) were seeded onto AC-6.21 feeder 1 d before the treatments. mRNA of the treated CLPs was prepared with TurboCapture kit (QIAGEN), followed by cDNA synthesis via HiScript I reverse transcription (BIONOVAS) using random primer. The cDNA was then subjected to RT-qPCR using primers to Ifna4, Ifit3, Oas2, and Actb. 50,000 splenic cDCs and pDCs or CLP-derived cDCs and pDCs using AC-6.21 feeder co-culture system were sorted and subjected to RT-qPCR for Tcf4, Rag1, Id2, and Actb. Data were normalized to Actb. All the primer sequences are listed in Table S1.
Retroviral transduction.
Retroviral transduction of CLPs was performed as previously described (Onai et al., 2006). In brief, MigR1 or MigR1-mFlt3 (provided by S. Nutt, Walter and Eliza Hall Institute of Medical Research, Victoria, Australia) plasmids were co-transfected with two plasmids expressing retroviral gag-pol and env, respectively, into 293T cells using the calcium phosphate precipitation method. The viral supernatants were collected 2 d later, concentrated, and then spin-infected GP+E86 cells in DMEM containing 10% FBS and 8 µg/ml polybrene (Sigma-Aldrich) at 1,100 g for 50 min. After viral transduction for 48 h, GFPhiGP+E86 cells were sorted. For infection of CLPs, sorted WT and Ifnar1−/− CLPs were co-cultured with γ-irradiated (2,000 rad) GFPhiGP+E86 cells (1.5 × 105 cells/well of 24-well plate) for 18 h in the presence of 4 µg/ml polybrene and 100 ng/ml FL. Transduced CLPs were subjected to in vitro AC-6.21 feeder co-culture system as described in the In vitro DC development section.
Statistical analyses.
All values are shown as mean ± SD. Two-tailed, unpaired Student’s t test was used to assess statistical significance: *, P < 0.05; **, P < 0.01; and ***, P < 0.005.
Supplementary Material
Supplemental Material
Acknowledgments
We are grateful to Drs. Stephen Nutt, Markus Manz, Irving Weissman, Tzyy-Choou Wu, Lih-Hwa Hwang, and Chia-Chi Ku for providing reagents; and David Levy, Betty Wu-Hsieh, and John Kung for reading and/or helpful discussions of the manuscript. We also acknowledge the service provided by the Cell Sorting Core Facility of the First Core Laboratory, National Taiwan University College of Medicine.
This work was supported by the National Science Council, Taiwan (NSC 99-2320-B-002-010-MY3) and the National Health Research Institutes, Taiwan (NHRI-EX102-10256SI).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
cDC
conventional DC
CDP
common dendritic progenitor/common DC precursor
CLP
common lymphoid progenitor
CMP
common myeloid progenitor
EV
empty vector
FL
huFlt3 ligand-Ig
HGT
hydrodynamic gene transfer
IFNAR1
type I IFN receptor 1
IFN-I
type I IFN
lin−
lineage negative
pDC
plasmacytoid dendritic cell
References
- Asselin-Paturel C., Brizard G., Chemin K., Boonstra A., O’Garra A., Vicari A., Trinchieri G. 2005. Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J. Exp. Med. 201:1157–1167 10.1084/jem.20041930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.S., Wei P.C., Liu T., Kao C.H., Pai L.M., Lee C.K. 2009. STAT2 hypomorphic mutant mice display impaired dendritic cell development and antiviral response. J. Biomed. Sci. 16:22 10.1186/1423-0127-16-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicha L., Jarrossay D., Manz M.G. 2004. Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J. Exp. Med. 200:1519–1524 10.1084/jem.20040809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico A., Wu L. 2003. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J. Exp. Med. 198:293–303 10.1084/jem.20030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalod M., Salazar-Mather T.P., Malmgaard L., Lewis C., Asselin-Paturel C., Brière F., Trinchieri G., Biron C.A. 2002. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J. Exp. Med. 195:517–528 10.1084/jem.20011672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Gautier G., Humbert M., Deauvieau F., Scuiller M., Hiscott J., Bates E.E., Trinchieri G., Caux C., Garrone P. 2005. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J. Exp. Med. 201:1435–1446 10.1084/jem.20041964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliet M., Cao W., Liu Y.J. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8:594–606 10.1038/nri2358 [DOI] [PubMed] [Google Scholar]
- Hahm B., Trifilo M.J., Zuniga E.I., Oldstone M.B. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity. 22:247–257 10.1016/j.immuni.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Honda K., Sakaguchi S., Nakajima C., Watanabe A., Yanai H., Matsumoto M., Ohteki T., Kaisho T., Takaoka A., Akira S., et al. 2003. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc. Natl. Acad. Sci. USA. 100:10872–10877 10.1073/pnas.1934678100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky H., Merad M., Cozzio A., Weissman I.L., Manz M.G. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313 10.1084/jem.20030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Levy D.E., Marié I.J., Durbin J.E. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol. 1:476–486 10.1016/j.coviro.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.S., Gelbard A., Martinez G.J., Esashi E., Zhang H., Nguyen-Jackson H., Liu Y.J., Overwijk W.W., Watowich S.S. 2011. Cell-intrinsic role for IFN-α-STAT1 signals in regulating murine Peyer patch plasmacytoid dendritic cells and conditioning an inflammatory response. Blood. 118:3879–3889 10.1182/blood-2011-04-349761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.S., Yang C.Y., Nallaparaju K.C., Zhang H., Liu Y.J., Goldrath A.W., Watowich S.S. 2012. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 120:4363–4373 10.1182/blood-2012-07-441311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275–306 10.1146/annurev.immunol.23.021704.115633 [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397 10.1126/science.1171243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman R.S., Braun D., Pellegrini S., Rice C.M., Darnell R.B., Albert M.L. 2007. Dendritic-cell maturation alters intracellular signaling networks, enabling differential effects of IFN-alpha/beta on antigen cross-presentation. Blood. 109:1113–1122 10.1182/blood-2006-05-023465 [DOI] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497 [PubMed] [Google Scholar]
- Merad M. 2010. PU.1 takes control of the dendritic cell lineage. Immunity. 32:583–585 10.1016/j.immuni.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Proietto A.I., Wilson N.S., Dakic A., Schnorrer P., Fuchsberger M., Lahoud M.H., O’Keeffe M., Shao Q.X., Chen W.F., et al. 2005. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J. Immunol. 174:6592–6597 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Tussiwand R., Lanzavecchia A., Manz M.G. 2006. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J. Exp. Med. 203:227–238 10.1084/jem.20051645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Sathe P., Vremec D., Wu L., Corcoran L., Shortman K. 2013. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood. 121:11–19 10.1182/blood-2012-02-413336 [DOI] [PubMed] [Google Scholar]
- Schmid M.A., Kingston D., Boddupalli S., Manz M.G. 2010. Instructive cytokine signals in dendritic cell lineage commitment. Immunol. Rev. 234:32–44 10.1111/j.0105-2896.2009.00877.x [DOI] [PubMed] [Google Scholar]
- Shigematsu H., Reizis B., Iwasaki H., Mizuno S., Hu D., Traver D., Leder P., Sakaguchi N., Akashi K. 2004. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 21:43–53 10.1016/j.immuni.2004.06.011 [DOI] [PubMed] [Google Scholar]
- Stirewalt D.L., Radich J.P. 2003. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer. 3:650–665 10.1038/nrc1169 [DOI] [PubMed] [Google Scholar]
- Swiecki M., Wang Y., Vermi W., Gilfillan S., Schreiber R.D., Colonna M. 2011. Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J. Exp. Med. 208:2367–2374 10.1084/jem.20110654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., Nussenzweig M. 2008. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9:676–683 10.1038/ni.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock C.A., Tidmarsh G.F., Muller-Sieburg C., Weissman I.L. 1987. Bone marrow stromal cell lines with lymphopoietic activity express high levels of a pre-B neoplasia-associated molecule. Cell. 48:1009–1021 10.1016/0092-8674(87)90709-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material