Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae (original) (raw)
Abstract
The role of chemotaxis in the virulence of gastrointestinal pathogens is ill defined. Counterintuitively, nonchemotactic mutants of the polarly flagellated pathogen Vibrio cholerae greatly out-compete the wild-type strain during infection of the small intestine. We show that the out-competition phenotype is dependent on the direction of flagellar rotation and independent of Toxin Co-regulated Pilus function. Specifically, the out-competition associated with the loss of chemotaxis required the presence of counterclockwise-biased flagellar rotation and smooth straight runs by the bacteria. In contrast, a nonchemotactic strain with clockwise-biased flagellar rotation was confined to small-scale net movement and was attenuated for infection. The significance of the out-competition phenotype was examined and was shown to correlate with a true increase in infectivity. Counterclockwise-biased mutants are aberrantly distributed throughout the infant mouse small intestine and we find that the expression of virulence factors occurs normally in all segments. Thus, alteration of the chemotactic properties of V. cholerae allows it to exploit additional niches in the host intestine.
The Gram-negative bacterium Vibrio cholerae is the causative agent of the epidemic disease cholera. Cholera patients are afflicted with a profuse watery diarrhea resulting from the action of the ADP-ribosylating cholera toxin (CT). This organism is highly motile by means of a single polar sheathed flagellum and is believed to use the processes of motility and chemotaxis to travel from the lumen of the small intestine to its preferred intestinal niche on the intestinal epithelium. In this niche V. cholerae expresses a number of virulence factors including CT and the toxin co-regulated type IV pilus (TCP). The latter has been shown to be essential for colonization of humans (1), as well as in the infant mouse model of infection (2).
Within the V. cholerae genome are multiple paralogues of chemotaxis genes (3). Despite the presence of three chemotaxis operons, only one of these operons is required for chemotaxis (4). The function of the remaining chemotaxis operons remains unknown. Chemotaxis in many organisms is achieved by modulating change in the direction of flagellar rotation from the default direction of counterclockwise (CCW) to clockwise (CW) rotation. In the peritrichously flagellated bacterium Escherichia coli, CCW rotation results in the bacterium swimming smoothly in a mostly straight line, whereas CW rotation causes the cell to turn abruptly in a process known as tumbling (5). Because of the presence of a single polar flagellum, V. cholerae does not tumble per se but instead reverses direction briefly, thereby allowing the bacterium to randomly reorient itself and swim in a new direction.
Although motility and chemotaxis are believed to guide V. cholerae to its preferred colonization site within the small intestine, the precise role of each of these processes during infection has not been clearly established. Previous work by Freter et al. (6) revealed that motility is required for the establishment of an infection. However, subsequent work by two other groups indicated that motility might in fact be dispensable for infection (7, 8). Much of this prior work was done by using undefined nonmotile mutants, and we have since shown that defined nonmotile mutants of the El Tor biotype, whether flagellated or aflagellated, are severely attenuated in the infant mouse model of infection (9). In contrast to the attenuation seen with nonmotile mutants, the loss of chemotactic ability by itself does not inhibit V. cholerae infection. On the contrary, motile but nonchemotactic El Tor biotype V. cholerae dramatically out-compete the wild type during infection of the infant mouse (9, 10).
A similar competitive advantage was also seen with fresh cholera stool bacteria. Stool V. cholerae (El Tor, Inaba serotype) out-competed in vitro grown V. cholerae from 10- to 100-fold in the infant mouse small intestine, and this competitive advantage was maintained after incubation of stool bacteria in local pond water for several hours (11). However, this advantage was lost after subsequent growth in LB and was therefore transient. Despite their being motile, transcriptional profiling of stool V. cholerae showed repression of all cheW and cheR paralogues compared with a stool isolate grown in vitro (11). In E. coli (12) and V. cholerae (data not shown), both cheW and cheR are required for chemotaxis, and mutations in either gene result in a CCW-biased, smooth swimming phenotype. These data suggest that V. cholerae exit the human host in a transiently nonchemotactic state. Such a state would augment the infectivity of stool V. cholerae in the infant mouse model and possibly aid rapid epidemic spread of cholera in humans.
In this study we have determined that the out-competition phenotype during infection (in vivo) requires the presence of a CCW-biased flagellum, because nonchemotactic CW-biased V. cholerae were attenuated in vivo. In addition, we show that nonchemotactic CCW-biased V. cholerae induce virulence gene expression throughout the length of the infant mouse small intestine. Finally, we demonstrate that the out-competition phenotype corresponds with a one-order-of-magnitude decrease in the infectious dose that may have relevance for understanding fecal-oral spread of cholera.
Materials and Methods
Bacterial Strains and Plasmids. V. cholerae strains used in this work are derivatives of the El Tor biotype strain C6709-1. All strains and plasmids used in this study are listed in Table 1. Bacterial strains were grown in LB broth with aeration at 37°C or on LB agar plates at 37°C. L agar (without salt) was supplemented with 10% sucrose for counter selection of the pCVD442-lac plasmid derivatives. Chemotaxis assays were done in swarm agar (1% tryptone/0.5% NaCl/0.3% agar), and swarm plates were incubated at 37°C overnight. Antibiotics were used at the following concentrations: ampicillin (Ap), 50 μg·ml-1; kanamycin (Kn),150 μg·ml-1; streptomycin (Sm), 100 μg·ml-1; and tetracycline (Tc), 1 and 3 μg·ml-1.
Table 1. Bacterial strains and plasmids used in this study.
| Strain/plasmid | Relevant genotype (phenotype) | Source/ref. |
|---|---|---|
| V. cholerae | ||
| AC-V51 | C6709-1, SmR (chemotactic [che+]) | 34 |
| AC-V66 | C6709-1 lacZ::res-tet-res, SmR TcR (che+) | 35 |
| AC-V1033 | AC-V66 cheYD60N (che−, CCW-biased) | 9 |
| AC-V1323 | AC-V66 Δ_cheY_ (che−, CCW-biased) | This work |
| AC-V1035 | AC-V1033 _ctxA::tnpR_mut135 merodiploid, SmR TcR ApR | 9 |
| AC-V1036 | AC-V1033 _tcpA::tnpR_mut135 merodiploid, SmR TcR ApR | This work |
| AC-V1634 | AC-V51 Δ_tcpA_ (che+) | This work |
| AC-V1635 | AC-V1033 Δ_tcpA_ (che−, CCW-biased) | This work |
| AC-V1399 | AC-V66 cheYD16KY109W (che−, CW-biased) | This work |
| AC-V1636 | AC-V1399 Reverted to wild-type cheY (che+) | This work |
| MKW107 | 0395 (CTXcalc-KnΦ), SmR KnR | 20 |
| E. coli | ||
| DH5αλ_pir_ | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 λ::pir | 36 |
| SM10λ_pir_ | thi recA thr leu tonA lacY supE RP4-2-Tc::Mu λ::pir | 35 |
| Plasmids | ||
| pCVD442-lac | Parent allelic exchange vector | 13 |
| pMMB67EH | Low-copy plasmid | 37 |
| p_cheY_ | pMMB67EH::cheY, ApR | This work |
| pSB27 | pCVD441-lac::ΔcheY, ApR | This work |
| pSB29 | pCVD442-lac::cheYD16K, ApR | This work |
| pSB34 | pCVD442-lac::cheYY109W, ApR | This work |
| pSB66 | pCVD442-lac::ΔtcpA, ApR | This work |
| pSB67 | pCVD442-lac::cheY, ApR | This work |
Strain Constructions. The alleles for construction of deletion and point mutation strains were generated by splicing overlap extension PCR with PFU polymerase (Stratagene). PCR products were cloned into the pCR-Script Amp SK vector (Stratagene). Outer primers contained _Sac_I and Sph_I restriction sites subsequently used to subclone the fragment into the pCVD442-lac. E. coli Sm10λ_pir carrying each plasmid were mated with V. cholerae. The deletions and point mutations were introduced into the V. cholerae genome by allelic exchange with pCVD442-lac as described (13). PCR and DNA sequencing were used to confirm the presence of the correct mutations.
Video Tracking. V. cholerae strains were observed under ×40 dark-field magnification by using a Zeiss Axioplan 2 microscope. Images of the bacteria were captured by using openlab 3.1.5 (Improvision, Lexington, MA). For each strain a total of 150 bacteria were recorded at 1-s intervals, and the frequency of direction change was calculated as the mean number of direction changes per second.
Complementation Analysis. cheY (bases -14 to +435) was amplified by PCR, cloned into PCR-Script Amp SK, and subcloned into the low-copy-number vector pMMB67EH by using _Sac_I and Sph_I restriction sites incorporated into the primers used for amplification. The complementing plasmid p_cheY and the pMMB67EH vector were introduced into V. cholerae by electroporation. The pMMB67EH vector with and without cheY was stable during growth in vitro and in vivo.
Competition Assays. The competitive index (CI) was determined after intragastric inoculation of 5-day-old CD-1 mice as described (2). Briefly, each mouse was inoculated with ≈105 colony-forming units (cfu) of a 1:1 mixture of wild-type (LacZ+) and mutant (LacZ-) overnight cultures. In addition, 2 ml of LB was inoculated with ≈104 cfu of this mixture and grown at 37°C overnight. At 24 h postinoculation, animals were euthanized and the small intestines were removed. These were homogenized in LB + 20% glycerol, and serial dilutions were plated on LB agar supplemented with Sm and 40 μg·ml-1 of 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-Gal). The CI is the ratio of LacZ- to LacZ+ bacteria after 24 h of infection divided by the ratio of LacZ- to LacZ+ bacteria after growth overnight in LB.
Intestinal CTXΦ Transduction Assay. This assay was done as described (14). Briefly, 5-day-old mice were inoculated intragastrically with 106 cfu of a 1:1 mixture of phage donor (MKW107, LacZ+, SmR, KnR) and recipient (LacZ-, SmR, TcR) strain from overnight cultures. At 10 h postinoculation the small intestines were removed, homogenized, and plated on LB agar plus Sm and X-Gal, and LB agar plus Kn and Tc. The frequency of transduction was calculated as the total number of transductants (KnR, TcR) divided by the total number of recipient cells (SmR, LacZ-) in the small intestine.
In Vivo Resolution Assays. Strains harboring tnpR fusions (cheYD60N ctxA::tnpR and cheYD60N tcpA::tnpR) were grown overnight at 37°C with aeration in LB broth supplemented with Sm, Ap, and Tc. Approximately 106 cfu of each strain was inoculated intragastrically into 5-day-old mice. At 7 h postinoculation the small intestine was removed and divided into four segments of equal length. Each segment was homogenized and plated, and the percent TcS cfu was calculated as described (14).
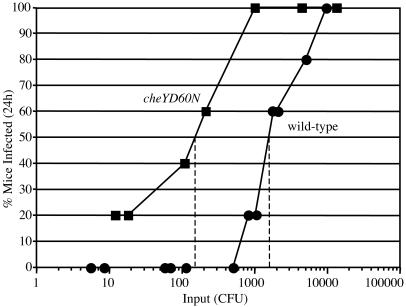
Determination of ID50. Five-day-old CD-1 mice were inoculated intragastrically with doses ranging from ≈10 to 104 cfu. Small intestines were removed at 24 h, homogenized, and plated to determine the number of cfu per small intestine. The limit of detection was ≈10 cfu, and values equal to or greater than this were recorded as a positive for infection. The ID50 was determined graphically by using the curves in Fig. 3.
Fig. 3.
Determination of the ID50 for wild-type and cheYD60N strains. Groups of five mice were infected with varying doses (x axis) of the wild type (filled circles) or the cheYD60N mutant (filled squares). Each data point represents the percentage of the five mice that were infected (y axis) after a 24-h period. The limit of detection of the mouse output was 10 cfu per small intestine; however, amongst mouse outputs with detectable numbers of V. cholerae, 1,000 cfu was the lowest output observed. The ID50 value for each strain was determined graphically.
Results
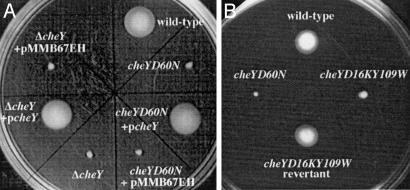
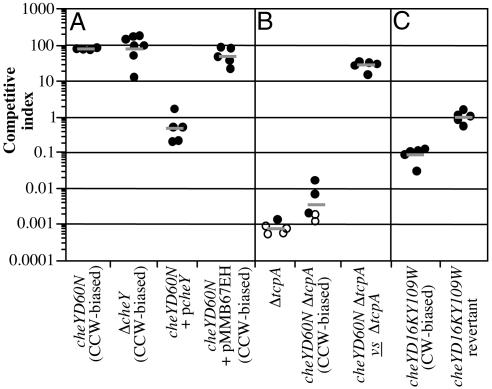
Loss of CheY Function Leads to an Out-Competition Phenotype in Vivo. We had previously shown that the nonchemotactic cheY-3D60N (hereafter cheYD60N) mutant, in which the conserved phosphorylation site is rendered nonphosphorylatable (15, 16), out-competes the wild-type strain 70-fold in vivo (9). To show that the out-competition phenotype of this strain was not due to an aberrant activity associated with the mutated CheY-3 protein, we constructed an in-frame deletion of cheY-3 (hereafter cheY). The resulting Δ_cheY_ strain was defective for chemotaxis as determined in a swarm-plate assay, and this defect was fully complemented by the introduction of the wild-type allele on a low-copy-number plasmid but not by the pMMB67EH vector alone (Fig. 1_A_). The Δ_cheY_ strain out-competed the wild-type strain to a similar extent as the cheYD60N point mutant when tested in vivo, and this out-competition was fully complemented by supply of cheY in trans (Fig. 2_A_). Thus, it is the lack of CheY function that is responsible for the out-competition phenotype.
Fig. 1.
Analysis of chemotactic ability by using swarm plates. Genetic backgrounds are indicated next to each swarm location.
Fig. 2.
Competition assays in infant mice between wild-type and mutant V. cholerae strains. Mutant strain backgrounds for each experiment are indicated on the x axis. Each data point represents the competitive index (CI) from one mouse. The CI is given as the ratio of mutant to wild type after infection divided by the ratio of mutant to wild type after overnight growth in LB. Horizontal bars indicate the geometric mean of the CIs for each experiment. (A) Competition between the cheYD60N, Δ_cheY_, cheYD60N (p_cheY_), and cheYD60N (pMMB67EH) strains (all LacZ-) and the wild-type strain (LacZ+). (B) Competition between either the cheYD16KY109W mutant or cheYD16KY109W reversion strain (both LacZ-) and wild type (LacZ+). (C) Competition between either the Δ_tcpA_ mutant or the cheYD60N_Δ_tcpA double mutant and wild type, and between the cheYD60N_Δ_tcpA mutant (LacZ-) and the Δ_tcpA_ mutant (LacZ+). Open symbols indicate that the mutant was below the limit of detection.
Out-Competition Is Independent of TCP. Whereas wild-type V. cholerae colonize primarily within the distal half of the infant mouse small intestine, the cheYD60N mutant has been shown to be evenly distributed throughout the length of the small intestine (9). This aberrant distribution is largely responsible for the out-competition phenotype observed with the cheYD60N and Δ_cheY_ strains. Because TCP is normally required for colonization of the small intestine, we wished to examine whether there are any differences in TCP expression between the wild-type and cheYD60N strains in vivo. In addition to its requirement for infection, TCP is also the receptor for CTXΦ, the lysogenic bacteriophage whose genome contains the structural genes for CT (17). However, the requirement of TCP for infection is independent of its function as the CTXΦ receptor (18). CTXΦ is secreted from V. cholerae via the extracellular protein secretion (EPS) type II secretion system (19), and the expression of TCP can be examined by measuring the frequency of transduction of the kanamycin-resistance-marked derivative CTXcalc-KnΦ from a donor strain to the recipient test strain during infection, as described (14, 20). Using this technique, we calculated the frequency of in vivo transduction to be 3.3% (±1.0%) for wild-type and 2.5% (±0.7%) for the cheYD60N mutant. Thus, there appears to be no gross difference between the two strains with respect to TCP production during infection.
Although TCP production was equivalent between wild-type and cheYD60N strains, we wanted to determine whether TCP is expressed by the cheYD60N mutant throughout the length of the small intestine. We also wished to examine whether additional virulence genes might be expressed in a similar manner. The tcpA gene encodes the pilin subunit of TCP (2), and ctxA encodes the enzymatic subunit of CT. We have previously shown that in wild-type V. cholerae these genes are fully induced by 7 h postinoculation by using a DNA recombinase (tnpR) transcriptional fusion reporter method (14), wherein expression of tnpR mediates excision and loss of a tetracycline resistance cassette located elsewhere in the genome. Thus, the level of conversion to tetracycline sensitivity is a direct result of transcriptional induction (for a recent review see ref. 21). Induction of both tcpA::tnpR and ctx::tnpR fusions in the cheYD60N mutant background was measured at 7 h postinoculation in four divided segments of small intestine (Table 2). The extensive induction of both fusions in the cheYD60N mutant observed at 7 h in the distal small intestine was equivalent to induction in the wild-type strain background (9), indicating that the cheYD60N mutation and loss of chemotaxis do not affect virulence gene expression at this time point. In addition, both fusions were induced in segments of the proximal small intestine, suggesting that the signals required for TCP and CT expression are present in these locations.
Table 2. Induction of gene expression during infection.
| Small intestinal segment | Tetracycline-sensitive cfu*, % | |
|---|---|---|
| cheYD60N tcpA::tnpR | cheYD60N ctxA::tnpR | |
| 1 (proximal) | 93 ± 4 | 96 ± 2 |
| 2 | 91 ± 3 | 96 ± 1 |
| 3 | 80 ± 7 | 96 ± 2 |
| 4 (distal) | 83 ± 7 | 89 ± 4 |
To determine whether nonchemotaxis and TCP act independently to aid colonization, an epistasis test was performed. An in-frame deletion of tcpA was constructed in the wild-type and cheYD60N mutant backgrounds. Introduction of this mutation in either strain background greatly reduced the level of colonization (Fig. 2_B_). However, although the double mutant was attenuated, this attenuation was not as severe as in the single Δ_tcpA_ mutant. This was confirmed by competing the cheYD60N_Δ_tcpA double mutant against the single Δ_tcpA_ mutant (Fig. 2_B_). Although TCP is clearly required for high levels of colonization, the ability of the cheYD60N_Δ_tcpA double mutant to out-compete the Δ_tcpA_ mutant indicates that chemotaxis and TCP act independently during infection.
CCW-Biased Flagellar Rotation Is Required for Out-Competition. The absence of phosphorylated CheY results in CCW-biased flagellar rotation in E. coli and Salmonella enterica serovar Typhimurium (22). However, a nonchemotactic state is also observed in the presence of excess levels of CheY-P, which results in CW-biased flagellar rotation (23, 24). We therefore tested whether a CW-biased V. cholerae mutant would give rise to the out-competition phenotype during infection. Although CheY has an intrinsic phosphatase activity, it is not adequate for the high turnover of CheY-P required for efficient chemotaxis (25). The protein CheZ stimulates the rate of CheY-P dephosphorylation (26, 27) and is required for chemotaxis in organisms such as Pseudomonas aeruginosa, E. coli, and S. enterica serovar Typhimurium (28, 29). Despite the presence of a single cheZ in the V. cholerae genome, this gene is not required for chemotaxis in V. cholerae (data not shown). We were therefore unable to use the cheZ deletion as a CW-biased nonchemotactic test strain. Instead we constructed a cheYD16KY109W double point mutant, because the homologous mutations in E. coli confer CW-biased flagellar rotation to the bacterium (15, 30, 31). The resulting V. cholerae cheYD16KY109W mutant was defective for chemotaxis in swarm agar (Fig. 1_B_).
We confirmed that the cheYD16KY109W mutant exhibited CW-biased flagellar rotation by using video tracking to determine the frequency with which the bacteria change their direction of swimming. By using video tracking (32), wild-type V. cholerae was calculated to change direction on average once every 7.3 s. In contrast, the cheYD16KY109W mutant changed direction on average once every 0.2 s, thus confirming that the double point mutation in cheY causes an increased CW-biased flagellar rotation. Finally, the cheYD60N mutant changed direction on average only once every 33 s, consistent with CCW-biased flagellar rotation.
In contrast to the cheYD60N and Δ_cheY_ strains, the cheYD16KY109W strain was attenuated in vivo (Fig. 2_C_) and had an intestinal distribution similar to that of a nonmotile V. cholerae mutant (data not shown). Neither the CCW- nor CW-biased mutant strains showed any competition defect in vitro, and both grow with identical kinetics to the wild-type strain during growth in broth culture. The attenuation observed with the cheYD16KY109W mutant in vivo is in stark contrast to the 70-fold out-competition seen in the presence of CCW-biased flagellar rotation. Expression of TCP in the cheYD16KY109W mutant was equivalent to that of the wild-type strain during infection (data not shown), and therefore the attenuation is not a result of a reduction in TCP production. Both the attenuation in vivo and the nonchemotactic phenotype were abolished on reversion of the cheYD16KY109W mutant to the wild-type sequence by allelic exchange. Complementation analysis could not be performed in this instance because of the dominant negative nature of the D16K mutation (15). Thus, the out-competition phenotype exhibited by cheY mutant strains, besides requiring loss of chemotaxis, is critically dependent on the nature of the flagellar rotation and therefore either the net movement capabilities of the bacteria or the mechanical properties of the bacteria as they contact the intestinal mucosa.
CCW-Biased Rotation Results in Increased Infectivity. Although CCW-biased mutants out-compete the wild-type strain in vivo, the significance of this out-competition phenotype with respect to virulence has not been established. Early work by Freter et al. (6, 10) proposed that the ability of nonchemotactic V. cholerae to out-compete wild-type was a result of an inability of the former to enter the intestinal crypts. It was proposed that a large percentage of wild-type V. cholerae are killed by innate immune defenses within the crypts and that the out-competition of nonchemotactic V. cholerae is a direct result of avoiding such killing. Our results suggest that the ability of nonchemotactic V. cholerae to colonize throughout the length of the infant mouse small intestine, as opposed to only the distal portion, may account for the out-competition phenotype. These non-mutually exclusive models could, in either case, manifest in a lower infectious dose for nonchemotactic strains. To examine this, we determined the ID50 of the wild-type and cheYD60N strains. The ID50 of the cheYD60N mutant (150 cfu) was approximately one order of magnitude lower than that of the wild-type strain (1,700 cfu) (Fig. 3). The CCW-biased cheYD60N mutant is therefore increased for infectivity compared to the wild-type strain.
Discussion
The competitive advantage associated with CCW-biased nonchemotactic V. cholerae mutants, as well as the potential existence of a transiently CCW-biased nonchemotactic state in stool V. cholerae, makes understanding this out-competition phenotype particularly important. We showed through mutation and complementation analyses that the out-competition phenotype is intricately linked to CCW-biased flagellar rotation, because a CW-biased mutant was attenuated in vivo. In addition, we demonstrated the functional independence of TCP and nonchemotaxis in colonization: the former being required for high levels of colonization, but the latter being able to cause an out-competition phenotype even in a TCP-minus strain background. Based on these results, we conclude that it is not the failure to chemotax per se that results in the out-competition phenotype but is instead a multifactorial phenomenon requiring motility, expanded host-tissue range in the small intestine, and “head-first” smooth swimming as a result of CCW-biased flagellar rotation.
The aberrant distribution of the cheYD60N CCW-biased flagellar rotation mutant throughout the small intestine indicates that it is the process of chemotaxis that is responsible for guiding in vitro grown wild-type V. cholerae to the distal half of the infant mouse small intestine. Despite this preference, it is apparent that V. cholerae in a nonchemotactic CCW-biased state are competent for colonizing sites within the proximal small intestine. The expression of TCP and CT was induced throughout the length of the small intestine in the cheYD60N mutant, indicating that signals for induction of virulence gene expression are present throughout and also that the ability of V. cholerae to sense and respond to these signals is independent of chemotaxis. Both cheY and cheA were previously isolated in a screen for in vivo regulators of the major virulence gene regulator ToxT (9). Although mutations in both genes resulted in defects with respect to the timing of ctxA transcriptional induction, induction of ctxA reached wild-type levels by 7 h postinfection. We believe that these genes were identified in our screen because of the delay in induction of virulence gene expression coupled with the competitive advantage associated transposon insertions in these genes.
The propensity of chemotactic V. cholerae to colonize mainly the distal portion of the small intestine despite having the ability to colonize throughout suggests that the bacteria are responding to either an unknown chemoattractant or chemorepellent gradient present in the proximal small intestine. Because the small intestine of 5-day-old mice is ≈13 cm in length and because there is unidirectional flow of luminal contents down the small intestine as a result of peristalsis, we believe that such a gradient (whether chemoattractant or chemorepellent) must form between the intestinal epithelium and the lumen, as opposed to over the length of the small intestine. In this model, the concentration of chemoattractant would be higher in the lumen and lower near the epithelium, or vice versa in the case of a chemorepellent gradient. In either case, V. cholerae would chemotax away from the epithelium and concentrate in the lumen to be transported to the distal small intestine. We have found wild-type V. cholerae to be chemotactically attracted to the lining of the proximal small intestine in vitro (data not shown). These data suggest that chemorepellents are absent from the surface of the proximal small intestine, and that V. cholerae may be responding to a chemoattractant gradient that is higher in the intestinal lumen than at the epithelial surface. If V. cholerae is responding to such a gradient in the proximal small intestine, it is likely that the chemoattractants may be nutrients. It is interesting that by the time the intestinal contents reach the distal small intestine, typically the majority of nutrients have been absorbed. This may account for the propensity of V. cholerae to colonize the distal small intestine.
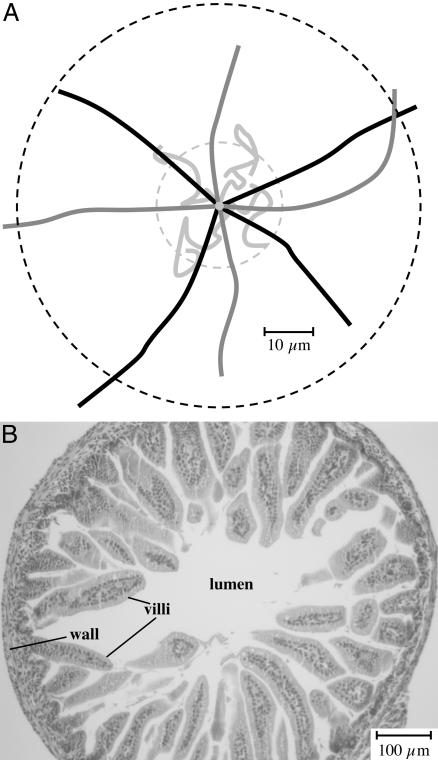
Jones et al. (33) showed that a CW-biased nonchemotactic mutant of S. enterica serovar Typhimurium was defective for adherence to epithelial cells in vitro and was attenuated for virulence in a murine infection model. In this case, the resulting unbundled peritrichous flagella are believed to act as a barrier to interaction between the bacterium and the host cell. We also observed an attenuation phenotype for V. cholerae with CW-biased flagellar rotation. However, because of the presence of but a single polar flagellum, we do not believe that steric hindrance is the explanation for the attenuation. Instead, we favor the alternative model that the frequent change of direction prevents individual bacteria from making any significant net progress from the lumen to the intestinal wall. This was confirmed by measuring the average distance and paths traveled by individual bacteria over time (Fig. 4_A_). The V. cholerae wild-type strain (black lines) and the CCW-biased mutant (dark gray lines) were both found to travel in straight runs an average distance of 39 ± 5 μm in a 1-s interval, whereas the CW-biased mutant (light gray lines) traveled in a zigzag pattern an average distance of only 13 ± 3 μm in 1 s. Taking into account the low-turn frequencies of the wild-type strain and CCW-biased mutant (one turn per 7.3 and 33 s, respectively), and extrapolating the net linear distances traveled over tens of seconds, it is clear that both of these strains are capable of easily traversing the ≈300-μm diameter of the small intestinal lumen of the infant mouse (see cross-section in Fig. 4_B_) to make contact with the intestinal villi. This is in contrast to the CW-biased strain (one turn per 0.2 s), which is confined to a small three-dimensional space within the lumen of the small intestine.
Fig. 4.
(A) Analysis of net movement by individual V. cholerae bacteria. Video images of motile V. cholerae were recorded under dark-field microscopy. The net linear distances traveled by individual bacteria over time were calculated, and the means and standard deviations are presented in the text. The mean net linear distance traveled over a 1-s interval by the CW-biased strain is indicated by the inner hatched circle, and that for the wild-type and CCW-biased strains (which were indistinguishable) is indicated by the outer hatched circle. The paths traveled by four representative bacteria of the wild-type (black lines), CCW-biased (dark gray lines), and CW-biased (light gray lines) strains are shown. Note that we have superimposed the original position of each bacterium at the center of the figure and have rotated and distributed the paths to facilitate their visualization. (B) Light micrograph (×10 magnification) of a hematoxylin/eosin (H&E)-stained cross section through the small intestine of a 5-day-old mouse. Note that the mucus gel normally overlaying the villi is not present in this micrograph.
These observations may be significant for understanding the chemotactic state and infective potential of stool V. cholerae.We have previously shown that cheW and cheR genes are repressed in stool V. cholerae, whereas other essential genes in the chemotaxis signaling pathway, including cheB paralogues, are expressed at levels comparable to that of in vitro grown chemotactic V. cholerae (11). This combination of gene expression is predicted to lead to an increase in CCW-biased flagellar rotation. In contrast, repression of cheB would have been predicted to result in a CW-biased flagellar rotation (12). In support of this model, when observed under a microscope, the motility pattern of stool V. cholerae is inconsistent with a CW-biased flagellar rotation state (data not shown). If stool V. cholerae have CCW-biased flagellar rotation, then this would explain the hyperinfectivity phenotype previously observed in the infant mouse model of infection (11). That the presence of a CCW-biased nonchemotactic state results in hyperinfectivity is highly unusual given the fact that chemotaxis is required for infection by other organisms such as Helicobacter pylori. It is intriguing to speculate that V. cholerae has evolved to use a transient CCW-biased nonchemotactic state to increase its infective potential. Whether fresh stool or CCW-biased mutant strains of V. cholerae are hyperinfectious in humans remains to be determined.
Acknowledgments
We thank S. H. Lee for constructing VC1036. This work was supported by National Institutes of Health Grants AI045746 and AI055058.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ap, ampicillin; CCW, counterclockwise; cfu, colony-forming units; CT, cholera toxin; CW, clockwise; Kn, kanamycin; Sm, streptomycin; Tc, tetracycline; TCP, toxin co-regulated type IV pilus.
References
- 1.Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R. K. & Levine, M. M. (1988) J. Exp. Med. 168**,** 1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor, R. K., Miller, V. L., Furlong, D. B. & Mekalanos, J. J. (1987) Proc. Natl. Acad. Sci. USA 84**,** 2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson, J. D., Umayam, L., et al. (2000) Nature 406**,** 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gosink, K. K., Kobayashi, R., Kawagishi, I. & Hase, C. C. (2002) J. Bacteriol. 184**,** 1767-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenbach, M. (1990) Mol. Microbiol. 4**,** 161-167. [DOI] [PubMed] [Google Scholar]
- 6.Freter, R., O'Brien, P. C. & Macsai, M. S. (1981) Infect. Immun. 34**,** 234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson, K. (1991) Infect. Immun. 59**,** 2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardel, C. L. & Mekalanos, J. J. (1996) Infect. Immun. 64**,** 2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, S. H., Butler, S. M. & Camilli, A. (2001) Proc. Natl. Acad. Sci. USA 98**,** 6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freter, R. & O'Brien, P. C. (1981) Infect. Immun. 34**,** 222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K. & Camilli, A. (2002) Nature 417**,** 642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkinson, J. S. & Houts, S. E. (1982) J. Bacteriol. 151**,** 106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnenberg, M. S. & Kaper, J. B. (1991) Infect. Immun. 59**,** 4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, S. H., Hava, D. L., Waldor, M. K. & Camilli, A. (1999) Cell 99**,** 625-634. [DOI] [PubMed] [Google Scholar]
- 15.Bourret, R. B., Hess, J. F. & Simon, M. I. (1990) Proc. Natl. Acad. Sci. USA 87**,** 41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders, D. A., Gillece-Castro, B. L., Stock, A. M., Burlingame, A. L. & Koshland, D. E., Jr. (1989) J. Biol. Chem. 264**,** 21770-21778. [PubMed] [Google Scholar]
- 17.Waldor, M. K. & Mekalanos, J. J. (1996) Science 272**,** 1910-1914. [DOI] [PubMed] [Google Scholar]
- 18.Kirn, T. J., Lafferty, M. J., Sandoe, C. M. & Taylor, R. K. (2000) Mol. Microbiol. 35**,** 896-910. [DOI] [PubMed] [Google Scholar]
- 19.Davis, B. M., Lawson, E. H., Sandkvist, M., Ali, A., Sozhamannan, S. & Waldor, M. K. (2000) Science 288**,** 333-335. [DOI] [PubMed] [Google Scholar]
- 20.Davis, B. M., Kimsey, H. H., Chang, W. & Waldor, M. K. (1999) J. Bacteriol. 181**,** 6779-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angelichio, M. J. & Camilli, A. (2002) Infect. Immun. 70**,** 6518-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenbach, M. (1996) Mol. Microbiol. 20**,** 903-910. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson, J. S. (1978) J. Bacteriol. 135**,** 45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanna, M. G., Swanson, R. V., Bourret, R. B. & Simon, M. I. (1995) Mol. Microbiol. 15**,** 1069-1079. [DOI] [PubMed] [Google Scholar]
- 25.Segall, J. E., Manson, M. D. & Berg, H. C. (1982) Nature 296**,** 855-857. [DOI] [PubMed] [Google Scholar]
- 26.Hess, J. F., Oosawa, K., Kaplan, N. & Simon, M. I. (1988) Cell 53**,** 79-87. [DOI] [PubMed] [Google Scholar]
- 27.Sanatinia, H., Kofoid, E. C., Morrison, T. B. & Parkinson, J. S. (1995) J. Bacteriol. 177**,** 2713-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao, R., Collins, E. J., Bourret, R. B. & Silversmith, R. E. (2002) Nat. Struct. Biol. 9**,** 570-575. [DOI] [PubMed] [Google Scholar]
- 29.Masduki, A., Nakamura, J., Ohga, T., Umezaki, R., Kato, J. & Ohtake, H. (1995) J. Bacteriol. 177**,** 948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scharf, B. E., Fahrner, K. A., Turner, L. & Berg, H. C. (1998) Proc. Natl. Acad. Sci. USA 95**,** 201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, X., Amsler, C. D., Volz, K. & Matsumura, P. (1996) J. Bacteriol. 178**,** 4208-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Homma, M., Oota, H., Kojima, S., Kawagishi, I. & Imae, Y. (1996) Microbiology 142**,** 2777-2783. [DOI] [PubMed] [Google Scholar]
- 33.Jones, B. D., Lee, C. A. & Falkow, S. (1992) Infect. Immun. 60**,** 2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camilli, A., Beattie, D. & Mekalanos, J. (1994) Proc. Natl. Acad. Sci. USA 91**,** 2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camilli, A. & Mekalanos, J. J. (1995) Mol. Microbiol. 18**,** 671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan, D. (1983) J. Mol. Biol. 166**,** 557-580. [DOI] [PubMed] [Google Scholar]
- 37.Morales, V. M., Backman, A. & Bagdasarian, M. (1991) Gene 97**,** 39-47. [DOI] [PubMed] [Google Scholar]