The GTPase-Activating Enzyme Gyp1p Is Required for Recycling of Internalized Membrane Material by Inactivation of the Rab/Ypt GTPase Ypt1p (original) (raw)
Abstract
Rab/Ypt GTPases are key regulators of membrane trafficking and together with SNARE proteins mediate selective fusion of vesicles with target compartments. A family of GTPase-activating enzymes (GAPs) specific for Rab/Ypt GTPases has been discovered, but little is known about their function and substrate specificity in vivo. Here we show that the GAP activity of Gyp1p, a yeast member of this family, is specifically required for recycling of the SNARE Snc1p and the membrane dye FM4-64, implying that inactivation of a Rab/Ypt GTPase may be necessary for recycling of membrane material. Interestingly, recycling of GFP-Snc1p in _gyp1_Δ cells is partially restored by reducing the activity of Ypt1p. Moreover, GFP-Snc1p accumulated intracellularly in wild-type cells expressing a GTP-locked, mutant form of Ypt1p (Ypt1p-Q67L), suggesting that GTP hydrolysis of Ypt1p is essential for recycling. Ypt6p is known to be required for the fusion of recycling vesicles to the late Golgi compartment. Interestingly, the deletions of GYP1 and YPT6 were synthetic lethal, raising the possibility that at least two distinct pathways are involved in recycling of membrane material.
Internalized proteins travel through two morphologically and biochemically distinct organelles called early and late endosomes. After arrival at the endosome, membrane proteins and lipids (solutes) are either directed to the lysosomal or vacuolar compartment where they are degraded or returned to the plasma membrane as in the case of recycling receptors and bulk membrane constituents. Recycling of many signaling receptors back to the plasma membrane allows multiple rounds of ligand binding and internalization. In the yeast Saccharomyces cerevisiae, several endocytosed proteins have been shown to undergo recycling. For example, pheromone-induced internalization of Ste3p results in its recycling back to the plasma membrane (8, 9), while the chitin synthase Chs3p translocates between sites of chitin deposition on the cell surface and an internal structure called the chitosome (42, 47). Moreover, recycling of the exocytic SNARE Snc1p, which is involved in fusion of Golgi compartment-derived secretory vesicles with the plasma membrane, allows reutilization of Snc1p for several round of secretion (22). Despite a detailed morphological analysis of cargo transport through endosomal compartments, the machinery and molecular mechanisms underlying plasma membrane protein sorting and recycling are still largely unknown.
Correct targeting of vesicles to the acceptor compartment by SNAREs and Rab/Ypt GTPases is of fundamental importance to ensure specificity among trafficking pathways (33). Available evidence suggest that Rabs and their recruited effectors function in multiple steps including cargo selection, vesicle budding, movement, docking, and fusion (32, 33, 46). The yeast genome encodes 11 Ypt GTPases compared to more than 60 in mammalian cells, and they are thought to regulate distinct trafficking steps. Rab/Ypt GTPases exist in an active GTP- and an inactive GDP-bound conformation, and this cycle is facilitated by guanine-nucleotide exchange factors that stimulate both GDP loss and GTP uptake as well as GTPase-activating enzymes (GAPs) that catalyze GTP hydrolysis. A family of Rab-specific GAPs termed GYPs (GAP for Ypts) was discovered in yeast, but homologues also exist in other species (1, 12, 37). Most GAPs of the GYP family display broad and overlapping substrate specificity when assayed in vitro. For example, Gyp1p increases GTP hydrolysis of Ypt1p, Sec4p, Ypt51p, and Ypt7p (1, 12, 37). Moreover, Gyp1p, Gyp5p and Gyp8p are all able to inactivate Ypt1p GTP in vitro (1, 11). The substrate specificity of Gyps is less clear in vivo. Thermosensitive growth defect of _gyp1_Δ cells is rescued by partial loss of function of YPT1, suggesting that Gyp1p functions as a GAP in early secretion for Ypt1p in vivo (13). Recently, it was shown that the double mutant gyp3/_msb3_Δ _gyp4/msb4_Δ exhibits secretion defects, most likely because Gyp3p and Gyp4p regulate the Ypt Sec4p (17). However, as yeast cells with deletions of any single GYP-GAPs are viable and exhibit only minor characterized trafficking defects, it is thought that the GYPs are functionally redundant in vivo (11) or, alternatively, that GTP hydrolysis may not be required for Rab/Ypt function (30).
In yeast, the Rab family GTPase Ypt6p and its heterodimeric guanine-nucleotide exchange factor Ric1p/Rgp1p appears to play key roles for recycling of Snc1p (34). Ypt6p may be regulated by the GAP Gyp2p (21) and a complex containing Rcy1p and Skp1p (16, 45). Ypt6p mediates fusion of endosomal vesicles with the Golgi compartment by recruiting a multisubunit tethering factor termed VFT (Vps Fifty Three)/GARP complex (34-36). Interestingly, the VFT subunit Vps51p is able to link this VFT complex to the SNARE Tlg1p (10, 36), which is required for recycling of Chs3p and Snc1p (20, 22). These results led those authors to conclude that the recruited VFT tethering factors promote SNARE assembly, and thereby directly link the tethering and fusion events. Finally, retrieval of Snc1p from early endosomes has recently been shown to be dependent on three sorting nexins Snx4p, Snx41p, and Snx42p (19).
In this study, we investigated the role of the known Gyp family members for a potential function in recycling pathways. Interestingly, we found that _gyp1_Δ cells are defective for recycling of GFP-Snc1p and FM4-64, most likely because they accumulate activated Ypt1p. Our results thus suggest that hydrolysis of Ypt1p may be important for efficient recycling of membrane material.
MATERIALS AND METHODS
Strais constructions and genetic manipulations.
Yeast strains are described in Table 1. Standard yeast growth conditions and genetic manipulations were used (18). Strains with deletions of GYP1 were constructed as described (24).
TABLE 1.
Yeast strains used in this study
| Strain name | Relevant genotype | Source |
|---|---|---|
| BY4741 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 | Euroscarf |
| YCL 248 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::KanMX4 | Euroscarf |
| YCL 246 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp2::KanMX4 | Euroscarf |
| YCL 250 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp3::KanMX4 | Euroscarf |
| YCL 252 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp4::KanMX4 | Euroscarf |
| YCL 281 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp5::KanMX4 | Euroscarf |
| YCL 254 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp6::KanMX4 | Euroscarf |
| YCL 282 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp7::KanMX4 | Euroscarf |
| YCL 283 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp8::KanMX4 | Euroscarf |
| YCL 154 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 ypt6::KanMX4 | Euroscarf |
| YCL 267 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 vps5::KanMX4 | Euroscarf |
| YCL 316 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gos1::KanMX4 | Euroscarf |
| YCL 222 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::HIS3 ypt7::KanMX4 | This study |
| YCL 223 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::HIS3 ypt10::KanMX4 | This study |
| YCL 230 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::HIS3 ypt11::KanMX4 | This study |
| YCL 224 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::HIS3 ypt51::KanMX4 | This study |
| YCL 225 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::HIS3 ypt52::KanMX4 | This study |
| YCL 226 | matahis3_Δ_1 leu2_Δ_0 met15_Δ_0 ura3_Δ_0 gyp1::HIS3 ypt53::KanMX4 | This study |
| YCL 192 | _mat_α ura3-52 his3_-Δ_200 leu2-3,112 | P. Novick |
| YCL 193 | _mat_α ura3-52 his3_-Δ_200 leu2-3,112 ypt1-2 | P. Novick |
| YCL 209 | _mat_α ura3-52 his3_-Δ_200 leu2-3,112 ypt1-2 gyp1::HIS3 | This study |
| YCL 210 | _mat_α ura3-52 his3_-Δ_200 leu2-3,112 gyp1::HIS3 | This study |
| YCL 262 | matahis4-539 lys2-801 ura3-52 | N. Segev |
| YCL 263 | matahis4-539 lys2-801 ura3-52 ypt1-Q67L | N. Segev |
| YGY 06 | _mat_α gyp1::_natMX mf_α::MF_α_1pr-LEU2 can1::MFA1pr-HIS3 lys_Δ_0 | This study |
DNA manipulations.
Plasmids used in this study are described in Table 2. Standard procedures were used for recombinant DNA manipulations (3). PCRs were performed with the Expand polymerase kit as recommended by the manufacturer (Roche). Oligonucleotides were synthesized by Proligo (France); sequences are available upon request.
TABLE 2.
Plasmids used in this study
| Plasmid name | Relevant characteristics | Source |
|---|---|---|
| pJMG 118 | GFP-SNC1 URA3 CEN | H. Pelham |
| pJMG 122 | GFP-SNC1-pem URA3 CEN | H. Pelham |
| pCL 174 | GYP1 LEU2 CEN | P. Novick |
| pCL 175 | GYP1(212-637) LEU2 CEN | P. Novick |
| pCL 178 | GYP1(212-620) LEU2 CEN | P. Novick |
| pCL 180 | GYP1(212-637 R243K) LEU2 CEN | P. Novick |
| pCL 140 | GFP-TLG1 LEU2 | H. Pelham |
| pCL 141 | GFP-TLG2 URA3 | H. Pelham |
| Yep 352fF | End-FUR4 URA3 2μm | R. Haguenauer-Tsapis |
Antibodies, Western blots, and microscopy.
Standard procedures were used for yeast cell extract preparation and immunoblotting (15). Monoclonal antibodies against GFP, Pgk1p (Roche), Vps10p and carboxypeptidase Y (CPY) (Molecular Probes) were used as recommended by the manufacturer. Immunoblots were quantified using the NIH image software.
For microscopy, cells were grown at 30°C to early log phase and visualized on a Zeiss Axiophot fluorescence microscope with a Photometrics charge-coupled device camera. GFP-tagged proteins were visualized using a Chroma GFPII filter (excitation wavelength, 440 to 470 nm). For quantification, at least 200 cells were counted. Where indicated, the actin polymerization inhibitor Latrunculin-A (LAT-A) (200 μM final concentration in dimethyl sulfoxide [DMSO]) or the solvent DMSO was added 10 min before crude extract preparation. Aliquots of the culture were treated with rhodamine-phalloidin (Molecular Probes) to check the disruption of actin cytoskeleton upon LAT-A addition. For FM4-64 localization experiments, yeast cells expressing or not expressing GFP-Snc1p were allowed to internalize FM4-64 for 12 min at 24°C and were then washed three times with ice-cold synthetic dextrose (SD) medium (18). The cells were then incubated for 0 to 10 min in SD medium at 30°C, and internalized FM4-64 was visualized using a rhodamine filter (Zeiss FS1). Vacuolar staining was obtained by incubating cells grown to early log phase with 0.1% CMAC (Molecular Probes) at 30°C for 1 h and the cells were then visualized with a DAPI (4′,6′-diamidino-2-phenylindole) filter (Zeiss FS20).
Uracil uptake assay and Fur4p degradation.
The endocytosis and subsequent vacuolar degradation of Fur4p was measured as described previously (14). At time zero exponentially growing cells transformed with Yep352fF (end-Fur4 on a 2μm/URA3 plasmid) were treated with cycloheximide (CX) (final concentration, 50 μg/ml). The uracil uptake was measured, and crude extracts were prepared each 30 min after addition of CX. Uracil uptake was measured as previously described. Yeast culture (1 ml) were incubated with 5 μM [14C]uracil (Amersham) for 20 s at 30°C and quickly filtered through Whatman GF/C filters. Filters were washed twice with ice-cold water and then counted for radioactivity. We used a polyclonal antibody raised against the last 10 residues of Fur4p to detect the permease by western immunoblotting.
Determination of Vps10p half-life.
Cultures were grown to early log phase in rich medium at 30°C, at which time CX (Sigma) was added to a final concentration of 50 μg/ml. Aliquots were collected at the times indicated, and protein levels were analyzed by immunoblotting with specific antibodies.
Detection of secreted CPY.
For the detection of secreted CPY, 10 μl of exponentially growing cells were spotted onto yeast-peptone-dextrose (YPD) medium and grown for 2 days in contact with a 0.45-mm-pore-size nitrocellulose membrane. The filter was removed and washed with distilled water, and CPY was immunologically detected as previously described (31).
FM4-64 recycling assays.
FM4-64 recycling assays were performed as described previously (45). Briefly, yeast cells were allowed to internalize FM4-64 for 12 min at 24°C, and washed three times with ice-cold SD medium. After the last wash, the cells are resuspended in 10 μl of SD medium and kept on ice. Prewarmed SD medium at 30°C was added to the cells and fluorescence was recorded during 600 s.
SGA analysis of GYP1.
Synthetic genetic array (SGA) analysis was performed with _gyp1_Δ cells essentially as described by Tong et al. (39) using a Beckman Biomek FX robot. All identified mutants were confirmed by random sporulation or tetrad analysis (39). Representation of the interactors was performed with the Osprey program (7). Classification of the interactors and additional interactions between them were added based on data from GRID (6) and were manually refined with published data.
RESULTS
The GAP Gyp1p is required for efficient recycling of the SNARE Snc1p and FM4-64.
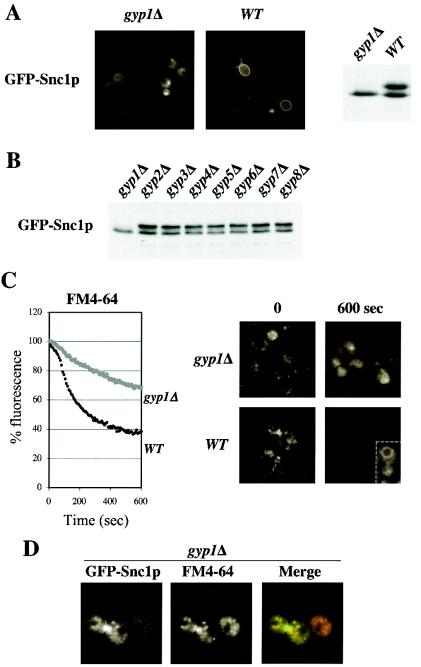
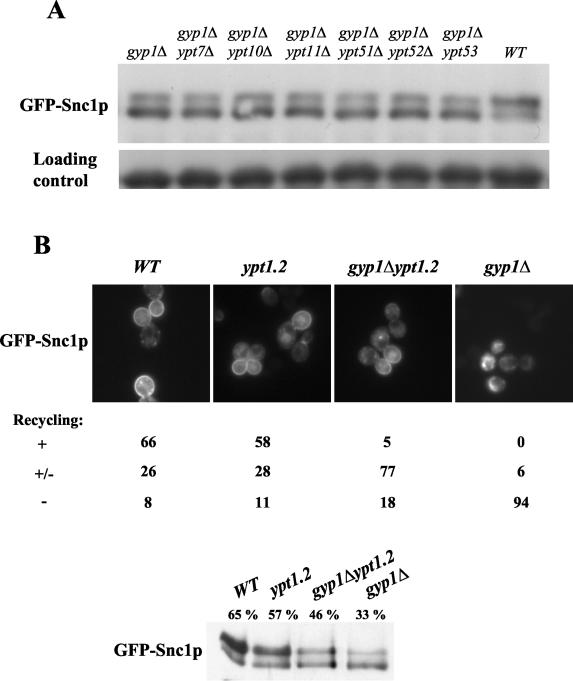
The localization and phosphorylation state of GFP-Snc1p provide a convenient marker of recycling. In wild-type cells GFP-Snc1p is mostly localized at the plasma membrane in a phosphorylated state, while it accumulates in internal structures in an underphosphorylated state in cells deficient for recycling (16, 22). Recycling can also be quantified by measuring the secretion of previously internalized fluorescent dye FM4-64 (45). We used these two assays to investigate possible functions of the known Rab/Ypt-GAPs for recycling of membrane material. Interestingly, GFP-Snc1p was predominantly intracellular and accumulated in its underphosphorylated form in _gyp1_Δ cells (Fig. 1A), while it was found mostly located at the plasma membrane in wild-type cells and strains with deletions of GYP2 to GYP8 (Fig. 1B and data not shown). In addition, _gyp1_Δ cells also exhibited a recycling defect of the fluorescent dye FM4-64 (Fig. 1C), suggesting a general role of Gyp1p in recycling. While in wild-type cells internalized FM4-64 was rapidly recycled, it accumulated in punctate structures in _gyp1_Δ cells (Fig. 1C, right panels), which could correspond to either endosomes or fragmented vacuoles (see below). After longer exposure, the remaining FM4-64 prominently stained the vacuolar membrane of wild-type cells (inset). Interestingly, GFP-Snc1p colocalized at least partially with FM4-64 at the beginning of the time course (Fig. 1D), suggesting that GFP-Snc1p is likely to accumulate in endosomes in _gyp1_Δ cells.
FIG. 1.
Gyp1p is required for recycling of the SNARE Snc1p and FM4-64. (A) Wild-type (BY4741) and _gyp1_Δ (YCL248) cells transformed with GFP-Snc1p were grown at 30°C in selective SD medium to early log phase and analyzed by fluorescence microscopy (left panels) and immunoblotting with GFP-antibodies (right panel). Note that GFP-Snc1p is intracellular and underphosphorylated in _gyp1_Δ cells, characteristic of a recycling defect. (B) The phosphorylation state of GFP-Snc1p was examined by immunoblotting with GFP-antibodies in the indicated strains grown at 30°C to early log phase in selective SD medium. The following strains were analyzed: _gyp1_Δ (YCL248), _gyp2_Δ (YCL246), _msb3_Δ/_gyp3_Δ (YCL250), _msb4_Δ/_gyp4_Δ (YCL252), _gyp5_Δ (YCL281), _gyp6_Δ (YCL254), _gyp7_Δ (YCL282), and _gyp8_Δ (YCL283). (C) Wild-type (BY4741) and _gyp1_Δ (YCL248) cells were analyzed for their ability to recycle the membrane dye FM4-64 as described in Materials and Methods. Fluorescence microscopy pictures were taken at the beginning and the end of the time course (right panels). The cell shown in the inset was exposed longer to visualize staining of FM4-64 at the vacuolar membrane. (D) _gyp1_Δ cells expressing GFP-Snc1p were incubated with FM4-64 for 12 min, washed and analyzed by fluorescence microscopy. Note that GFP-Snc1p partially colocalizes with structures stained by FM4-64.
The GAP activity of Gyp1p is required for its recycling function.
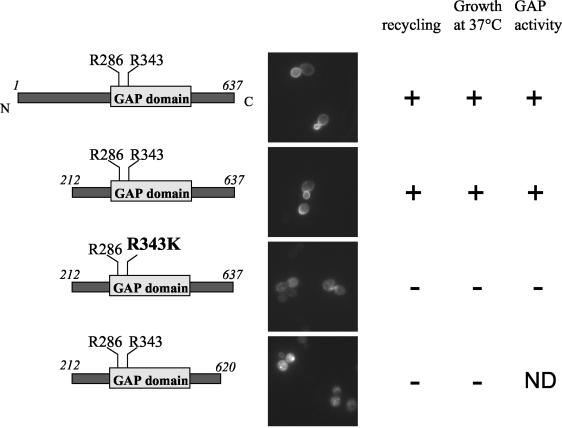
Gyp1p contains a large amino-terminal region and a GAP-domain at its carboxy terminus (amino acids 280 to 502) (Fig. 2), which contains a conserved arginine-residue (arginine 343) typically required for the catalytic activity of GAPs (arginine finger) (2). To determine which domains of Gyp1p are important for its recycling function, _gyp1_Δ cells expressing either wild-type or mutant versions of Gyp1p from the endogenous promoter were tested for their ability to recycle GFP-Snc1p (Fig. 2). Interestingly, truncated Gyp1p containing the catalytic GAP domain but missing the amino-terminal domain (Gyp1p-212-637) could rescue the recycling defect of _gyp1_Δ cells. In contrast, a single mutation affecting the arginine-finger (Gyp1p-R343K) within the GAP domain completely abolished the ability to complement the recycling defect (Fig. 2 and data not shown). In contrast, mutation of arginine 286 did not impair recycling, as it did not impair GAP activity (data not shown) (2). Surprisingly, the very C-terminal domain of Gyp1p (amino acids 620 to 637) was required for its recycling function. Based on the domain structure of Gyp1p (Fig. 2) it is unlikely that this domain is required for GAP activity. As this C-terminal domain is poorly conserved among yeast Gyps but displays good homology between likely Gyp1p orthologs (see Fig. A1), we speculate that it may be involved in the correct subcellular localization of Gyp1p or may contribute to its specificity for a given target Ypt GTPase. The ability of Gyp1p mutants to recycle GFP-Snc1p also correlated with their previously reported ability to rescue the temperature-sensitive growth defect of _gyp1_Δ cells (Fig. 2) (13), suggesting that the ability to grow at high temperature and the recycling function of Gyp1p may be linked. Taken together, we conclude that GAP activity of Gyp1p is essential for its recycling function, implying that catalyzed inactivation of an unknown GTPase is involved.
FIG. 2.
The GAP activity of Gyp1p is required for its recycling function. gyp1Δ (YCL248) cells expressing GFP-Snc1p (pJMG118) were transformed with plasmids encoding either wild-type or Gyp1p mutant proteins. The tested mutants are schematically indicated on the left. The cells were grown at 30°C in selective SD medium to early log phase, and analyzed by GFP microscopy. The ability of wild-type and the indicated Gyp1p-mutants to restore recycling of GFP-Snc1p was compared to their previously reported growth at 37°C and GAP activity in vitro (2, 13). ND, not determined. Symbols: +, wild-type activity; −, no activity.
Endocytosis and secretion are not affected in _gyp1_Δ cells.
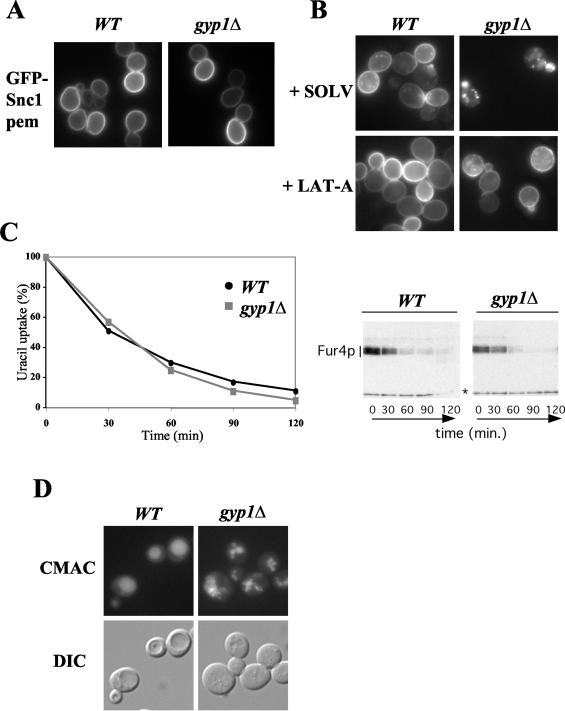
The intracellular accumulation of GFP-Snc1p could result from a defect in late secretion, from increased endocytosis, or from a defect in a recycling pathway between endosomes and the Golgi compartment. To distinguish between these possibilities, we first examined secretion using GFP-Snc1p-pem, a version of Snc1p mutated in its internalization signal (22). As shown in Fig. 3A, GFP-Snc1p-pem was found exclusively at the plasma membrane both in wild-type and _gyp1_Δ cells, indicating that it is efficiently targeted to the plasma membrane. Moreover, the targeting of Fur4p, Pma1p and Gas1p at the plasma membrane was normal in _gyp1_Δ cells (data not shown). Together, these results demonstrate that Gyp1p is not required for efficient secretion of various cargoes. Consistent with these results, treatment of _gyp1_Δ cells with low levels of the actin depolymerization drug LAT-A, which predominantly blocks endocytosis (42), resulted in an accumulation of GFP-Snc1p at the plasma membrane (Fig. 3B).
FIG. 3.
Secretion and endocytosis are not altered in _gyp1_Δ cells. (A) Wild-type (BY4741) and _gyp1_Δ (YCL248) cells transformed with a plasmid expressing internalization-defective GFP-Snc1p-pem (pJMG122) were grown at 30°C in selective SD medium to early log phase and analyzed by GFP microscopy. (B) The subcellular localization of GFP-Snc1p (pJMG118) was analyzed in wild-type (BY4741) and _gyp1_Δ (YCL248) cells, treated for 10 min with 200 μM LAT-A (+LAT-A) or DMSO (+SOLV). Note that GFP-Snc1p relocated to the plasma membrane in _gyp1_Δ cells treated with LAT-A. (C) Wild-type (BY4741) and _gyp1_Δ (YCL248) cells transformed with a multicopy plasmid encoding Fur4p under its own promoter (Yep352fF) were grown to early log phase. CX was added to a final concentration of 50 μg/ml, and the uracil uptake (left graph) and Fur4p degradation (right panel) were followed over time as described in Materials and Methods. For a loading control we used the marked unspecific band (*). (D) The morphology of the vacuole was examined by fluorescence microscopy in wild-type (BY4741) and _gyp1_Δ (YCL248) cells incubated with CMAC for 1 h. The corresponding DIC images are shown below.
To exclude that the increased intracellular levels of GFP-Snc1p in _gyp1_Δ cells result from faster internalization, we measured the internalization kinetics of the fluid phase dye lucifer yellow (data not shown) and the uracil permease Fur4p. For this purpose, wild-type and _gyp1_Δ cells expressing Fur4p were treated with CX, which is known to trigger rapid internalization and subsequent degradation of the permease in the vacuole (14). As shown in Fig. 3C, the decrease in uracil uptake after CX addition was comparable in wild-type and _gyp1_Δ cells, implying that the internalization step is not affected. Moreover, the rate of vacuolar degradation of Fur4p was not altered in _gyp1_Δ cells, although _gyp1_Δ cells displayed strongly fragmented vacuoles (Fig. 3D), suggesting that trafficking of Fur4p to the vacuole occurs normally. We conclude from these results that Gyp1p is specifically involved in recycling of membrane material, and is not generally involved in secretion or endocytosis of membrane proteins.
_gyp1_Δ cells are impaired in retrograde traffic from endosomes to the Golgi compartment.
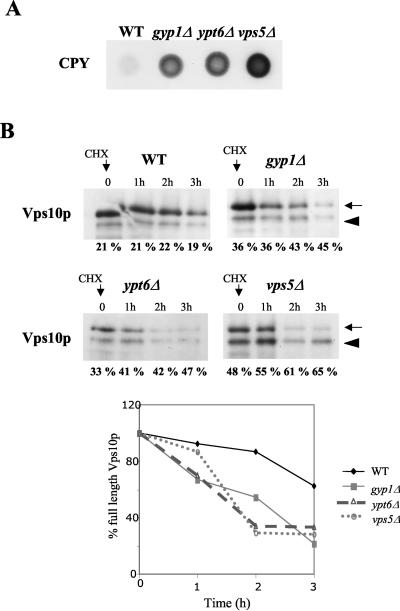
To test whether Gyp1p may be involved in trafficking to the Golgi compartment, we analyzed the sorting of the vacuolar CPY, which undergoes N glycosylation in the Golgi compartment before being transported to the vacuole. In cells defective for anterograde Golgi compartment-to-endosome or retrograde endosome-to-Golgi compartment trafficking, part of CPY is secreted in a nonmaturated form. Consistent with previous results (5, 13), CPY was secreted in _gyp1_Δ and _ypt6_Δ cells, although the effect was less pronounced compared to the retromer mutant _vps5_Δ (Fig. 4A). This observation could be explained by a defect in retrograde transport from the prevacuolar compartment (PVC) to the Golgi compartment, which leads to a mislocalization of Vps10p, the receptor of CPY. To test this possibility, we followed degradation of Vps10p after the addition of the translation inhibitor CX to the culture medium. As shown in Fig. 4B, the half-life of Vps10p was significantly reduced in _gyp1_Δ, _ypt6_Δ and _vps5_Δ cells. Quantification of these results revealed that the half-life of Vps10p was less than two hours (lower graph) in these mutant cells, while Vps10p remained almost stable over the entire time course in wild-type cells. Moreover, the cleaved form of Vps10p slightly accumulated in _gyp1_Δ, _ypt6_Δ, and _vps5_Δ cells compared to wild-type cells (expressed as a percentage of the total amount of Vps10p), a characteristic feature for increased vacuolar targeting (28). Based on these results, we conclude that the trafficking route from the Golgi compartment to the PVC and the retrograde transport from the PVC to the Golgi compartment are partially impaired in _gyp1_Δ and _ypt6_Δ cells.
FIG. 4.
_gyp1_Δ cells are impaired in endosome-to-Golgi compartment retrograde traffic. (A) Wild-type (BY4741) and _gyp1_Δ (YCL248) cells were spotted on a nitrocellulose membrane on YPD plates (105 cells/spot). After 48 h at 30°C, secreted CPY was detected with a monoclonal CPY antibody as described in Materials and Methods. (B) The half-life of Vps10p was determined by CX (CHX) chase in wild-type (BY4741), _gyp1_Δ (YCL248), _vps5_Δ (YCL267), and _ypt6_Δ (YCL154) cells. Cells were grown in YPD medium to mid-log phase, and CX was added (time zero) to a final concentration of 50 μg/ml. Proteins extracts were prepared at the indicated times after CX addition and analyzed by SDS-PAGE and immunoblotting with a monoclonal antibody directed against Vps10p. The arrow marks the position of full-length Vps10p, while the arrowhead points to the truncated form of Vps10p, which is cleaved in the vacuole. The bands were quantified, and the level of the lower band was expressed as a percentage of the total Vps10p signal. The decrease of full-length Vps10p was quantified and plotted as the ratio of residual intensity compared to the initial one expressed in percentage over time (in hours).
The ypt1-2 mutant allele partially rescues the recycling defect of _gyp1_Δ cells.
Since the GAP activity of Gyp1p is required for its recycling function, it is likely that hyperactivation of one or several Ypt GTPases is responsible for this defect. We thus expected that reduction of this activated Ypt GTPase should suppress the recycling defect of _gyp1_Δ cells. In vitro, Gyp1p was shown to display GAP activity on Sec4p, Ypt1p, Ypt7p, and Ypt51p (2, 12). Note that Ypt52p, Ypt53p, Ypt10p, and Ypt11p have not been tested in Gyp1p GAP assays in vitro. Deletion of neither one of the above nonessential Ypts was able to rescue the recycling defect of _gyp1_Δ cells (Fig. 5A), suggesting that their hyperactivation was not singly responsible for the observed recycling defect. To test a possible role of the essential Rab GTPase Ypt1p, we deleted GYP1 in a strain harboring the partial loss of function allele _ypt1_-2. This mutant exhibits a defect in GTP binding and _ypt1_-2 cells are partially defective for CPY secretion (4). Moreover, the _ypt1_-2 allele has been shown to rescue the temperature-sensitive growth defect of _gyp1_Δ cells on synthetic medium (13). Importantly, recycling of GFP-Snc1p in _gyp1_Δ ypt1-2 double mutants was partially rescued compared to _gyp1_Δ cells (Fig. 5B). Quantification of cells expressing GFP-Snc1p by microscopy revealed that recycling was at least partially restored in over 80% of _gyp1_Δ ypt1-2 cells, implying that the recycling defect observed in _gyp1_Δ cells could be caused by increased levels of Ypt1p-GTP.
FIG. 5.
The ypt1-2 mutant allele partially rescues the recycling defect of _gyp1_Δ cells. (A) _gyp1_Δ (YCL248), _gyp1_Δ _ypt7_Δ (YCL222), _gyp1_Δ _ypt10_Δ (YCL223), _gyp1_Δ _ypt11_Δ (YCL230), _gyp1_Δ _ypt51_Δ (YCL224), _gyp1_Δ _ypt52_Δ (YCL225), _gyp1_Δ _ypt53_Δ (YCL226), and wild-type (BY4741) cells were transformed with a plasmid expressing GFP-Snc1p (pJMG118), grown in selective SD medium to early log phase, and the phosphorylation state of GFP-Snc1p was examined by Western blot using an antibody raised against GFP. Note that GFP-Snc1p is hyperphosphorylated only in wild-type cells. Pgk1p was used as a loading control. (B) Wild-type (YCL192), _ypt1_-2 (YCL193), _gyp1_Δ ypt1-2 (YCL209), and _gyp1_Δ (YCL210) cells expressing GFP-Snc1p (pJMG118), were analyzed by GFP-fluorescence microscopy and by Western blotting. For each strains, the cells were quantified by microscopy and expressed as percentage of the total number of cells according to their complete (+), partial (±), or absent (−) GFP-Snc1p recycling. Moreover, the bands obtained by Western blot were quantified, and the level of the upper band was expressed as a percentage of the total GFP-Snc1p signal. Note that the plasma membrane localization of GFP-Snc1p is partially recovered in _gyp1_Δ ypt1-2 compared to _gyp1_Δ cells.
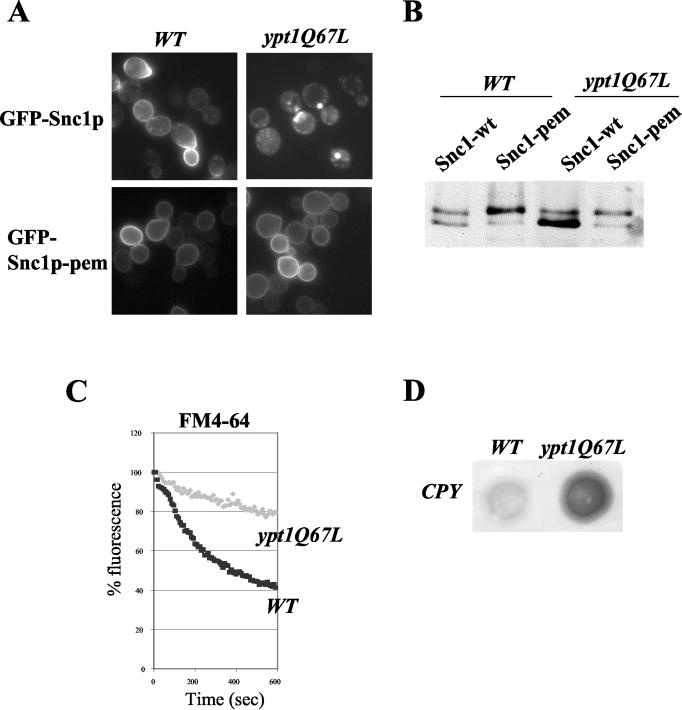
Cells expressing Ypt1p-Q67L exhibit a recycling defect.
If hyperactive Ypt1p interferes with recycling, we predict that expression of a hyperactive, GTPase deficient mutant form of Ypt1p (Ypt1p-Q67L) (30) should similarly cause a defect in recycling. Indeed, _ypt1_Δ cells expressing Ypt1p-Q67L were unable to recycle GFP-Snc1p (Fig. 6A and B) and the membrane dye FM4-64 (Fig. 6C). Like _gyp1_Δ, we observed that cells expressing Ypt1p-Q67L secrete comparatively more CPY into the medium than wild-type (Fig. 6D), consistent with a defect in retrograde transport of Vps10p from the late endosome to the Golgi compartment. However, GFP-Snc1p-pem accumulated efficiently at the plasma membrane (Fig. 6A and B), demonstrating that secretion is not affected in these cells as previously reported based on endoplasmic reticulum (ER)-to-Golgi compartment trafficking of CPY (30). In contrast, GFP-Snc1p was efficiently recycled in cells expressing Sec4p-Q79L (data not shown), suggesting that the observed trafficking phenotype was specific and not a general consequence of activated Ypt/Rab GTPases implicated in secretion. Together these results imply that in addition to its known role in ER-to-Golgi compartment transport, Ypt1p may also be involved in recycling of membrane material.
FIG. 6.
Ypt1pQ67L expressing cells exhibit a recycling defect. (A and B) The subcellular localization (panel A) and phosphorylation state (panel B) of GFP-Snc1p (pJMG118) and GFP-Snc1p-pem (pJMG122) was analyzed in wild-type (YCL262) and ypt1Q67L (YCL263) cells, grown in selective SD medium to early log phase. (C) Wild-type (YCL262) and ypt1Q67L (YCL263) cells were analyzed for their ability to recycle the membrane dye FM4-64. (D) Wild-type (YCL262) and ypt1Q67L (YCL263) cells were grown on YPD plates (105 cells by spot) for 48 h in contact with nitrocellulose membrane. Secreted CPY was detected on the membrane by probing with a monoclonal antibody directed against CPY.
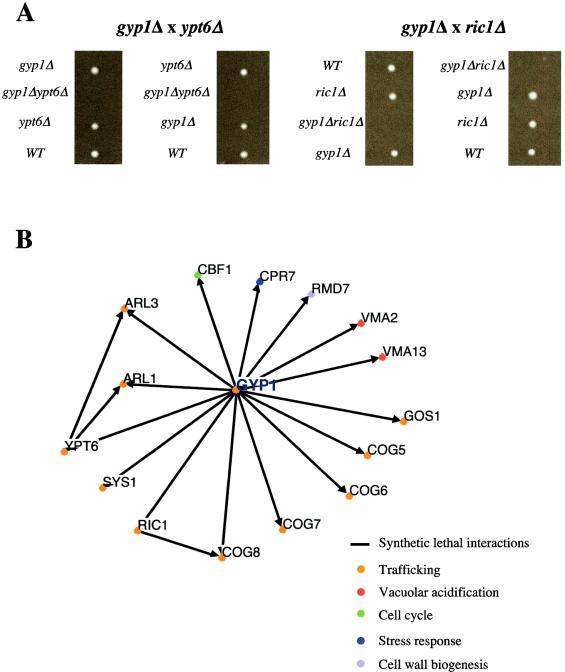
GYP1 is synthetic-lethal with multiple components involved in recycling.
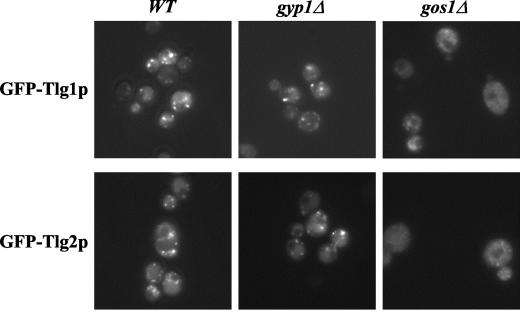
_gyp1_Δ and _ypt6_Δ cells are viable, suggesting that either recycling of membrane proteins is not an essential process or, alternatively, that redundant trafficking routes may exist. We thus searched for genes that are required for viability in the absence of GYP1. For this purpose, _gyp1_Δ cells were crossed with all viable strains of the available yeast knockout collection (Euroscarf), and the resulting double mutants were selected and analyzed as described previously by Tong et al. (39). A total of 201 positive hits were identified and synthetic-lethality with 15 interactors (Fig. 7) and synthetic-sickness with 11 interactors (data not shown) was subsequently confirmed by random sporulation and/or tetrad analysis (Fig. 7A and data not shown). The identified synthetic-lethal interactions with _gyp1_Δ are shown in Fig. 7B (a complete list is presented in Tong et al. [40]). Interestingly, we found that GYP1 and YPT6 deletions were synthetically lethal, implying that these two genes may function in distinct trafficking pathways. GYP1 deletion was also synthetic lethal with deletions of RIC1, ARL1, and ARL3, which encode for proteins that function as activators or downstream effectors of Ypt6p (27, 34). These genetic results suggest that Gyp1p must have functions that are not shared by the Ypt6p pathway, indicating that Gyp1p may function at a novel step during recycling of membrane proteins. In addition, _gyp1_Δ showed synthetic-lethal interactions with deletions of other genes coding for proteins involved in trafficking, including several components of the COG complex (Sec34/35 complex), which was shown to be required for recycling of Snc1p (43). Likewise, _gyp1_Δ is synthetic lethal with the recycling mutant _gos1_Δ (Fig. 7B) (35) in which the SNAREs Tlg1p and Tlg2p are mislocalized (35) (Fig. 8). However, the localization of Tlg1p and Tlg2p was not altered in _gyp1_Δ cells (Fig. 8), suggesting that the recycling defect of _gyp1_Δ cells is not due to mislocalization of the SNAREs.
FIG. 7.
Synthetic lethal interactions with GYP1. (A) Dissection of tetrads from heterozygous diploids revealed synthetic-lethal interactions between GYP1 and YPT6 or RIC1 deletions. The genotype of the germinating spores was determined by plating them on media containing Nourseothricin (Nat) or Geneticin (G418). (B) Genetic interaction network of the synthetic-lethal interactions identified by the SGA analysis. The genes that were identified as synthetic lethal with GYP1 are represented as nodes. Analysis and representation of the interactors was performed with the use of the Osprey program.
FIG. 8.
GFP-Tlg1p and GFP-Tlg2p are correctly localized in _gyp1_Δ cells. The subcellular localization of GFP-Tlg1p and GFP-Tlg2p was analyzed by fluorescence microscopy in wild-type (BY4741), _gyp1_Δ (YCL248), and _gos1_Δ (YCL316) cells. Note the punctate staining of Tlg1p and Tlg2p in _gyp1_Δ and wild-type cells compared to the hazy staining observed in _gos1_Δ cells.
DISCUSSION
We report here that Gyp1p is required for recycling of GFP-Snc1p and the lipophilic dye FM4-64, most likely because of the accumulation of activated Ypt1p in _gyp1_Δ cells. Our data imply that both activation and inactivation of Rabs may be crucial for some steps in membrane trafficking and suggest that other Gyps may similarly regulate specific trafficking routes.
The role of Gyps for trafficking.
Surprisingly, the major trafficking routes are normal in cells lacking any single GAP of the Gyp family. The absence of dramatic trafficking phenotypes has been explained by the fact that the Gyps function in a redundant manner. Indeed, only cells with deletions of MSB3 and MSB4 exhibit a strong secretion defect, probably because of the accumulation of Sec4p GTP (17). Consistent with overlapping functions, Gyp1p, Gyp5p, and Gyp8p are all able to stimulate GTP hydrolysis of Ypt1p in vitro (11, 12). However, the results presented here demonstrate that the GAP activity of Gyp1p is specifically required for recycling of membrane material in vivo. Moreover, previous findings suggest a specific role of Gyp2p as a negative regulator of Ypt6p (21), indicating that the Gyps may be more specific in vivo than in vitro. Gyp1p localizes predominantly to the Golgi compartment (11, 13), while Gyp5p is cytosolic and Gyp8p localizes to few large structures that do not colocalize with Gyp1p (11). It is thus possible that subcellular compartmentalization of Gyp GAPs may contribute to their substrate specificity in vivo. Given that GAPs of the Gyp family remained highly conserved during evolution (see Fig. A1), it will be interesting to test whether human Gyp1p homologues are also required for recycling.
FIG. A1.
Sequence alignment of Gyp1p with its closest homologues in other species. Sequence homology between Gyp1p from S. cerevisiae (Sc) and putative orthologs in S. pombe, SPBC530.01 (Sp); C. elegans, F32B6.8 (Ce); mouse, MGC61359 (Mm); and human, C22orf4 (Hs). Sequences were aligned with ClustalW and displayed using Boxshade. Residues are shaded if identical (black) or conserved (grey) in at least three proteins. The bracket indicates the GAP domain; the catalytic arginine (arginine finger) is also marked (R*).
Ypt1p is the critical substrate of Gyp1p for recycling.
Several lines of evidence suggest that the recycling defect observed in _gyp1_Δ cells is caused by an accumulation of hyperactivated Ypt1p. First, Gyp1p has previously been shown to activate GTP hydrolysis of Ypt1p in vitro (2, 12). Second, the recycling defect of _gyp1_Δ mutant cells, like their thermosensitive growth defect (13), can be partially rescued by decreasing the activity of Ypt1p. Finally, wild-type cells expressing a mutationally GTP-locked form of Ypt1p (Ypt1p-Q67L) exhibit a recycling defect (Fig. 6), but are not affected for secretion of membrane proteins (30). These results imply that GTP hydrolysis is specifically required for the recycling function of Ypt1p. It is possible that GTP hydrolysis is necessary to spatially or temporally limit its activity or to allow rapid reuse of Ypt1p. In the latter case, Ypt1p GTP may titrate out an effector, which may be limiting for recycling but not for the ER-to-Golgi compartment role of Ypt1p.
What is the function of Ypt1p during recycling?
In addition to the well established role of Ypt1p for trafficking of secretory vesicles from the ER to the Golgi compartment (25, 33), and a possible function at a late step during secretion (26), our results implicate Ypt1p in recycling of membrane proteins. At least two possibilities may account for these observations. First, it remains possible that deletion of GYP1 or expression of GTP-locked Ypt1p-Q67L may indirectly disturb recycling by delocalizing essential recycling factors. For example, _gos1_Δ cells fail to recycle GFP-Snc1p because the SNAREs Tlg1p/Tlg2p are mislocalized from the late Golgi compartment to transport vesicles (35). Although Tlg1p and Tlg2p were correctly localized to the Golgi compartment in _gyp1_Δ cells (Fig. 8), we cannot exclude that other recycling components may be mislocalized.
Alternatively, Ypt1p may function in the Golgi compartment to promote fusion of endosomal vesicles. Interestingly, the COG or Sec34p/35p complex has been described as an effector of Ypt1p (38). This complex exerts several functions together with Ypt1p, and has been proposed to function in ER sorting and as a tethering factor at the Golgi compartment for both anterograde transport from ER-to-Golgi compartment and retrograde intra-Golgi compartment transport (25, 38, 43, 44). Moreover, the COG complex could be involved in tethering endosomal vesicles to the Golgi compartment, since _dor1/cog8_Δ cells are defective for recycling of GFP-Snc1p (43). We propose that Gyp1p-catalyzed GTP hydrolysis of Ypt1p is essential for tethering endosome-derived recycling vesicles to the Golgi compartment.
Yeast cells may use two distinct recycling routes.
In mammalian cells, several pathways of recycling to the plasma membrane have been described, including a direct pathway from early endosomes and a slower route through early and recycling endosomes (46). In yeast, the only well characterized recycling route is the one followed by the SNARE Snc1p which travels from early endosomes to the Golgi compartment and back to the plasma membrane (22). In this pathway, activated Ypt6p recruits the VFT tethering complex, which in turn interacts with the SNAREs Tlg1p and Tlg2p, thereby promoting fusion of vesicles derived from early endosomes to the Golgi compartment (34-36). Cells with deletions of YPT6 or its regulators RIC1/RGP1 exhibit a recycling defect of GFP-Snc1p (34), and accumulate vesicles thought to be on their way to the Golgi compartment (41). However, _ypt6_Δ, _ric1_Δ, or _rgp1_Δ cells are viable, implying that either recycling is not essential for cell viability, or that recycling is an essential process but ensured by at least two redundant pathways. Interestingly, we found that GYP1 deletion is synthetic-lethal with YPT6, RIC1, ARL1 and ARL3 deletions. Arl1p functions in the Golgi compartment and was previously shown to interact with the VFT complex through Vps53p (27). In addition to known components of the Ypt6p pathway, _gyp1_Δ was also found to be synthetic-lethal with deletions of genes encoding multiple components of the COG complex including COG5, -6, -7, and -8, and other proteins previously implicated in trafficking (Fig. 7B). Similarly, _ric1_Δ has recently been found to be synthetic-lethal with _cog8_Δ (43). Interestingly, these four COG components are not crucial for the association of the core proteins, suggesting that these four subunits may be peripheral components of the COG complex (29). One hypothesis to explain their synthetic lethality with _gyp1_Δ would be that the remaining COG complex is still partially functional, but requires correct regulation of Ypt1p to perform its role during recycling. Although the molecular basis of the observed synthetic-lethal interactions are not clear, it is tempting to speculate that GFP-Snc1p can recycle by two distinct routes: first, from early endosomes using the Ypt6p-VFT pathway for fusion of the vesicles with the late Golgi compartment, and second, using the Ypt1p-pathway for fusion of endosomal vesicles with the _cis_-Golgi compartment. Consistent with this hypothesis, it was previously shown that overexpression of Ypt1p is able to suppress the growth defect of temperature-sensitive ypt6 mutants (23). Based on the observations reported here, it may be worthwhile to analyze recycling and other trafficking pathways in the mutants that are synthetic-lethal with GYP1, which previously have not been functionally connected to trafficking (Fig. 7B). Taken together, our results suggest that recycling of membrane proteins may be an essential process regulated by at least two distinct trafficking routes.
Acknowledgments
We thank P. Novick, N. Segev, H. Pelham, H. Riezman, and S. Alberts for plasmids and yeast strains. We are grateful to Monika Gersbach for expert technical assistance, C. Boone's laboratory for help in the synthetic-lethal analysis, Nicolas Pagé for robot expertise, members of the Haguenauer-Tsapis and Peter groups for stimulating discussion, and Marc Sohrmann for critical reading of the manuscript.
J.-M.G. is supported by the CNRS, and work in the M.P. laboratory is supported by the Swiss National Science Foundation, the Functional Genome Center Zurich, and the ETH/Zurich.
Appendix
Sequence comparisons between various GAPs of the GYP family revealed that Gyp1p is conserved among species (Fig. A1).
REFERENCES
- 1.Albert, S., and D. Gallwitz. 1999. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem. 274**:**33186-33189. [DOI] [PubMed] [Google Scholar]
- 2.Albert, S., E. Will, and D. Gallwitz. 1999. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 18**:**5216-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 4.Bacon, R. A., A. Salminen, H. Ruohola, P. Novick, and S. Ferro-Novick. 1989. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J. Cell Biol. 109**:**1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bensen, E. S., B. G. Yeung, and G. S. Payne. 2001. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell 12**:**13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitkreutz, B. J., C. Stark, and M. Tyers. 2003. The GRID: the General Repository for Interaction Datasets. Genome Biol. 4**:**R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz, B. J., C. Stark, and M. Tyers. 2003. Osprey: a network visualization system. Genome Biol. 4**:**R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., and N. G. Davis. 2000. Recycling of the yeast a-factor receptor. J. Cell Biol. 151**:**731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., and N. G. Davis. 2002. Ubiquitin-independent entry into the yeast recycling pathway. Traffic 3**:**110-123. [DOI] [PubMed] [Google Scholar]
- 10.Conibear, E., J. N. Cleck, and T. H. Stevens. 2003. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 14**:**1610-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Antoni, A., J. Schmitzova, H. H. Trepte, D. Gallwitz, and S. Albert. 2002. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J. Biol. Chem. 277**:**41023-41031. [DOI] [PubMed] [Google Scholar]
- 12.Du, L. L., R. N. Collins, and P. J. Novick. 1998. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J. Biol. Chem. 273**:**3253-3256. [DOI] [PubMed] [Google Scholar]
- 13.Du, L. L., and P. Novick. 2001. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol. Biol. Cell 12**:**1215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galan, J. M., V. Moreau, B. Andre, C. Volland, and R. Haguenauer-Tsapis. 1996. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271**:**10946-10952. [DOI] [PubMed] [Google Scholar]
- 15.Galan, J. M., and M. Peter. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96**:**9124-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan, J. M., A. Wiederkehr, J. H. Seol, R. Haguenauer-Tsapis, R. J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21**:**3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, X. D., S. Albert, S. E. Tcheperegine, C. G. Burd, D. Gallwitz, and E. Bi. 2003. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J. Cell Biol. 162**:**635-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press Inc., San Diego, Calif.
- 19.Hettema, E. H., M. J. Lewis, M. W. Black, and H. R. Pelham. 2003. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22**:**548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holthuis, J. C. M., B. J. Nichols, and H. R. B. Pelham. 1998. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol. Biol. Cell 9**:**3383-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafourcade, C., J. M. Galan, and M. Peter. 2003. Opposite roles of the F-Box protein Rcy1p and the GTPase-activating protein Gyp2p during recycling of internalized proteins in yeast. Genetics 164**:**469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, M. J., B. J. Nichols, C. Prescianotto-Baschong, H. Riezman, and H. R. Pelham. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11**:**23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, B., and J. R. Warner. 1998. Genetic interaction between YPT6 and YPT1 in Saccharomyces cerevisiae. Yeast 14**:**915-922. [DOI] [PubMed] [Google Scholar]
- 24.Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14**:**953-961. [DOI] [PubMed] [Google Scholar]
- 25.Morsomme, P., and H. Riezman. 2002. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell 2**:**307-317. [DOI] [PubMed] [Google Scholar]
- 26.Mulholland, J., A. Wesp, H. Riezman, and D. Botstein. 1997. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretory vesicle. Mol. Biol. Cell 8**:**1481-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panic, B., J. R. Whyte, and S. Munro. 2003. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 13**:**405-410. [DOI] [PubMed] [Google Scholar]
- 28.Piper, R. C., A. A. Cooper, H. Yang, and T. H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131**:**603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ram, R. J., B. Li, and C. A. Kaiser. 2002. Identification of Sec36p, Sec37p, and Sec38p: components of yeast complex that contains Sec34p and Sec35p. Mol. Biol. Cell 13**:**1484-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson, C. J., S. Jones, R. J. Litt, and N. Segev. 1998. GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol. Cell. Biol. 18**:**827-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, C. J., C. K. Raymond, C. T. Yamashiro, and T. H. Stevens. 1991. Methods for studying the yeast vacuole. Methods Enzymol. 194**:**644-661. [DOI] [PubMed] [Google Scholar]
- 32.Seabra, M. C., E. H. Mules, and A. N. Hume. 2002. Rab GTPases, intracellular traffic and disease. Trends Mol. Med. 8**:**23-30. [DOI] [PubMed] [Google Scholar]
- 33.Segev, N. 2001. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13**:**500-511. [DOI] [PubMed] [Google Scholar]
- 34.Siniossoglou, S., S. Y. Peak-Chew, and H. R. Pelham. 2000. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19**:**4885-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siniossoglou, S., and H. R. Pelham. 2001. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 20**:**5991-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siniossoglou, S., and H. R. Pelham. 2002. Vps51p links the VFT complex to the SNARE Tlg1p. J. Biol. Chem. 277**:**48318-48324. [DOI] [PubMed] [Google Scholar]
- 37.Strom, M., P. Vollmer, T. J. Tan, and D. Gallwitz. 1993. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature 361**:**736-739. [DOI] [PubMed] [Google Scholar]
- 38.Suvorova, E. S., R. Duden, and V. V. Lupashin. 2002. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157**:**631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294**:**2364-2368. [DOI] [PubMed] [Google Scholar]
- 40.Tong, A. H., G. Lesage, G. D. Bader, et al. 2004. Genetic interaction networks: large-scale mapping of synthetic genetic interactions in yeast. Science 303**:**808-813. [DOI] [PubMed] [Google Scholar]
- 41.Tsukada, M., E. Will, and D. Gallwitz. 1999. Structural and functional analysis of a novel coiled-coil protein involved in Ypt6 GTPase-regulated protein transport in yeast. Mol. Biol. Cell 10**:**63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valdivia, R. H., D. Baggott, J. S. Chuang, and R. W. Schekman. 2002. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell 2**:**283-294. [DOI] [PubMed] [Google Scholar]
- 43.Whyte, J. R., and S. Munro. 2001. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell 1**:**527-537. [DOI] [PubMed] [Google Scholar]
- 44.Whyte, J. R., and S. Munro. 2002. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115**:**2627-2637. [DOI] [PubMed] [Google Scholar]
- 45.Wiederkehr, A., S. Avaro, C. Prescianotto-Baschong, R. Haguenauer-Tsapis, and H. Riezman. 2000. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149**:**397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zerial, M., and H. McBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2**:**107-117. [DOI] [PubMed] [Google Scholar]
- 47.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9**:**1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]