Temperature dependence of metabolic rates for microbial growth, maintenance, and survival (original) (raw)
Abstract
Our work was motivated by discoveries of prokaryotic communities that survive with little nutrient in ice and permafrost, with implications for past or present microbial life in Martian permafrost and Europan ice. We compared the temperature dependence of metabolic rates of microbial communities in permafrost, ice, snow, clouds, oceans, lakes, marine and freshwater sediments, and subsurface aquifer sediments. Metabolic rates per cell fall into three groupings: (i) a rate, μg(T), for growth, measured in the laboratory at in situ temperatures with minimal disturbance of the medium; (ii) a rate, μm(T), sufficient for maintenance of functions but for a nutrient level too low for growth; and (iii) a rate, μs(T), for survival of communities imprisoned in deep glacial ice, subsurface sediment, or ocean sediment, in which they can repair macromolecular damage but are probably largely dormant. The three groups have metabolic rates consistent with a single activation energy of ≈110 kJ and that scale as μg(T):μm(T):μs(T) ≈ 106:103:1. There is no evidence of a minimum temperature for metabolism. The rate at -40°C in ice corresponds to ≈10 turnovers of cellular carbon per billion years. Microbes in ice and permafrost have metabolic rates similar to those in water, soil, and sediment at the same temperature. This finding supports the view that, far below the freezing point, liquid water inside ice and permafrost is available for metabolism. The rate μs(T) for repairing molecular damage by means of DNA-repair enzymes and protein-repair enzymes such as methyltransferase is found to be comparable to the rate of spontaneous molecular damage.
There is now both direct and indirect evidence that microorganisms live inside glacial and sea ice and permafrost at temperatures well below the freezing point of pure water (1-11). The thermodynamic stability of ion-rich liquid veins at triple grain-boundaries in polycrystalline ice (12) and of thin films of unfrozen water on microbial surfaces in permafrost (13, 14) enables transport of nutrients to and waste from them. Certain impurities such as mineral acids or salts can reduce the freezing point of water in narrow intergranular veins to as low as -90°C. Acidophilic psychrophiles in a Greenland mine (15) survive the cold winters at -30°C, probably by taking advantage of such a habitat. It has been conjectured that microbial life may have arisen under similar conditions in subsurface Martian permafrost or in warm diapirs in Europa's ice.
After some brief remarks about metabolism in a natural microbial community, we cite several examples of microbes in ice or permafrost at subfreezing temperatures that we cannot analyze quantitatively, either because information on both microbial concentration and metabolic rate in the same locations was not provided or because only isolates in pure cultures were studied. We then discuss methods and present quantitative data from the literature and from our unpublished work that enable us to calculate metabolic rates per cell. We display the results in Fig. 1 and, finally, discuss the implications.
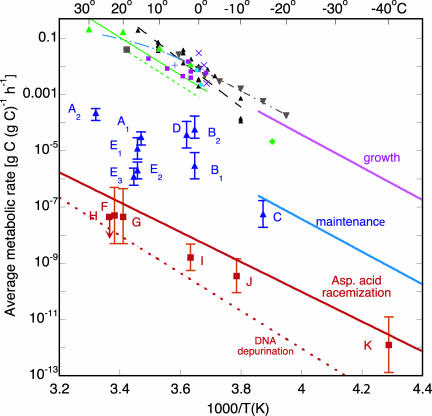
Fig. 1.
Comparison of metabolic rates for prokaryotes as a function of temperature in various ecosystems (see text). In the top group, labeled “growth,” lines join measurements by the same investigator at different temperatures: filled black triangles and dashed line, permafrost (27); filled brown inverted triangles and dot-dash line, permafrost (26); dashed triple-dot line, lake (31); solid green line, marine bacteria (33); short-dash green line, freshwater bacteria (33). Symbols without lines are labeled as follows: purple X, polar ocean and ice (35, 36, 38); +, permanently ice-covered Antarctic lake (39); ♦, South Pole ice and snow (28); •, cloud (29); filled brown squares, ocean (32); filled green triangles, salt-marsh tidal creek (37); and filled purple squares, marine sediment (30, 34). To avoid clutter, error bars are not shown. Points labeled with letters in the middle and bottom groups are keyed to paragraphs in the text. Lines at the bottom are extrapolated from rates of racemization of aspartic acid (solid line, ref. 55) and rates of DNA depurination (dashed line, ref. 56), both measured at high temperature.
In a culture containing unlimited nutrients, energy sources, room for growth, and no predators, some microbial species can double in about an hour at room temperature. However, low growth rate, or intermittent periods of rapid growth rate interspersed with prolonged periods of nongrowth and starvation, is the norm for microorganisms in nature, for which the mean doubling time can be as long as millennia. Microbes grow until they deplete the supply of nutrient or accumulate toxic byproducts of metabolism; then they adopt defensive measures. Some engage in cannibalism or develop ultramicrocells; some synthesize stress proteins or express starvation genes; some become dormant; some form spores. To survive at low temperature, microbes can reduce their cell size; reduce their capsular polysaccharide coat thickness; change their fatty acid and phospholipid composition; decrease the fractional volume of cellular water; increase the fraction of ordered cellular water; or extract energy by catalyzing redox reactions of ions in aqueous veins in ice or in thin aqueous films in permafrost.
- Abyzov et al. (1) filtered melt water from a section of an ice core from depths 1665 to 2750 m at Vostok Station (Antarctica), counted stained cells, incubated with 14C-labeled protein hydrolysate in the dark at 12°C and 18°C for 4-40 h, and measured the activities. Incubating for 4-7 h was sufficient for cell reactivation and consumption of labeled carbon whereas an 18- to 40-h incubation did not result in an increase in labeled carbon. They concluded that the cells were able to maintain vital functions but were not able to reproduce. We excluded these data because the incubations were not at in situ temperature and because they did not measure cell size.
- Priscu et al. (2) reported 103 to 3 × 104 microbial cells per milliliter in samples of accretion ice (refrozen from subglacial Lake Vostok water below the overlying glacial ice) at a depth of 3,590 m in an ice core from Vostok Station, Antarctica; and Karl et al. (3) detected 200 to 300 per milliliter at 3,603 m. The high concentration was in a region containing rock fragments, and the low concentration was in ice without fragments.
- Willerslev et al. (4) identified 57 distinct taxa of eukaryotes from the Hans Tausen ice core, northern Greenland; Gruber and Jaenicke (5) estimated a concentration of ≈3,000 per milliliter in the same core; and Ma et al. (6) identified fungi from the GISP2 (Greenland Ice Sheet Project) ice core.
- Christner et al. (7) extracted bacteria from cores in glacial ice from Greenland, Antarctica, Tibet, and Bolivia. They were successful in cultivating only when it took place in a low-nutrient medium over a long time, from which they inferred that cells in low-nutrient media have sufficient time for cell repair before growth begins, whereas cells in rich-nutrient media attempt to grow before they have repaired all of the accumulated cell damage.
- Junge et al. (8) studied prokaryotes inside brine channels in Arctic sea ice with staining and epifluorescence microscopy on a cold stage at -2°C to -20°C. A large fraction of the microbes were attached to particles or to ice-crystal boundaries and were actively respiring even at -20°C.
- Gilichinsky (16) reviewed research on viable microbes in permafrost at depths corresponding to ages of up to several million years. As indirect evidence of active metabolism, he cited the ability of immobilized enzymes in permafrost to become instantly activated in the laboratory; the simultaneous presence of nitrifying bacteria and nitrites and of methanogenic archaea and methane; and microcalorimetry experiments. As we will see later, this list can be expanded to include, for polar ice, the presence of N2O, CH4, and CO2 in concentrations that correlate quantitatively with microbial concentration as a function of depth. Using a culture medium containing a cryoprotectant, Gilichinsky et al. (17, 18) observed cell growth at temperatures as low as -5°C after 6 months and at -10°C after 1.5 yr. Gilichinsky et al. (19) detected assimilation of [14C]glucose at temperatures as low as -15°C by bacteria in cryopegs (brine lenses) found in Siberian permafrost. They concluded that psychrophiles survive at -15°C in cryopegs, probably repairing cell membranes and DNA damage resulting from background soil radiation and maintaining other vital functions, but without being able to reproduce.
- Using 14C-carbonate and 14C-acetate, Rivkina et al. (20) studied evolution of methane by a community of methanogenic archaea from permafrost at temperatures from5°C down to -16.5°C.
- Bakermans et al. (21) studied reproduction and metabolism of a number of species of bacteria isolated from Siberian permafrost. One psychrophilic isolate grew at -10°C at a specific rate of 7 × 10-4 per hour, as measured by culture turbidity and plate counts. The others showed the ability to reduce blue resazurin to pink resorufin but grew at a rate that was too small to measure with their techniques.
- Kappen et al. (22) detected CO2 exchange by polar lichens at -20°C and found that photosynthetic carbon production and respiratory loss were measurable at -18°C to -12°C for certain species. The lichens recovered after having been stored 10 yr at -20°C.
- Johnston and Vestal (23) estimated average turnover rates of ≈4 × 10-9 per hour for photosynthetic carbon incorporation in Antarctic cryptoendolithic microbial communities at ambient temperatures that were subzero for most of the year. The annual temperature cycle of these communities varies so much with sun angle that we are unable to compute metabolic rate as a function of temperature.
Methods
To determine growth metabolism, we used data for communities, not isolates, whose growth was measured in time-course observations at in situ pressures and temperatures. Isolates in pure cultures are usually not representative of community behavior: they have bell-shaped growth curves with lowest, optimal, and highest temperature that depend on taxa, and they are not challenged by the factors that would be present in the natural environment. Methods for studying metabolism and growth include
- incorporation of [3H]thymidine and [3H]leucine ([3H]Leu) into nucleic acids and proteins, respectively. When data with both methods were available, we used the thymidine data.
- [2-3H]adenine incorporation, which measures RNA and DNA synthesis.
- [2-14C]acetate or other labeled organic C for incorporation into glycolipids and for 14CO2 respiration.
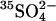 assimilation as a measure of total microbial community protein mass.
assimilation as a measure of total microbial community protein mass.- fluorescently labeled oligonucleotide probes of quantity of rRNA and DNA.
- growth of turbidity of the medium or direct counts of dividing cells.
An exponential increase in the measured quantity with time is often used as evidence that growth is being measured.
To determine maintenance metabolism, i.e., metabolism for energy exclusive of growth, is a more difficult task. When activities of natural communities of bacteria are measured in terms of quantities such as oxygen consumption, sulfate respiration, or electron transport system activity, respiration independent of growth is measured (24). Radiolabeling can be used to detect respiration in which the labeled substrate is excreted to 14CO2, 14CH4, or 35SH2. A commonly used technique involves feeding a radiolabeled nutrient in an amount just sufficient to incorporate radioactivity at a detectable level but insufficient for growth. In some experiments, too rich a nutrient was used, which led to growth. We excluded such data as well as batch culture and chemostat data. We selected published experiments in which substrate had not been added and the observed quantity grew linearly, not exponentially.
The geochemical technique is a powerful method for measuring metabolism in an environment so hostile that rates are far too low to be detected on a laboratory timescale. This method requires that a microbial community has existed for a known time interval in a closed system and that the waste products of its metabolism, usually gaseous, are above a background level due to abiotic sources. This method is so sensitive that it can measure survival metabolism of a dormant community. The production rate μs of excess gas of type j is given by
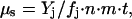 |
1 |
|---|
where _Y_j is the yield of biogenic gas of type j; _f_j is the fraction of reactions going to gas of type j; n is the concentration of microbes; m is the mean mass per microbe; and t is the age of a closed system of ice or permafrost containing the microbes. In all geochemical cases that we analyzed, the products were the gases CO2, CH4, and N2O, which were measured with standard gas chromatographic techniques. Note that the geochemical method measures average rate: it does not take into account changes in the nutrient supply with time or changes in physiological state of the microbial community. We use the symbol μs, with the subscript “s” for survival, to suggest that metabolism measured in this way may be different from that measured in time-course experiments.
For growth metabolism, maintenance metabolism, and survival metabolism, when information on average carbon mass per cell was not provided, or when we could not calculate mass from electron micrographs, we used the values 10 fg for aquatic sediments, 20 fg for planktonic prokaryotes, 65 fg for marine sediments, 100 fg for soil, and 86 fg for a terrestrial aquifer, recommended in ref. 25.
Cell concentrations were determined by direct counts on stained cells rather than by cultivation. As a consequence, the metabolic rates represent averages over active, dormant, and even some dead cells.
Data
Fig. 1 summarizes the results of our study. Points labeled with the symbols given in the legend show carbon-specific growth rates μg in grams of carbon incorporated into cell material per gram of total biomass carbon per hour. The rates are for communities taken from permafrost, sea ice, snow, cloud, oceans, lakes, fresh and marine waters, and sediments, and measured in the laboratory under in situ conditions. Those data fall on a band near the top of the figure. To reduce clutter, several data sets with the greatest number of points were fitted with smooth curves without symbols, as indicated in the legend.
Points labeled A through E in blue show carbon-specific maintenance metabolic rates, μm, for communities taken from glacial ice, Antarctic lake ice, soil, and marine sediment, and measured in the laboratory.
Points labeled F through K in red show rates, μs, for in situ geochemical data for immobile communities in unperturbed closed systems: glacial ice, ocean sediments, and deep subsurface sandy and clayey aquifer sediments. The participating carbon is that converted into CO2, CH4, 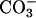 , or other waste.
, or other waste.
A brief description of the growth data and nongrowth data follows.
- Rivkina et al. (26) monitored the incorporation of 14C-labeled acetate into glycolipids of a bacterial community from Siberian permafrost at seven temperatures from -5°C to -20°C for times up to 550 days. The brown dot-dashed curve near the top of Fig. 1 is a fit to their data for the exponential growth phase of those organisms that took up acetate (4). The nutrient uptake rate in the exponential phase reflects the growth potential under ideal conditions, which do not exist in nature, because of limited nutrient availability and limited rate of transport through the thin water film that exists between soil grains and ice in permafrost at low temperature.
- Jakosky et al. (27) monitored turbidity of the medium as a measure of growth rates of Siberian permafrost bacteria at temperatures from 16°C to -10°C. The black long-dashed curve is an exponential fit to their data.
- Carpenter et al. (28) found concentrations of 200 to 5,000 bacterial cells per milliliter in surface snow and firn at the South Pole; 10-20% of them were members of the Deinococcus-Thermus group. From their scanning electron microscopy photos, we estimated a mass of 160 fg per cell. They measured uptake rates of [3H]thymidine and [3H]Leu by the community at -17°C, the snow temperature at the time of their sample collection. Their result for [3H]thymidine is plotted.
- Sattler et al. (29) used [3H]thymidine and [14C]Leu uptake rates at 0°C to measure cell multiplication rates and protein synthesis by bacteria in cloud droplets and snow. We plotted their [3H]thymidine results. They suggested that airborne bacteria in clouds grow and divide at ambient air temperatures as low as -9°C and that there is sufficient dissolved organic carbon in cloud droplets to support bacterial growth.
- Sulfate reducers active in polar sediments can metabolize at low temperatures. Using
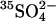 tracer together with lactate or acetate in anoxic containers, Knoblauch et al. (30) calculated specific growth rates for a community of sulfate reducers from marine sediments off the coast of Svalbard. Assuming oxidation of 2 moles organic carbon per mole of reduced sulfate, we converted their rates to carbon-specific growth rates for psychrophiles at -1.8°C and 2.6°C and included data for mesophiles from temperate sediments at 4°, 8°C, and 13°C.
tracer together with lactate or acetate in anoxic containers, Knoblauch et al. (30) calculated specific growth rates for a community of sulfate reducers from marine sediments off the coast of Svalbard. Assuming oxidation of 2 moles organic carbon per mole of reduced sulfate, we converted their rates to carbon-specific growth rates for psychrophiles at -1.8°C and 2.6°C and included data for mesophiles from temperate sediments at 4°, 8°C, and 13°C. - Coveney and Wetzel (31) assayed specific growth rates of bacterioplankton as a function of temperature in oligotrophic Lawrence Lake, Michigan, using [3H]thymidine. The blue triple-dot curve shows the rates they found for exponential growth with unlimited nutrient availability. They inferred lower rates for in situ growth under conditions when C, N, and P were limiting.
- Simon and Azam (32) used [3H]thymidine and [3H]Leu to measure specific growth rates in the exponential phase for marine bacterioplankton from the Southern California Bight at 19°C.
- White et al. (33) analyzed published pelagic bacterial growth rate data from 57 studies that had been conducted in fresh, marine, and estuarine/coastal waters mainly with [3H]thymidine. They determined best-fit equations for specific growth rate as a function of temperature, algal chlorophyll-a concentration, and cell size. Our green short-dashed curve shows the dependence on in situ temperature from 2-20°C for freshwater bacteria, with cell size = 0.083 μm3. Our green solid curve shows the temperature dependence for marine bacteria, with chlorophyll-a = 62 μg·liter-1 (nutrient-unlimited).
- Novitsky (34) added [2-3H]adenine to sediment slurries taken at 1-cm intervals at six depths from the water-sediment interface to a depth of 5 cm from a coastal marine sediment in Halifax Harbor and measured specific growth rates of the aerobic microbial communities at the in situ temperature 2°C. The average growth rates for the interface and for the depth interval 0-5 cm are plotted.
- Bird and Karl (35) used [3H]Leu to measure the exponential growth rate of bacteria from shallow water near the Antarctic peninsula, at a temperature of ≈0°C. They filtered out grazers and phytoplankton and obtained rates with and without amino acid amendments.
- Smith and Clement (36) used [3H]thymidine augmented with glucose to study exponential growth rate of bacteria from the bottom few centimeters of sea ice in the Canadian Arctic, at the in situ temperature of -1.5°C.
- Shiah and Ducklow (37), using [3H]thymidine, found that the specific growth rate of heterotrophic bacterioplankton in a salt-marsh tidal creek near Chesapeake Bay depended strongly on water temperature. Results at four in situ temperatures from 2-30°C are plotted.
- Kottmeier et al. (38), using [3H]thymidine, measured specific growth rate of bacteria in sea ice in McMurdo Sound, Antarctica, and of bacterioplankton in the Southern Ocean at a temperature of ≈-1.9°C.
- Ward and Priscu (39) used turbidity measurements to study growth of denitrifying bacteria from various depths in the permanently ice-covered Lake Bonney, Antarctica, at in situ temperatures and salinities of -0.5°C to 6°C and 60-99 g/liter.
- Anderson and Domsch (40) determined maintenance carbon requirements of the microbial communities in “starving” agricultural and forest soils. They measured the amount of carbon required to keep a community at a constant maintenance level, with no net microbial carbon gain or loss. They analyzed the CO2 output of 100 g of soil in a closed container to which glucose had been added. Point A1 was an average metabolic rate for three soil types after incubation for 2 weeks at 15°C, and point A2 was for 1 week at 28°C. The rates were lower by a factor ≈103 than the glucose consumption rate in pure culture where growth was allowed.
- In sealed anoxic containers at 1°C, Franzmann et al. (41) measured 14CH4 yields resulting from uptake of NaH[14C]O3, [14C]formate, and [2-14C]acetate by microbes that were living at 1°C in Ace Lake, Antarctica. We calculated the microbial concentrations from data of Mancuso et al. (42), who measured the composition of phospholipids as a function of depth in water-column and sediment samples. Franzmann et al. concluded that methanogenesis occurs both in the sediments and in the water column. Point B1 is our estimate of the maintenance metabolic rate in the sediment for methanogenesis from acetate. Point B2 is the maximum maintenance rate they found, corresponding to methanogenesis from CO2 at a 20-m water depth.
- Christner (43) studied the metabolism of psychrotrophs from polar ice cores. He exposed them to [3H]thymidine and [3H]Leu for 100 days at -15°C. Taking into account the high concentration (≈107 per milliliter), the genome size (3 Mbp), and average protein size (36 kDa), he inferred that <1% of the genome had been replicated and only ≈100 protein molecules had been synthesized per cell. He concluded that bacteria can synthesize macromolecules in ice at -15°C at a rate that is very low but sufficient to repair macromolecular damage. From his data, we infer that the rate of cell reproduction was too slow for the cells to have gone beyond the lag phase into an exponential growth phase. He conjectured that cells remain metabolically active in solute-enriched water films on the surfaces of mineral grains in ice or in liquid veins at triple junctions (12) in ice.
- Karl et al. (3) studied metabolism in accretion ice from subglacial Lake Vostok. Incubating the ice melts with 14C-labeled acetate and glucose for up to 18 days at 3°C (slightly above the in situ temperature) in the dark led to production of 14CO2, which indicated respiration by metabolizing cells. The rate for acetate respiration (point D) was ≈800 times higher than for glucose respiration. Rates of 14C incorporation into nucleic acids and protein were not above background rates.
- Parkes et al. (44) determined anaerobic respiration rates of bacteria in marine sediment from the Peru Margin at 16°C, using [35S]sulfate and [14C]bicarbonate to measure rates of sulfate reduction and methanogenesis. Points E1, E2, and E3 show the rates for depths of 0.12—3.5 m, 8—40 m, and 90 m, respectively. The metabolic rate decreases with depth due to nutrient deprivation.
- Chapelle and Lovley (45) measured metabolism in deep aquifers in the South Carolina coastal plain where aerobic and anaerobic reactions, mostly leading to CO2 production, take place in different regions. From measurements of concentrations of dissolved organic carbon as a function of distance along the flowpaths of each aquifer, they estimated a production rate of ≈10-2 μmol of CO2/liter per year by a community of 3 × 106 bacteria per gram of sediment during 80 million years, with an uncertainty of a factor 10.
- Phelps et al. (46) studied microbial respiration in the same aquifers. They extracted groundwater from boreholes at five points along the flowpath of the Middendorf aquifer at depths 250-400 m, at temperatures of 23-25°C, in complete isolation from atmospheric CO2. Dissolved oxygen concentration along the flow path decreased from near saturation in the recharge zone to anaerobic 80 km downstream at the last borehole. The rate of microbial respiration was calculated from groundwater chemistry, cell counts, and groundwater age determined from the 14C activity of the dissolved inorganic carbon. Comparing metabolic rates inferred from geochemical methods and from radioisotope time-course experiments, they concluded that radiotracer results overestimated in situ rates by factors 103 to 106. They concluded that radioisotope methods are unsuitable for estimating in situ metabolic rates in the deep subsurface, mainly because of the much lower specific activity, but perhaps also because of failure to reproduce the in situ distribution of nutrients and various microorganisms in the radiolabeling experiments.
- Both Phelps et al. (46) and Chapelle and Lovley (45) found microbial concentrations to be a factor ≈103 lower and specific metabolic rates to be much lower in clayey sediments than in sandy sediments. In clays, pore size is so small that microbes are immobile and must depend on diffusion of gaseous or liquid nutrients toward them.
- D'Hondt et al. (47) used depth profiles of methane and sulfate to make an in situ assessment of the metabolic activity of sulfate reducers and methanogens in marine sediments down to several hundred meters below sea floor at various ocean-margin sites and open-ocean sites. From these profiles and from cell counts by R. J. Parkes and B. A. Cragg, they determined subsurface respiration rates of CO2 per cell and mean
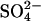 reduction rates per cell as a function of depth. The specific metabolic rates at ≈2°C in sediment at sulfate-rich open-ocean sites were typically three orders of magnitude lower than at methane-rich ocean-margin sites, and both rates were found by the geochemical method to be orders of magnitude lower than had been found with radiotracers. They concluded that sub-seafloor metabolic activity is greatly concentrated in relatively narrow zones of sulfate-reducing methane oxidation along ocean margins, and that microorganisms in sub-seafloor sediments are adapted for extraordinarily low metabolic activity. Point J1 gives the average metabolic rate for three open-ocean sites: equatorial Pacific, Lau Basin, and Izu-Bonin Trench.
reduction rates per cell as a function of depth. The specific metabolic rates at ≈2°C in sediment at sulfate-rich open-ocean sites were typically three orders of magnitude lower than at methane-rich ocean-margin sites, and both rates were found by the geochemical method to be orders of magnitude lower than had been found with radiotracers. They concluded that sub-seafloor metabolic activity is greatly concentrated in relatively narrow zones of sulfate-reducing methane oxidation along ocean margins, and that microorganisms in sub-seafloor sediments are adapted for extraordinarily low metabolic activity. Point J1 gives the average metabolic rate for three open-ocean sites: equatorial Pacific, Lau Basin, and Izu-Bonin Trench. - Tison et al. (9) found huge increases in CO2 (up to 130,000 ppm by volume) and CH4 (up to 6,000 ppm by volume) in the bottom 6 m of the European borehole [Greenland Ice Core Project (GRIP)] at Summit, Greenland, within a few meters of subglacial till. We suspected that those strong gradients were due to in situ microbial metabolism. For the present article, we analyzed CH4 in the basal ice from the U.S. borehole (GISP2, near GRIP) and found even higher concentrations: 1.2% CH4 in air extracted from the ice at a depth of 3,044 m, at which the in situ temperature was -9°C. In collaboration with one of us (T.S.), V. Miteva examined GISP2 basal ice. She found 6-9 × 107 cells per milliliter of melt water at the same depth (11) and estimated a live:dead ratio of ≈5:1. Point J shows the average metabolic rate, which we calculated for an average carbon mass of 15 fg for the dwarf cells observed by Miteva. Based on her successful cultivation of anaerobes at -2°C, it is likely that methanogens were mainly responsible for the excess methane.
- Sowers (10) reported peaks in the concentrations of N2O and δ15N of N2O and dips in the values of δ18O of N2O occurring at the same depths in Vostok ice where Abyzov et al. (1) found peaks in bacterial concentration and where Petit et al. (48) found peaks in dust concentration. The ice temperature at that depth is -40°C. The remarkable correlation strongly suggests that, during the penultimate glacial maximum ≈140,000 years ago, dust and microorganisms were windborne with greater than normal efficiency and deposited onto Antarctic ice. The observed high δ15N and low δ18O of N2O indicated nitrification, in contrast to denitrification, which would have resulted in a high δ15N and high δ18O of N2O. Sowers inferred in situ N2O production by nitrifying bacteria that oxidize ammonia present in the ice. At low but finite oxygen concentration, nitrifiers such as Nitrosomonas can convert ≈10% of the total fixed ammonia to N2O. The N2O is produced both by the denitrification of the
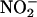 product of
product of 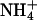 oxidation and by the oxidation of NH2OH intermediate. This interpretation is consistent with studies of N2O production in Antarctic lake ice (Lake Bonney) at depths where there was a strong excess of nitrifiers (49). Nitrifiers require an aqueous, dark medium, at low O2 pressure: conditions that exist in liquid veins deep in the Vostok ice. Carbon for growth is obtained from fixation of CO2 by means of the Calvin cycle. From the measurements in Lake Bonney (49), together with calculations (50), we used a ratio (C fixed)/(N2O produced) ≈ 0.04 to convert from the rate of nitrification to the rate of carbon metabolism.
oxidation and by the oxidation of NH2OH intermediate. This interpretation is consistent with studies of N2O production in Antarctic lake ice (Lake Bonney) at depths where there was a strong excess of nitrifiers (49). Nitrifiers require an aqueous, dark medium, at low O2 pressure: conditions that exist in liquid veins deep in the Vostok ice. Carbon for growth is obtained from fixation of CO2 by means of the Calvin cycle. From the measurements in Lake Bonney (49), together with calculations (50), we used a ratio (C fixed)/(N2O produced) ≈ 0.04 to convert from the rate of nitrification to the rate of carbon metabolism.
We add to this list the fascinating work of Campen et al. (51), who reported very large excesses of CO2 (≈32× tropospheric value), CH4 (8×), and N2O (up to 240×) at depths corresponding to ages of 14,000 yr (for CO2 and N2O) and 15,400 yr (for CH4) in air extracted from an ice core from the Sajama (Bolivia) glacier. They concluded that the excesses of CO2, CH4, and N2O were products of metabolism due to microbes probably situated in liquid veins along triple junctions of ice grains (12). Unfortunately, not knowing the microbial concentrations at those depths, we cannot estimate metabolic rates.
Discussion
Morita (52) proposed two levels of metabolism for starving, nongrowing microbes. Maintenance energy is the energy required for osmotic regulation, maintenance of intracellular pH, futile cycles, turnover of macromolecules, motility, and energy dissipation by proton leak and ATP hydrolysis, exclusive of biomass production; and survival energy is the energy required just for repair of macromolecular damage. Remarkably, in Fig. 1 the metabolic data for microbial communities do fall into three groupings: the rate for exponential growth (uppermost band) is orders of magnitude greater than most of the rates for maintenance without growth (blue letters), and the rates for communities trapped in glacial ice or deep subsurface sediments (red letters) are lower by another three orders of magnitude. The clear separation justifies using Morita's term, survival metabolism, to distinguish the extremely weak metabolism of immobile, probably dormant communities from maintenance metabolism of communities more accessible to nutrient and free to move but still at too low a level for growth. Maintenance metabolism depends, of course, on nutrient availability as well as on temperature. The data of Parkes et al. (3) provide an example of the dependence on nutrient availability: maintenance metabolic rate decreases with sample depth in marine sediment depth as shown by points E1, E2, and E3.
We added lines in Fig. 1 to denote the three groupings into which the data fall. The upper solid line, in purple, for exponential growth of a community of permafrost bacteria (fed in the laboratory) has a slope corresponding to an “effective” activation energy _U_g ≈ 110 kJ/mole. The solid line in blue through the points labeled A-E is drawn parallel to the purple line; the errors on the middle grouping of points are too large to enable us to determine the effective activation energy accurately, but it is not inconsistent with the value for growth: _U_m ≈ _U_g, with μg(T)/μm(T) ≈ 103. _U_g and _U_m are not activation energies in the chemical sense, but are measures of the temperature response of microbial communities. They are higher than activation energies of ≈40-75 kJ/mole for growth of individual species (53, 54) measured on the low-temperature side of the maximum in the growth curve.
Instead of drawing a line through points F-K that would be parallel to the purple and blue lines, we draw attention to the remarkable similarity of the rates in the bottom grouping to empirical rates of spontaneous macromolecular damage. Brinton et al. (55) recently determined the rate and activation energy for spontaneous aspartic acid racemization of microbes in wet samples of Siberian permafrost. The solid red line is an extrapolation of their data into the temperature range of our study. Rates for the other amino acids are much lower. The short-dashed red line is an extrapolation of data on the rate of depurination of DNA (56). The errors in the extrapolated rates for racemization and depurination are roughly a factor two.
We know that DNA-repair enzymes in a living organism efficiently reverse the effects of spontaneous degradation because fewer than 10-3 of the accidental base changes in DNA cause a mutation (57). Some damaged proteins can also be repaired and replaced. Almost all cells contain a methyltransferase that can recognize and repair aging proteins such as the l-isoaspartyl residue and the racemized d-aspartyl residue (58) by means of a methyl esterification reaction. Other protein-repair enzymes may also exist. Brinton et al. (55) have presented evidence that permafrost organisms can maintain active recycling of d-amino acids for at least 30,000 yr.
The approximate agreement of the data on rates of aspartic acid racemization (55) and DNA depurination (56) with points F-K for metabolic rates for survival of immobilized, starved, possibly dormant microbes suggests that rates of repair of DNA and protein damage in living microbes are similar to rates of damage. We suggest a concrete definition of dormancy as the state of a microorganism in which it expends energy at the rate μs(T) solely to repair molecular damage.
Nutrients can be transported to immobile and mostly dormant microbes as H2 gas (59, 60) or as water-soluble organic compounds such as humic and fulvic acids (61, 62). For glacial ice (12), the access paths for gaseous and liquid nutrients and waste would be micrometer-diameter veins; for shale and clay (61, 62), they would be sub-micrometer-size pores.
The extremely low expenditures of survival energy enable microbial communities in extreme environments to survive indefinitely. Nitrifying bacteria with metabolic rate ≈10-12 per hour at -40°C (point L) have been encased in liquid veins in Vostok ice for >140,000 yr. At a rate of 10-12 per hour, it takes ≈108 yr to turn over the carbon in their cells without growth. Even more impressive is the ability of microbes to survive while deeply buried and immobilized in confining clays and shale for ≈108 yr (45, 46).
Our results disprove the view (63) that the lowest temperature at which life is possible is ≈-17°C in an aqueous environment, as well as the remark (27) that “the lowest temperature at which terrestrial and presumably martian life can function is probably near -20°C.” Our data show no evidence of a threshold or cutoff in metabolic rate at temperatures down to -40°C. A cell resists freezing, due to the “structured” water in its cytoplasm (64). Ionic impurities prevent freezing of veins in ice and thin films in permafrost and permit transport of nutrient to and products from microbes. The absence of a threshold temperature for metabolism should encourage those interested in searches for life on cold extraterrestrial bodies such as Mars and Europa.
Acknowledgments
We thank Jeffrey Bada, Cecilia Bartolucci, Kramer Campen, Ed Carpenter, Francis Chapelle, Brent Christner, Steven Clarke, Imre Friedmann, David Gilichinsky, Alex Glazer, Vanya Miteva, Richard Morita, T. C. Onstott, Tom Phelps, and Roland Psenner for discussions. This research was supported in part by National Science Foundation Grants OPP-0085400 (to P.B.P.) and ATM-9618067 (to T.S.).
References
- 1.Abyzov, S. S., Mitskevich, I. N. & Poglazova, M. N. (1998) Microbiologiya 67**,** 547-555. [Google Scholar]
- 2.Priscu, J. C., Adams, E. E., Lyons, W. B., Voytek, M. A., Mogk, D. W., Brown, R. L., McKay, C. P., Takacs, C. D., Welch, K. A., Wolf, C. F., et al. (1999) Science 286**,** 2141-2144. [DOI] [PubMed] [Google Scholar]
- 3.Karl, D. M., Bird, D. F., Bjorkman, K., Houlihan, T., Shackelford, R. & Tupas, L. (1999) Science 286**,** 2144-2147. [DOI] [PubMed] [Google Scholar]
- 4.Willerslev, E., Hansen, A. J., Christensen, B., Steffensen, J. P. & Arctanger, P. (1999) Proc. Natl. Acad. Sci. USA 96**,** 8017-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber, S. & Jaenicke, R. (2001) Meddelelser om Grønland Geosci. 39**,** 161-163. [Google Scholar]
- 6.Ma, L.-J., Rogers, S. O., Catranis, C. M. & Starmer, W. T. (2000) Mycologia 92**,** 286-295. [Google Scholar]
- 7.Christner, B. C., Mosley-Thompson, E., Thompson, L. G., Zagorodnov, V., Sandman, K. & Reeve, J. N. (2000) Icarus 144**,** 479-485. [Google Scholar]
- 8.Junge, K., Eicken, H. & Deming, J. W. (2004) Appl. Environ. Microbiol. 70**,** 550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tison, J.-L., Souchez, R., Wolff, E. W., Moore, J. C., Legrand, M. R. & de Angelis, J. (1998) J. Geophys. Res. Atmos. 103**,** 18885-18894. [Google Scholar]
- 10.Sowers, T. (2001) J. Geophys. Res. Atmos. 106**,** 31903-31914. [Google Scholar]
- 11.Sheridan, P. P., Miteva, V. I. & Brenchley, J. E. (2003) Appl. Environ. Microbiol. 69**,** 2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price, P. B. (2000) Proc. Natl. Acad. Sci. USA 97**,** 1247-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson, D. M. (1967) Nature 216**,** 563-566. [Google Scholar]
- 14.Anderson, D. M. (1973) Ecol. Stud. 4**,** 107-124. [Google Scholar]
- 15.Langdahl, B. R. & Ingvorsen, K. (1997) FEMS Microbiol. Ecol. 23**,** 275-283. [Google Scholar]
- 16.Gilichinsky, D. (2002) in Encyclopedia of Environmental Microbiology, ed. Bitton, G. (Wiley, New York), pp. 2367-2385.
- 17.Gilichinsky, D., Wagener, S. & Vishnevetskaya, T. A. (1995) Permafrost Periglacial Processes 6**,** 281-291. [Google Scholar]
- 18.Gilichinsky, D. A., Soina, V. S. & Petrova, M. A. (1995) Origins Life Evol. Biosphere 23**,** 65-75. [DOI] [PubMed] [Google Scholar]
- 19.Gilichinsky, D., Rivkina, E., Shcherbakova, V., Laurinavichus, K. & Tiedje, J. (2003) Astrobiology 3**,** 331-341. [DOI] [PubMed] [Google Scholar]
- 20.Rivkina, E. M., Laurinavichus, K. S., Gilichinsky, D. A. & Shcherbakova, V. A. (2002) Doklady Biol. Sci. 383**,** 179-181. [DOI] [PubMed] [Google Scholar]
- 21.Bakermans, C., Tsapin, A. I., Souza-Egipsy, V., Gilichinsky, D. A. & Nealson, K. H. (2003) Environ. Microbiol. 5**,** 321-326. [DOI] [PubMed] [Google Scholar]
- 22.Kappen, L., Schroeter, B., Scheidegger, C., Sommerkorn, M. & Hestmark, G. (1996) Adv. Space Res. 18**,** 119-128.11538952 [Google Scholar]
- 23.Johnston, C. G. & Vestal, J. R. (1991) Appl. Environ. Microbiol. 57**,** 2308-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaksen, M. F. & Jørgensen, B. B. (1996) Appl. Environ. Microbiol. 62**,** 408-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitman, W. B., Coleman, D. C. & Wiebe, W. J. (1998) Proc. Natl. Acad. Sci. USA 95**,** 6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivkina, E., Friedmann, E. I., McKay, C. P. & Gilichinsky, D. (2000) Appl. Environ. Microbiol. 66**,** 3230-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakosky, B. M., Nealson, K. H., Bakermans, C., Ley, R. E. & Mellon, M. T. (2003) Astrobiology 3**,** 343-350. [DOI] [PubMed] [Google Scholar]
- 28.Carpenter, E. J., Lin, S. & Capone, D. G. (2000) Appl. Environ. Microbiol. 66**,** 4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sattler, B., Puxbaum, H. & Psenner, R. (2001) Geophys. Res. Lett. 28**,** 239-242. [Google Scholar]
- 30.Knoblauch, C., Jørgensen, B. B. & Harder, J. (1999) Appl. Environ. Microbiol. 65**,** 4230-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coveney, M. F. & Wetzel, R. G. (1995) Limnol. Oceanogr. 40**,** 1187-1200. [Google Scholar]
- 32.Simon, M. & Azam, F. (1989) Mar. Ecol. Prog. Ser. 51**,** 201-213. [Google Scholar]
- 33.White, P. A., Kalff, J., Rasmussen, J. B. & Gasol, J. M. (1991) Microbiol. Ecol. 21**,** 99-118. [DOI] [PubMed] [Google Scholar]
- 34.Novitsky, J. A. (1987) Appl. Environ. Microbiol. 53**,** 2368-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bird, D. F & Karl, D. M. (1999) Aquatic Microbiol. Ecol. 19**,** 13-27. [Google Scholar]
- 36.Smith, R. E. H. & Clement, P. (1990) Polar Biol. 10**,** 351-357. [Google Scholar]
- 37.Shiah, F.-K. & Ducklow, H. W. (1995) Limnol. Oceanogr. 40**,** 55-66. [Google Scholar]
- 38.Kottmeier, S. T., McGrath Grossi, S. & Sullivan, C. W. (1987) Mar. Ecol. Prog. Ser. 35**,** 175-186. [Google Scholar]
- 39.Ward, B. B. & Priscu, J. C. (1997) Hydrobiologia 347**,** 57-68. [Google Scholar]
- 40.Anderson, T.-H. & Domsch, K. H. (1985) Biol. Fert. Soils 1**,** 81-89. [Google Scholar]
- 41.Franzmann, P. D., Roberts, N. J., Mancuso, C. A., Burton, H. R. & McMeeking, T. A. (1991) Hydrobiologia 210**,** 191-201. [Google Scholar]
- 42.Mancuso, C. A., Franzmann, P. D., Burton, H. R. & Nichols, P. D. (1990) Microbiol. Ecol. 19**,** 73-95. [DOI] [PubMed] [Google Scholar]
- 43.Christner, B. C. (2002) Appl. Environ. Microbiol. 68**,** 6435-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkes, R. J., Cragg, B. A., Fry, J. C., Herbert, R. A. & Wimpenny, J. W. T. (1990) Philos. Trans. R. Soc. London 331**,** 139-153. [Google Scholar]
- 45.Chapelle, F. H. & Lovley, D. R. (1990) Appl. Environ. Microbiol. 56**,** 1865-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phelps, T. J., Murphy, E. M., Pfiffner, S. M. & White, D. C. (1994) Microbiol. Ecol. 28**,** 335-349. [DOI] [PubMed] [Google Scholar]
- 47.D'Hondt, S., Rutherford, S. & Spivack, A. J. (2002) Science 295**,** 2067-2070. [DOI] [PubMed] [Google Scholar]
- 48.Petit, J. R., Jouzel, J., Raynaud, D., Barkov, N. I., Barnola, J.-M., Basile, I., Bender, M., Chappellaz, J., Davis, M., Delaygue, G., et al. (1999) Nature 399**,** 429-436. [Google Scholar]
- 49.Priscu, J. C., Downes, M. T. & McKay, C. P. (1996) Limnol. Oceanogr. 41**,** 1544-1551. [DOI] [PubMed] [Google Scholar]
- 50.Poughon, L., Dussap, C.-G. & Gros, J.-B. (2001) Biotechnol. Bioeng. 72**,** 416-433. [PubMed] [Google Scholar]
- 51.Campen, R. K., Sowers, T. & Alley, R. B. (2003) Geology 31**,** 231-234. [Google Scholar]
- 52.Morita, R. Y. (1997) Bacteria in Oligotrophic Environments (Chapman & Hall, New York).
- 53.Sagemann, J., Jørgensen, B. B. & Greeff, O. (1998) Geomicrobiol. J. 15**,** 85-100. [Google Scholar]
- 54.Knoblauch, C. & Jørgensen, B. B. (1999) Environ. Microbiol. 1**,** 45-467. [DOI] [PubMed] [Google Scholar]
- 55.Brinton, K. L. F., Tsapin, A. I., Gilichinsky, D. & McDonald, G. D. (2002) Astrobiology 2**,** 77-82. [DOI] [PubMed] [Google Scholar]
- 56.Lindahl, T. & N. Nyberg. (1972) Biochemistry 11**,** 3610-3618. [DOI] [PubMed] [Google Scholar]
- 57.Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K. & Watson, J. D. (1994) Molecular Biology of the Cell (Garland, New York), 3rd Ed.
- 58.Clarke, S. (2003) Ageing Res. Rev. 2**,** 263-285. [DOI] [PubMed] [Google Scholar]
- 59.Morita, R. Y. (2000) Microbiol. Ecol. 38**,** 307-320. [DOI] [PubMed] [Google Scholar]
- 60.Stevens, T. O. & McKinley, J. P. (1995) Science 270**,** 450-454. [Google Scholar]
- 61.Krumholz, L. R., McKinley, J. P., Ulrich, G. A. & Suflita, J. M. (1997) Nature 386**,** 64-66. [Google Scholar]
- 62.Lovley, D. R., Coates, J. D., Blunt-Harris, E. L., Phillips, E. J. P. & Woodward, J. C. (1996) Nature 382**,** 445-448. [Google Scholar]
- 63.Finegold, L. (1996) Adv. Space Res. 18**,** 87-95. [Google Scholar]
- 64.Pollack, G. H. (2001) Cells, Gels and the Engine of Life (Ebner, Seattle).