CREB Binds to Multiple Loci on Human Chromosome 22 (original) (raw)
Abstract
The cyclic AMP-responsive element-binding protein (CREB) is an important transcription factor that can be activated by hormonal stimulation and regulates neuronal function and development. An unbiased, global analysis of where CREB binds has not been performed. We have mapped for the first time the binding distribution of CREB along an entire human chromosome. Chromatin immunoprecipitation of CREB-associated DNA and subsequent hybridization of the associated DNA to a genomic DNA microarray containing all of the nonrepetitive DNA of human chromosome 22 revealed 215 binding sites corresponding to 192 different loci and 100 annotated potential gene targets. We found binding near or within many genes involved in signal transduction and neuronal function. We also found that only a small fraction of CREB binding sites lay near well-defined 5′ ends of genes; the majority of sites were found elsewhere, including introns and unannotated regions. Several of the latter lay near novel unannotated transcriptionally active regions. Few CREB targets were found near full-length cyclic AMP response element sites; the majority contained shorter versions or close matches to this sequence. Several of the CREB targets were altered in their expression by treatment with forskolin; interestingly, both induced and repressed genes were found. Our results provide novel molecular insights into how CREB mediates its functions in humans.
The cyclic AMP (cAMP)-responsive element-binding protein (CREB) is a key transcription factor that stimulates the expression of numerous genes in response to growth factors, hormones, neurotransmitters, ion fluxes, and stress signals. The CREB signaling pathway is activated by extracellular ligands that bind to cell surface receptors. Various intracellular second messengers then relay signals through kinase pathways to the nuclear resident CREB. Upon induction of the pathway, CREB is phosphorylated and activated at Ser-133. At least four types of kinases have been proposed to phosphorylate this residue: cAMP-dependent protein kinase, multiple mitogen-activated protein kinases (MAPKs), ribosome S6 kinase, Ca2+- and calmodulin-dependent kinases (CAMKs), and possibly Akt (reviewed in reference 22). Phosphorylation of CREB at Ser-133 leads to the recruitment of CREB binding protein (CBP) or its paralog p300 and subsequent transcriptional activation. A number of CREB targets have been identified, mostly through studies of individual genes (17, 26, 32). However, a full range of targets has not been explored. Given CREB's role in many important cellular processes, it is important to identify as many potential targets as possible.
CREB functions either by itself or with the related family members CREM and ATF1; CREB, CREM, and ATF1 homo- and heterodimerize through basic leucine zipper (bZIP) domains (27, 40). A number of studies indicate that CREB is constitutively bound to chromatin even in the absence of agonists. CREB usually binds to cAMP response elements (CREs) near TATA boxes (26). However, not all DNA binding regions match the conserved motif. Moreover, binding may occur within the first introns of genes with multiple promoter systems or in the introns of alternatively spliced genes, such as the phosphodiesterase PDE4 (7, 39), brain-derived neurotrophic growth factor (33, 36, 38), cytochrome P450 aromatase (reference 43 and references therein), and the inducible cAMP early repressor transcription factor (reviewed in reference 32). To date, a systematic and unbiased analysis of where CREB binds relative to its target genes has not been performed.
In this study we mapped the distribution of CREB1 binding regions over an entire human chromosome with an unbiased in vivo approach. CREB1 binding regions were identified by performing chromatin immunoprecipitations and using the immunoprecipitated DNA to probe a genomic DNA microarray that contained all nonrepetitive chromosome 22 sequences. We recently described an array containing most of the unique sequences of human chromosome 22 (31). This reagent provides a unique opportunity to examine the binding of CREB1 along an entire chromosome. Specifically, we sought to determine what types of genes CREB1 binds near, the locations of its binding sites within a gene locus, and its binding position relative to consensus CRE sites.
With our approach, we found that CREB1 has many binding sites on the chromosome and binds near many previously unappreciated targets. Moreover, CREB1 binding sites were situated not only at 5′ ends but also in internal introns and unannotated regions outside of known genes. Thus, our study provides substantial information about the diversity of putative CREB1 targets as well as the locations of potential regulatory elements within genes.
MATERIALS AND METHODS
Cell culture, nuclear extracts and immunoprecipitations.
JEG-3 cells (American Type Culture Collection number HTB-36) were grown in alpha minimal essential medium (αMEM) (Invitrogen) supplemented with 10% fetal calf serum. To test the anti-CREB1 antibodies in immunoprecipitations, JEG-3 cells were washed in Dulbecco's phosphate-buffered saline and harvested in hypotonic lysis buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40). All buffers contained 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail for mammalian cells (Sigma), and lysates were maintained at 4°C. Nuclear pellets were lysed in 1× radioimmunoprecipitation assay (RIPA) buffer (Upstate Biotechnology) and sheared three times with a 21-gauge needle. Lysates were clarified for 15 min at 15,000 × g and then cleared with protein A-protein G-Sepharose (Pierce) for 45 min. Lysates were incubated with anti-CREB NT antibody (Upstate Biotechnology, 06-863) for 3 h and then with protein A-protein G-Sepharose for 1 h. Immunocomplexes were recovered and analyzed by Western blotting with the same anti-CREB NT antibody (1:1,000 dilution), followed by a horseradish peroxidase-conjugated secondary antibody (1:2,500 dilution). Blots were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Chromatin immunoprecipitations.
Cells were cultured in 15-cm plates to approximately 80 to 90% confluence, and 14 to 15 plates were used per immunoprecipitation, with equivalent plate numbers used for the anti-CREB1 immunoprecipitation and the control immunoprecipitation without antiserum. For forskolin induction, cells were treated with 10 μM forskolin (Sigma) dissolved in dimethyl sulfoxide for 45 min. Cells were fixed with 1% formaldehyde in αMEM medium lacking fetal bovine serum for 10 min and then collected (13). Nuclei were pelleted in hypotonic lysis buffer and lysed in 1× RIPA as described above for immunoprecipitations. Sonication conditions were tested to yield DNA fragments averaging 600 bp as assessed by agarose gel electrophoresis. Lysates were clarified and cleared as described above for the immunoprecipitations. For each CREB1 immunoprecipitation, 20 μg of the CREB NT antibody was added to lysate prepared from the 15 15-cm plates of JEG-3 cells. After incubation with antibody for 3 to 4 h at 4°C, 300 μl of a 50% protein A-G slurry was added for an additional 1 h of incubation. The beads were washed four times: twice in 1× RIPA buffer, once in LiCl detergent wash (250 mM LiCl, 0.25% deoxycholic acid, 0.5% NP-40, 1 mM EDTA, 10 mM Tris, pH 8.0) and once in 1× TBS (150 mM NaCl, 20 mM Tris, pH 7.6). CREB1-DNA complexes were eluted and purified as described previously (13).
Expression analysis.
Polyadenylated RNA was isolated from JEG-3 cells treated with either 10 μM forskolin (dissolved in dimethyl sulfoxide) or cells treated with dimethyl sulfoxide alone after a 1.5-h induction, with the MicroPoly(A) pure and DNA-free kits (Ambion). Labeled cDNA was synthesized from 4 μg of polyadenylated RNA for each sample according to the Amino Allyl cDNA labeling kit (Ambion). Reverse transcription reactions were primed with both oligo(dT) and random decamers.
Chromatin immunoprecipitation probe labeling and microarray hybridization.
Chromatin-immunoprecipitated DNA and corresponding control samples lacking antiserum were labeled by incorporating amino-allyl dUTP with the Bioprime labeling system (Life Technologies) as described elsewhere (42). Labeled probes were purified from unconjugated dye by passage through Sepharose spin columns from Ambion's Amino Allyl cDNA labeling kit, concentrated by ethanol precipitation, and finally resuspended in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 25% formamide, 0.05% sodium dodecyl sulfate [SDS]).
Chromosome 22 microarray slides (31) were prehybridized in 5× SSC-25% formamide-10× Denhardt’s solution-0.1% bovine serum albumin in HybChambers (GeneMachines, San Carlos, Calif.) at 42°C for 1 h. The slides were dipped in water to remove the prehybridization buffer and coverslips and then dried by centrifugation. The probe was applied to a coverslip and placed on a microarray. Microarrays were hybridized at 42°C for 14 to 15 h and then washed twice in 2× SSC-0.05% SDS for 2 min, once in 0.1× SSC-0.05% SDS for 5 min, and four times in 0.1× SSC for 1 min.
Microarray signals were normalized with the ExpressYourself analysis program (http://bioinfo.mbb.yale.edu/ExpressYourself), with default parameters to correct for spatial, intensity-based, and dye-specific artifacts (23). Positively hybridizing fragments were identified as those with logged indodicarbocyanine (Cy5)/indocarbocyanine (Cy3) ratios more than 2.0 standard deviations above the mean for fragments with similar total intensities (30). Raw data are available at http://array.mbb.yale.edu/chr22.
PCR analysis.
CREB1-precipitated DNA was analyzed by real-time PCR with SYBR Green. For putative CREB1 targets and negative control regions on chromosome 22, primers were designed to tile across microarray fragments in 300-bp increments. Targets were initially chosen from an initial “top 500” list; ultimately, 40 of these lay below the cutoff that was used. For the human chorionic gonadotrophin alpha (hCGα) promoter, the primers spanned a 300-bp region around the tandem CREs (6, 15, 16). Only primer pairs that yielded quality PCR products with total lysate and that also did not form primer dimers, as assessed by dissociation curves in no-template control reactions, were used in assays with CREB1-precipitated DNA. Each PCR was repeated in triplicate in a 20-μl reaction volume with 1% of the total immunoprecipitated DNA and 1× SYBR Green DyNAmo master mix (Finnzymes). Fluorescence was detected each cycle with the ABI Prism 7000 sequence detection system (Applied Biosystems), and enrichment was determined according to Ct (changes in fluorescence per cycle number) measurements.
RESULTS
CREB1 chromatin immunoprecipitations.
To map and analyze CREB1 binding sites on chromosome 22, we selected a cAMP-inducible cell line so that CREB1 binding regions could be correlated with differential expression. Human JEG-3 choriocarcinoma (placental) cells can be activated in culture by incubation with forskolin. This treatment stimulates adenylate cyclase, which in turn activates CREB and induces the expression of a number of genes. One such gene is the alpha chain of the hormone chorionic gonadotrophin (hCGα), which is subject to CREB control via two CREs in its promoter (6, 15, 16). This gene served as a control for many of our experiments.
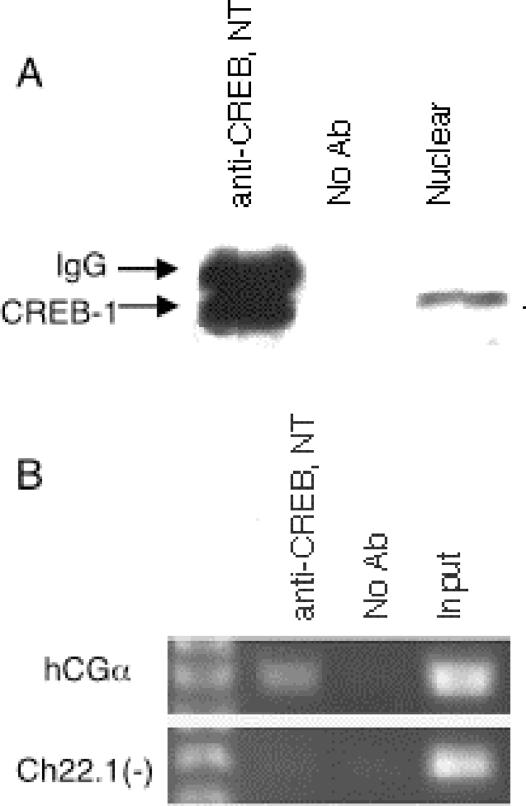
We first tested several commercially available antibodies to determine if any could specifically immunoprecipitate CREB1. One polyclonal antibody (CREB NT), raised to a synthetic peptide corresponding to residues 5 to 21 of human CREB1 (the non-DNA binding region), was found that specifically immunoprecipitated a single band at 43 kDa from nuclear extracts of untreated JEG-3 cells (Fig. 1A); this band was not observed in control experiments lacking antibody. Since the antibody was raised against a sequence that is unique to CREB1 expressed from chromosome 2 and not found in other members of the CREB transcription factor family, including CREM and the ATFs, only CREB1 is expected to be recognized.
FIG. 1.
Immunoprecipitation of CREB1. (A) CREB1 immunoprecipitated as a single 43-kDa band with the CREB NT antibody in JEG-3 cells. The CREB NT antibody was used for both the immunoprecipitation (lanes 1 and 2) and the immunoblot (lane 3) analysis. (B) PCR analysis of a control CREB1 target region after chromatin immunoprecipitation with the CREB NT antibody. The region around the tandem CREs in the hCGα promoter was selectively amplified in DNA purified from CREB1 immunoprecipitations in JEG-3 cells, whereas no enrichment was seen in a genomic region not known to bind CREB.
We next tested whether the antibodies could be used for chromatin immunoprecipitation with JEG-3 cells. Since CREB1 is known to be constitutively bound to its targets, these experiments were performed with untreated cells. JEG-3 cells were formaldehyde cross-linked to preserve protein-DNA complexes, and nuclear extracts were prepared. The chromatin was sheared by sonication, yielding DNA fragments ≈600 bp in length, and the CREB1-DNA complexes were immunoselected with the anti-CREB1 antibody. After reversal of the cross-links and DNA purification, each CREB1 chromatin immunoprecipitate was tested for specific binding site enrichment with PCR. As shown in Fig. 1B, a region surrounding the tandem CREs of the hCGα promoter was specifically enriched relative to the control lacking antibodies, and a chromosome 22 region that is not known to be involved in CREB binding was not enriched. Thus, the CREB NT antibody specifically immunoprecipitated its associated DNA in JEG-3 cells.
Many CREB1 binding regions lie on chromosome 22.
To determine the in vivo CREB1 binding regions on chromosome 22, a total of six independent chromatin immunoprecipitations were carried out in JEG-3 cells: three independent replicates of purified CREB1-associated DNA from uninduced cells, and three independent replicates of immunoprecipitated CREB1-associated DNA from forskolin-induced cells. Each immunoprecipitation preparation was performed in parallel with a control experiment lacking antibody. All six DNA isolations were independently labeled with Cy5 (anti-CREB1 samples) or Cy3 (no antiserum reference) and hybridized to the chromosome 22 array, which contains 21,024 PCR products spanning nearly all of the unique sequences of this chromosome (31). Microarray results were normalized with the ExpressYourself software by standard approaches (see Materials and Methods). No significant differences were observed in the results from forskolin-treated and untreated cells (see below); therefore, the results from all six experiments were pooled. Candidate CREB1 binding targets were identified as those enriched in at least four of the six replicates; most targets were found in five or more replicates (23, 30).
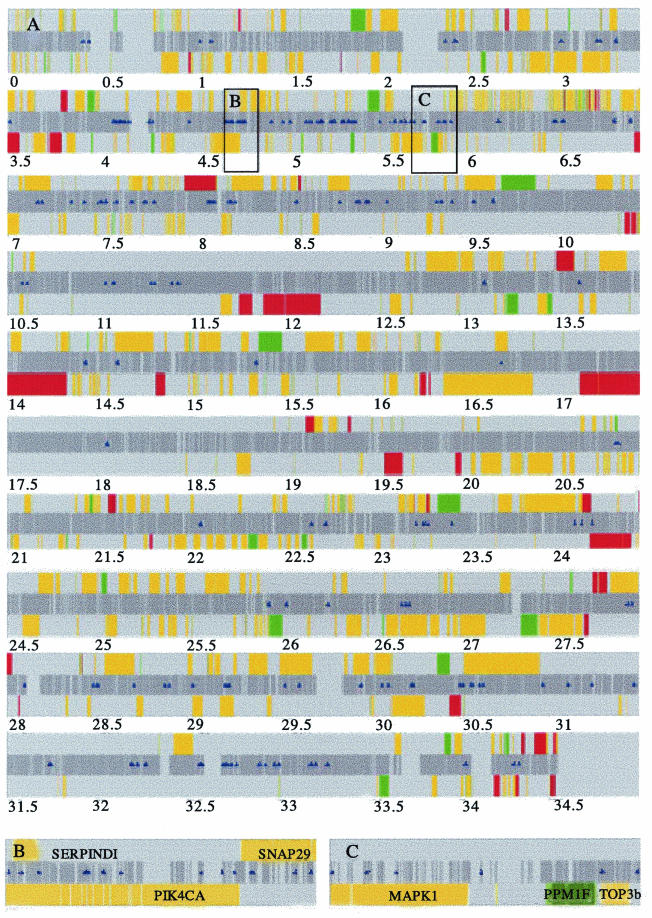
With this approach, we identified 215 hybridizing fragments as CREB1 binding targets (Fig. 2; also see Table 1 in our supplementary data at http://array.mbb.yale.edu/euskirchen). The CREB1 binding sites were distributed all along the chromosome. Since CREB1 cross-linked DNA had been sonicated to yield fragments ≈600 bp in length and the average size of the array fragment was ≈800 bp (range, 300 to 1,400 bp), it was expected that adjacent genomic fragments should hybridize on the array. This indeed was the case for 23 fragments, indicating that 192 chromosome 22 loci contain CREB1 binding sites. For the remaining fragments, it is likely that the binding site was centered in the array fragment or that the target bordered repetitive sequence that was not included on the array.
FIG. 2.
Map of CREB1 binding regions on chromosome 22q. (A) The sequence is ordered from centromere to telomere on the long arm of chromosome 22. The dark gray center band represents all of the nonrepetitive genomic sequence included on the DNA microarray. Sanger annotated genes are oriented 5′ to 3′ above the center band and 3′ to 5′ below the center band. Colored bars depict the differential expression analysis results in response to forskolin: upregulated genes are red, downregulated genes are green, and nondifferentially regulated genes are yellow. Blue triangles indicate the positions of CREB1-bound fragments. (B and C) Higher-resolution examples of genes with CREB1-bound fragments. (B) A total of 14 CREB1 binding regions occur within or the near the genes SERPINDI and SNAP29 (top strand) and PIK4CA (lower strand). (C) A total of six CREB1 binding regions occur within or near the genes for MAPK1, PPM1F, and TOP3b. All three genes are on the lower strand.
TABLE 1.
Functional categories of CREB1 targets
| Category | Examples |
|---|---|
| Transcription | PPARα, TCF20, Cabin 1, MKL1, CBX6 |
| Cell cycle or DNA replication | HIRA, CDC45L, TOP3b, SMC1L2, GTSE1 |
| Signal transduction | MAPK1, ARHGAP8, PPM1F, PIK4CA, dJ930L11.1, Em:AP000347.C22.3, BCR, ADRBK |
| Channels and cell surface receptors | CELSR1, dJ1170K4.C22.1, CACNA1I |
| Lipid and sterol metabolism | CERK, OSBP1, PPARα, CHKL, PIK4CA, Em:AP000557.C22.3 |
| Structural | β-Parvin, TUBAL2, CRYBB1 |
| Neuronal | PACSIN2, SYN3, ATAXIN10, SNAP29 |
| Tissue remodeling | TIMP3, MMP11, EMU1 |
| Secretory | ARFGAP1, SNAP29, CLTCL1 |
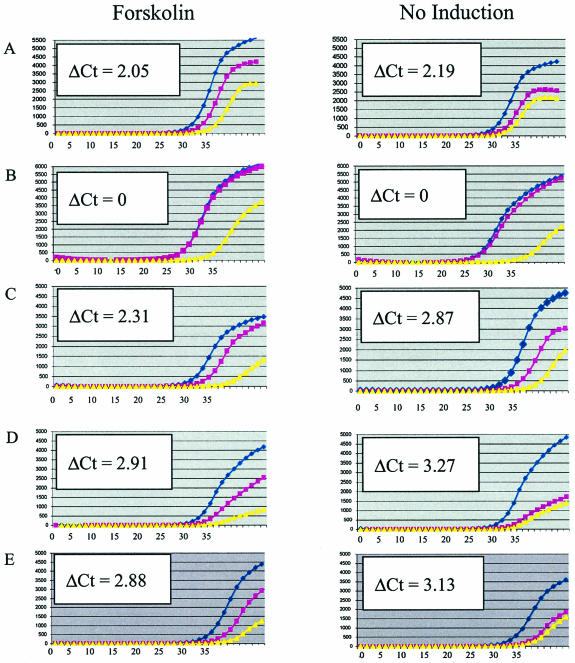
To determine the frequency of false-positives and false-negatives for the CREB1 binding sites, we performed real-time quantitative PCR assays on chromatin immunoprecipitates for a total of 71 fragments. Three examples are shown in Fig. 3 (and all data are summarized in the Table 2 of the supplementary data at the website cited above). Thirty-one CREB1-associated fragments, including three fragments immediately adjacent to the cutoff, were analyzed. In addition, 40 selected fragments that were below the final cutoff but that were initially near the cutoff were examined (see Materials and Methods). Twenty-eight of 31 fragments (89%) above the cutoff were enriched, including the three fragments just above the cutoff. Four of the 40 fragments (10%) that were below the cutoff were also found to be enriched by PCR. However, since all of these fragments were selected from near but below the cutoff, 10% is likely an overestimate of false negatives. Therefore, we conclude that most of the 215 identified CREB1 binding regions are bona fide sites in vivo.
FIG. 3.
Real-time PCR analysis of CREB1 chromatin immunoprecipitations. DNA from forskolin-treated and untreated JEG-3 cells was immunopurified with anti-CREB1 antibody and amplified (blue circles) and compared to control experiments lacking antibody (pink squares) and control reactions lacking template (yellow triangles). Graphs show averages of triplicate PCRs. Units are Δ_Rn_ (fluorescence) on the y axis and PCR cycle number on the x axis. One Δ_Ct_ (where Ct denotes threshold cycle) corresponds to approximately twofold enrichment. Enrichment of CREB1-bound regions is observed as an increase in fluorescence at earlier PCR cycle numbers than the no-antiserum controls. Examples shown are the positive control, hCGα promoter (A); the negative control, chromosome 22 nonspecific region (B); CACNA1I, 74.5-kbp fragment internal to the downregulated Ca2+ channel voltage-dependent α1I subunit (C); and dJ930L11.1, 69.7-kbp internal fragment (D). This gene is similar to KIAA0397, which has RabGAP domains and may be involved in membrane traffic. (E) Fragment >10 kb from the annotated region within chromosomal coordinates 23262970 to 23263520.
TABLE 2.
Chromosome 22 genes with CREB1 binding sites that are differentially regulated or not affected by forskolin treatmenta
To determine if CREB1 binding to the fragments varies with the presence and absence of forskolin, eight of the fragments were also analyzed for their enrichment in immunoprecipitates from forskolin-treated and untreated cells. In each case, similar enrichment was observed, indicating that CREB1 was bound in both cell populations (examples shown in Fig. 3), consistent with previous studies which demonstrated that CREB is bound to its targets constitutively.
CREB binds near many annotated genes as well as uncharacterized expressed regions.
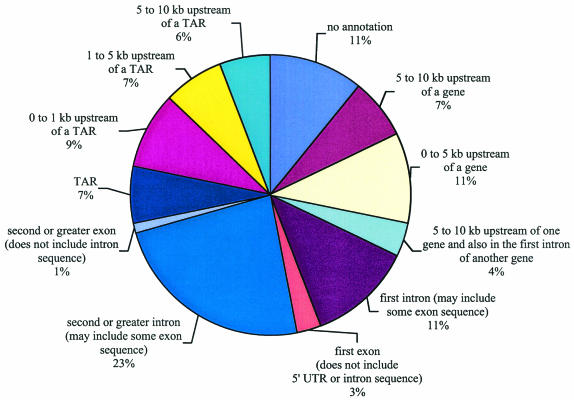
The 215 CREB1 binding fragments were compared with chromosome 22 features, including genes and pseudogenes. Human chromosome 22 contains 546 known, predicted, or partial protein coding genes (open reading frames of >300 bp), 234 pseudogenes, 31 non-protein-coding fragments, and 125 immunoglobulin lambda gene fragments, for a total of 936 loci (3, 12). The types of genes identified as putative CREB1 targets span a number of functional categories, including transcription factors and signaling components, as presented in Table 1 and discussed in more detail below. Further chromosome 22 annotation uncovered 1,302 transcriptionally active regions (TARs) that were not present in other annotations (31). Of the 215 CREB1 binding sites, 60% (130 fragments) mapped within or less than 10 kb upstream of one of the 936 annotated loci (Fig. 4). Of the remaining 85 fragments that were >10 kb from annotated loci, 62 CREB1 binding fragments either encoded or were less than 10 kb from a TAR (Fig. 4). Thus, 89% (130 plus 62) of the 215 CREB1 binding sites either overlapped or were less than 10 kb from a known transcribed region of human chromosome 22.
FIG. 4.
Position of CREB1 binding regions relative to chromosome 22 annotations. CREB1 binding sites were mapped relative to 5′ ends, introns, exons, internal introns and exons, and transcriptionally active regions (TARs).
We next examined the positions of the CREB1 binding fragments relative to annotated exons and introns. As summarized from the multiple categories shown in Fig. 4, 22% of CREB1 binding regions (47 fragments) lay within 10 kb of the 5′ end of the gene, and 15% (33 fragments) and 24% (52 fragments) of the CREB1 binding regions lay within a first intron or other intron, respectively. Six CREB1 binding fragments intersected an annotated first exon without any adjacent 5′ untranslated region or intron sequence mapping to the fragment. Three of these six fragments were in genes for which limited transcript data are available: two are expressed sequence tag (EST) clusters, and one is based on a gene prediction. The remaining three fragments landed in pseudogenes and likely reflect either archaic usage of CREB1 or binding of CREB1 to a homologous, functional copy of the parent gene. Curiously, two CREB1 binding fragments included internal exons and excluded noncoding sequence, exon 2 of MGAT3/GNT-III and exon 3 of MIF (encoding macrophage inhibition factor).
Consensus sites.
The 215 genomic hits were searched for CREs as well as TATA boxes and other transcription-regulatory sequences that may contribute to CREB binding. Searches for the strict canonical CREB binding site TGACGTCA revealed that six of the CREB targets had this exact motif. CREB has also been suggested to bind to the “half-site” sequence CGTCA (26); 65 of the fragments had this motif. Finally, nearly all of the fragments (83%) included or were within 500 bp of the strict canonical CREB binding site, a CREB half-site sequence, or two other consensus CREB binding sites, TGACGTMW and TGANNTCA, where M is A or C, W is A or T, and N is any base (17). We did not observe an enrichment, presumably because such half-site sequences are prevalent throughout the array (approximately one every 1,000 bp).
Identification of forskolin-regulated genes.
In order to determine potential CREB1 targets whose expression might be regulated by cAMP, we also identified genes whose expression was altered by treatment with forskolin. As noted above, forskolin activates adenylate cyclase and thereby is expected to affect CREB signaling. Polyadenylated RNA was harvested from JEG-3 cells treated with forskolin for 1.5 h and from untreated cells; 1.5 h was chosen because it was reported previously that synthesis of the inducible CRE repressor-inducible cAMP early repressor occurs between 1.5 and 2 h in JEG-3 cells and inactivation of CREB by dephosphorylation of Ser-133 takes place within 2 to 4 h of continuous stimulation (24, 26). Therefore, the most direct and peak effects of forskolin induction are expected at 1.5 h. Cy5- and Cy3-labeled probes were prepared to two independent polyadenylated RNA isolations. A Cy5-labeled probe to RNA from the forskolin-treated cells was mixed with a Cy3 probe to RNA from the untreated cells and hybridized to the chromosome 22 array; the dye labels were also reversed, so that Cy3 probes from the forskolin-treated cells and Cy5 probes from the untreated cells were hybridized to the arrays. A total of four replica hybridizations to the chromosome 22 array were performed.
A total of 99 genes on chromosome 22 (listed in Table 3 of the supplementary data at the website given above) had significantly altered expression levels after forskolin treatment (see Materials and Methods): 51 were upregulated and 48 were downregulated, and 16 of the 99 loci were pseudogenes (2 upregulated and 14 downregulated pseudogenes), including three downregulated pseudogenes that were related to γ-glutamyltransferase. We presume that either the pseudogenes are expressed or they cross-hybridize to transcripts of functional copies.
CREB1 binding sites were found 5′ or internal to eight of the upregulated genes and seven of the downregulated genes (Table 2). Examples of upregulated genes with CREB1 binding sites are the calcineurin binding protein Cabin 1, adenylosuccinate lyase (ADSL), ceramide kinase, and choline kinase-like. Examples of downregulated genes with CREB1 binding sites include protein phosphatase 1F (PPM1F), a potassium channel-related gene, a gene for a calcium channel subunit (CACNA1I), and a phosphatidylinositol-4-kinase 230-related gene.
CREB1 also bound near 85 genes whose expression was not affected by treatment with forskolin. Many of these were expressed at high levels in both the presence and absence of forskolin; these genes may already be activated by other factors, and CREB activation many not enhance their expression further.
Eighty-four genes exhibited alteration in mRNA levels after forskolin treatment but did not contain CREB1 binding sequences. Since CREB1 is not unique among transcription factors in activation by forskolin, genes lacking CREB1 sites presumably either are regulated by other factors or lie downstream and are indirect targets of CREB signaling.
Functional categories of potential CREB1 targets.
A number of interesting types of genes were discovered as potential CREB1 targets (Table 1). These include transcription factors (e.g., PPARα and MKL1) and genes that function in neurons (e.g., PACSIN2 and SYN3).
Of particular interest were a variety of genes involved in signal transduction, many of which encode proteins that are likely to modulate CREB signaling pathways, and a number of these putative targets are regulated by forskolin treatment (Tables 1 and 2; also see Discussion). These included two potential regulators of cAMP signaling, ADRBK2 (encoding a β-adrenergic receptor kinase) and ADSL (encoding adenylosuccinate lyase, which catalyzes two key steps in AMP synthesis); three genes involved in Ca2+ signaling, Cabin1 (a calcineurin binding protein that we speculate modulates CREB activity) (1, 11, 18, 21), PPMF1 (encoding a protein phosphatase that dephosphorylates the calcium/calmodulin-dependent protein kinase II (CaMKII) in vitro (37), and CACNA1I (encoding a subunit of a low-voltage-activated T-type calcium channel), and genes important for lipid signaling, the cytoskeleton, and vesicular trafficking. CREB1 binding regions were found in the genes for a catalytic subunit of phosphatidylinositol 4-kinase (PIK4CA) and Em:AP000557.C22.3 (a gene related to phosphatidylinositol 4-kinase 230). The phosphatidylinositol 4-kinases convert phosphatidylinositol into phosphatidylinositol-4-phosphate, an important intermediate for the activation of protein kinase C, cytoskeletal reorganization, and secretion. CREB1 binding regions were also found in genes for PACSIN2 (protein kinase C and casein kinase substrate in neurons 2), ARHGAP8 (rho GTPase-activating protein 8), Em:AP000347.C22.3 (calponin-like actin binding), β-parvin (α-actinin related), and CLTCL1 (clatherin heavy polypeptide-like 1). Finally, there were Ras/MAPK1 pathway genes, including MAPK1 itself (also known as ERK2), and two incompletely characterized genes, dJ930L11.1 and Em:AP000347.C22.3 (encoding proteins homologous to KIAA0397 and RALGDS, respectively). Each of these proteins potentially modulates the CREB pathway in vivo, as presented in the Discussion.
CREB1 regulatory regions occur in gene-dense areas of chromosome 22.
Many CREB1 binding regions were found in areas of overlapping genes. Of the 130 fragments that either lay within genes or within <10 kb of an annotated gene, 25 fragments overlapped or lay near two or more genes: 16 fragments included genes on opposite strands, and nine fragments were present in or near two genes on the same strand.
The positions of CREB1 binding regions relative to overlapping or adjacent genes may contribute to their coexpression. One example shown in Fig. 2 has six binding sites that lie in or near three genes, PPM1F, MAPK1, and TOP3b. All three genes are expressed in many tissues and at high levels. One CREB1 binding region lay on a fragment that overlapped the last intron and exon of TOP3b and was 4.3 kb upstream from the ATG of PPM1F. A second region was found 7.8 kb upstream of MAPK1. MAPK1 had three CREB1 binding regions: one region that was 49.3 kb internal and in the first intron, a second region that was 67.9 kb internal, and a third region mapping 92.2 kb internal. Interestingly, PPM1F was downregulated in response to forskolin, whereas MAPK1 and TOP3B exhibited no change in expression; both are constitutively expressed.
A similarly complex regulatory situation was found with PIK4CA, SNAP29, and SERPIND1, in which the genes lie on opposite strands (Fig. 2). PIK4CA is a large gene on the minus strand with 54 exons over 130 kb. All 13.6 kb of SERPINDI overlap PIK4CA. SNAP29 is also on the plus strand and is divergently transcribed from PIK4CA, with only 0.4 kb separating the ATG of SNAP29 and the ATG of PIK4CA. Thirteen CREB1 binding regions were found in PIK4CA, of which two were in the 5′ region of SERPINDI and four were in the 5′ region of SNAP29. Expression levels of PIK4CA, SERPINDI, and SNAP29 were not affected in forskolin-treated JEG-3 cells.
DISCUSSION
CREB1 binding regions lie throughout the human genome.
In this study we used an unbiased genomic DNA array containing all of the nonrepetitive sequences of human chromosome 22 to identify the distribution of CREB1 throughout an entire chromosome. We found 215 binding sites corresponding to 192 different loci and 100 annotated potential gene targets; most sites lay near known annotated genes and uncharacterized transcriptionally active regions (TARs).
Previously, most CREB targets were identified by studying genes one at a time. In many cases the targets were not conclusively verified. While we were preparing this work for publication, a study was published that identified CREB binding sites by searching for evolutionarily conserved sequences in the promoter regions of mouse and human DNA for matches to the most stringent CREB consensus sequence. From the entire genome, they identified 78 potential binding sites and verified approximately 10 (4).
Our study used an unbiased approach to comprehensively survey an entire chromosome and identified 215 targets. Assuming a similar distribution for the rest of the human genome, this extrapolates to an estimate of 19,000 binding sites throughout all nonrepetitive regions of the genome. This figure is on the same order of magnitude as an estimated 40,000 to 50,000 molecules of CREB per cell in PC12 cells (10), and it highlights several findings. First, a genomic DNA microarray is capable of experimentally finding many regions associated with a factor of interest and does not rely on either knowledge of consensus sites or positional information. Second, we can find sites that contain close matches to consensus sequences as well as those that are more divergent. For CREB and other human factors that we have studied (25), most binding sites are not perfect matches to the consensus. Third, we find binding sites that lie throughout a gene; indeed, the majority of binding sites do not lie close to the 5′ end. Genomic DNA microarrays may also provide insights into chromosomal looping, as linearly separated chromatin regions actually be adjacent in three-dimensional nuclei. Thus, a genomic DNA array is ideally suited for the comprehensive identification of all transcription factor targets.
Many CREB1 binding regions lie in complex regions in which transcription units lie near one another or even overlap. For example, we found 25 binding sites that lay near two or more annotated genes. The binding of CREB near multiple genes provides the opportunity for a single regulatory region to control the expression of multiple genes.
Many of the CREB1 binding sites lie near TARs, and two of the TARs were altered in response to forskolin treatment. The functional significance of the TARs remains to be established, but they have been speculated to have regulatory roles (31). Thus, control of TARs by CREB could have important regulatory consequences.
We recently completed a study analyzing the binding distribution of NF-κB along human chromosome 22 (25). Interestingly, 13 fragments for NF-κB are identical to a CREB1 binding region and another six NF-κB binding regions lay within 500 bp of a CREB1 region. Thus, these factors may jointly regulate the expression of several genes, consistent with the observation that cAMP levels can alter NF-κB transcriptional activity (5). Nonetheless, the majority of CREB1 and NF-κB gene targets are distinct, suggesting that they largely function independently.
CREB1 binds near genes that are upregulated, downregulated, and not affected by forskolin treatment.
CREB1 was found to bind near genes that were regulated by forskolin treatment as well as those that were not. Of the 100 genes that contained CREB1 sites near or within them, only 15% exhibited altered expression upon forskolin treatment. There are at least two explanations for why CREB target genes are not affected in their expression. First, they may already be expressed in the absence of forskolin, and forskolin induction may do little to enhance their expression further. This is likely in the case of many genes, such as MAPK1, which are expressed in most cell types. Second, expression may not be affected because additional factors may be required to work with CREB to activate gene expression. Combinatorial gene regulation with multiple factors is typical for most eukaryotic genes. This is a likely explanation for the neuronal genes that we found, since these are not expected to be expressed in placental cells. Nonetheless, it is possible to identify many such targets for a factor that is constitutively bound, such as CREB with a single cell type. Different targets in this set may be regulated by CREB in other cell types or tissues.
Although CREB1 is typically described as a transcriptional activator, we observed that a number of CREB1 binding regions were found in genes that are downregulated in response to forskolin. Several studies indicate that CREB might serve as a repressor. Binding of the CREB/CREM complex at the interleukin-2 promoter represses interleukin-2 production in anergic T cells (29). CREB has also been reported as a transcriptional repressor in the promoter for the vascular endothelial growth factor receptor (14).
There are several explanations for how CREB might repress transcription. First, CREB1 repression may occur through heterodimerization or interaction with another transcription factor. The transcription factor YY1 is thought to obstruct contact between DNA-bound CREB and transcription machinery (8). Second, repression might also be due to an inhibitory CREB1 covalent modification. CaMKII phosphorylates CREB at Ser-133 and also at a second site, Ser-142. In vitro, this second-site phosphorylation at Ser-142 blocks the activation of CREB that would otherwise result when Ser-133 alone is phosphorylated (28, 35). In rats, the addition of O-linked β_-N_-acetylglucosamine interferes with CREB's ability to associate with TAF(II)130 and consequently represses transcription in vitro (19). Lastly, CREB1 repression may arise through CBP and p300, both of which interact with CREB. SUMO (small ubiquitin-like modifier) conjugation to p300 leads to recruitment of a histone deacetylase and repression (9). Histone deacetylase 1 association with CREB was found in JEG-3 cells at times when the promoter was not active (2). Thus, repression of gene expression may occur by a variety of methods.
Eighty-four forskolin-regulated genes did not contain CREB1 binding sites. Since cAMP can also affect transcription through other transcription factors, including other CREB family members (e.g., CREM and ATF1), NF-κB, and some of the nuclear receptors (reviewed in reference 5), presumably these factors altered the expression of the non-CREB targets. Alternatively, such targets may lie downstream of CREB1; since several potential targets of CREB1 are themselves transcription factors, it is likely that many genes will be regulated by other components of the cascade.
Involvement of putative CREB1 targets in signal transduction.
One of the most interesting findings of this study is that many putative CREB1 targets are in signal transduction pathways. A number of these putative targets have the potential to up-regulate or down-regulate the CREB signaling pathway and suggest a number of new modes of pathway regulation (Fig. 5). CREB1 binds near two genes that may regulate cAMP pathway signaling, ADRBK2 and ADSL. The ADRBK2 β-adrenergic receptor kinase modulates G-protein-coupled receptor signaling, which is often linked to activation of adenylate cyclase; thus, the regulation of ADRBK2 via CREB1 may affect cAMP levels. ADSL, which we found was upregulated by forskolin treatment, catalyzes two steps in AMP synthesis and presumably indirectly affects cAMP levels. The discovery that CREB1 binds near putative targets that may modulate cAMP signaling raises the possibility of autoregulation by CREB.
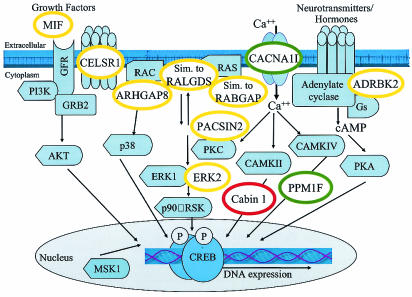
FIG. 5.
Model of CREB pathway and possible positions of proteins whose genes contain CREB1 binding sites. Proteins whose mRNAs are upregulated, downregulated, or not affected by forskolin are indicated in the red, green, and yellow circles, respectively. Components of signaling pathways (light blue shapes) that converge on CREB are adapted from reference 22. MIF, macrophage migration inhibitory factor; CELSR1, cadherin epidermal growth factor LAG seven-pass G-type receptor; ARHGAP8, rho GTPase-activating protein 8; dJ930L11.1, similar to the RabGAP domain-containing gene KIAA0397; Em:AP000347.C22.3, similar to mouse guanine nucleotide dissociation stimulator RALGDS; Cabin 1, calcineurin binding protein; PACSIN2, protein kinase C and casein kinase substrate in neurons 2; ERK2/MAPK1, mitogen-activated protein kinase 1; PPM1F, protein phosphatase 1F; ADRBK2, adrenergic beta receptor kinase 2.
Three potential CREB1 targets encode proteins involved in Ca2+ signaling. Cabin1 is a large, 240-kDa calcineurin binding protein that may serve as a scaffold to link calcineurin with its protein targets (11, 18). Cabin 1 may interfere with the dephosphorylation of CREB in a reaction catalyzed by protein phosphatase 1 and promoted by calcineurin (1, 21). Cabin 1 was upregulated by forskolin, suggesting that CREB1 binding to the Cabin 1 gene might lead to the persistence of activated CREB. PPM1F, which exhibited reduced expression upon forskolin treatment, encodes a protein phosphatase that dephosphorylates the CaM kinase CaMKII in vitro (37). CaMKII phosphorylates CREB at both Ser-133 and Ser-142; phosphorylation at Ser-142 inhibits the transactivation of CREB (35). Therefore, similar to CREB's effect on Cabin1, CREB1 binding to PPM1F might enhance CREB activity. Expression of the CACNA1I T-type calcium channel subunit was repressed upon forskolin treatment. In some cell types, stimulation of Ca2+ influx through T-type calcium channels is mediated through the cAMP-dependent protein kinase pathway upon activation of adenylate cyclase (20). Thus, repression of CACNA1I potentially results in downregulation of the CREB pathway.
CREB1 has multiple binding sites in genes important for lipid signaling, the cytoskeleton, and vesicular trafficking. The PIK4CA phosphatidylinositol 4-kinase and related phosphatidylinositol 4-kinase 230 gene Em:AP000557.C22.3 potentially regulate phosphatidylinositol-4 levels which in turn are likely to affect the activity of protein kinase C, cytoskeletal reorganization, and secretion. Consistent with a role in membrane trafficking and signaling, CREB1 binding regions were found in genes for PACSIN2, the Rho GTPase-activating protein ARHGAP8, Em:AP000347.C22.3 (encoding a calponin-like actin binding protein), β-parvin, and clatherin heavy polypeptide-like 1 gene.
Finally, CREB1 binds both 5′ and within the MAPK1 gene (Table 2) and other Ras/MAPK signaling components (Fig. 5). MAPK I activation leads to phosphorylation of Ser-133 of CREB. dJ930L11.1 and Em:AP000347.C22.3 encode proteins homologous to the RabGAP domain-containing gene KIAA0397 and RALGDS, respectively. Although the levels of expression of MAPK1, dJ930L11.1, and Em:AP000347.C22.3 did not vary in response to forskolin treatment, the expression of these genes may change in response to other stimuli.
In conclusion, a wide variety of signaling components are putative targets of CREB1. Since many of these are regulated by treatment with forskolin, it is expected that they will affect the dynamics of CREB signaling. Presumably such control is important in activating and downregulating the pathway at different times during the CREB activation response and in different cell types.
Many putative CREB1 targets are involved in neuronal function.
CREB has a well-established role in regulating memory and behavior in animals (22). Interestingly, although we worked with placenta-derived cells, several potential CREB1 targets are involved in neuronal function. These include SYN3, SNAP29, PACSIN2, and ATAXIN10. Syntaxin 3 and SNAP29 are important for synaptic transmission. Not only did we find many signal transduction components, but CREB1 also bound near genes important for cytoskeletal reorganization, including small GTPase effectors (e.g., ARHGAP8), and several actin-associated proteins (Em:AP000354.C22.2 and bK414D7.C22.1). We speculate that a number of potential CREB1 targets may be important for regulated secretion or may link signal transduction with axon growth or neuronal migration, especially considering that cytoplasmic levels of cAMP can switch growth cones between attraction and repulsion (34).
Finally, we note that many of the same signaling pathways underlie both neuronal navigation and leukocyte migration and that CREB has an emerging role in the immune system (17). We found that CREB1 targets the macrophage migration inhibitory factor lymphokine (Table 2). Additionally, in rats the migration inhibitory factor promoter has a functional CRE and binds CREB (41).
In this work, we identified the in vivo binding sites of CREB1 across an entire human chromosome. Our studies support the constitutive binding of CREB to chromatin. Only a modest percentage of sites were found near or within genes whose expression was altered by forskolin, a cAMP agonist. CREB1 binds upstream of genes and in introns. Furthermore, we found significant binding near transcriptionally active regions with no previous annotation, perhaps indicating the existence of unidentified genes in these regions. Lastly, the distribution CREB1 binding sites, as gathered from an unbiased approach, presents interesting issues of chromosomal looping and suggests that the number of potential regulatory sequences recognized by CREB has been underestimated. In summary, a large number of potential gene targets for CREB1 have been identified, raising the possibility that it has important roles in a variety of cellular processes.
Acknowledgments
We thank Eric White and Daniel Gelperin for discussion and critical reading of the manuscript. George Goad helped with construction of the human chromosome 22 microarray.
G.E. is supported by National Institutes of Health Postdoctoral Fellowship F32 HG02446-01. R.M. is supported by a National Institutes of Health predoctoral training grant. This work was supported by National Institutes of Health grant HG02357.
REFERENCES
- 1.Bito, H., K. Deisseroth, and R. W. Tsien. 1996. CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell 87**:**1203-1214. [DOI] [PubMed] [Google Scholar]
- 2.Canettieri, G., I. Morantte, E. Guzman, H. Asahara, S. Herzig, S. D. Anderson, J. R. Yates, and M. Montminy. 2003. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat. Struct. Biol. 10**:**175-181. [DOI] [PubMed] [Google Scholar]
- 3.Collins, J. E., M. E. Goward, C. G. Cole, L. J. Smink, E. J. Huckle, S. Knowles, J. M. Bye, D. M. Beare, and I. Dunham. 2003. Reevaluating human gene annotation: a second-generation analysis of chromosome 22. Genet. Res. 13**:**27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conkright, M. D., E. Guzman, L. Flechner, A. I. Su, J. B. Hogenesch, and M. Montminy. 2003. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell 11**:**1101-1108. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, P. B., W. H. Walker, and J. F. Habener. 1998. Cyclic AMP signaling and gene recognition. Annu. Rev. Nutr. 18**:**353-383. [DOI] [PubMed] [Google Scholar]
- 6.Delegeane, A. M., L. H. Ferland, and P. L. Mellon. 1987. Tissue-specific enhancer of the human glycoprotein hormone alpha-subunit gene: dependence on cyclic AMP-inducible elements. Mol. Cell. Biol. 7**:**3994-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Sa, C., L. M. Tolbert, M. Conti, and R. S. Duman. 2002. Regulation of cAMP-specific phosphodiesterases type 4B and 4D PDE4 splice variants by cAMP signaling in primary cortical neurons. J. Neurochem. 81**:**745-757. [DOI] [PubMed] [Google Scholar]
- 8.Galvin, K. M., and Y. Shi. 1997. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17**:**3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11**:**1043-1054. [DOI] [PubMed] [Google Scholar]
- 10.Hagiwara, M., P. Brindle, A. Harootunian, R. Armstrong, J. Rivier, W. Vale, R. Tsien, and M. R. Montminy. 1993. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol. Cell. Biol. 13**:**4852-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han, A., F. Pan, J. C. Stroud, H. Youn, J. O. Liu, and L. Chen. 2003. Sequence-specific recruitment of transcriptional co-repressor Cabin 1 by myocyte enhancer factor-2. Nature 422**:**730-734. [DOI] [PubMed] [Google Scholar]
- 12.Harrison, P. M., H. Hegyi, S. Balasubramanian, N. M. Luscombe, P. Bertone, N. Echols, T. Johnson, and M. Gerstein. 2002. Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 12**:**272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horak, C. E., M. C. Mahajan, N. M. Luscombe, M. Gerstein, S. M. Weissman, and M. Snyder. 2002. GATA-1 binding sites mapped in the β-globin locus by using mammalian chIP-chip analysis. Proc. Natl. Acad. Sci. USA 99**:**2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Illi, B., P. L. Puri, L. Morgante, M. C. Capogrossi, and C. Gaetano. 2000. Nuclear factor-κB and cAMP response element binding protein mediate opposite transcriptional effects on the Flk-1/KDR gene promoter. Circ. Res. 86**:**e110-e117. [PubMed] [Google Scholar]
- 15.Jameson, J. L., R. C. Jaffe, S. L. Gleason, and J. F. Habener. 1986. Transcriptional regulation of chorionic gonadotropin alpha- and beta-subunit gene expression by 8-bromo-adenosine 3′,5′-monophosphate. Endocrinology 119**:**2560-2570. [DOI] [PubMed] [Google Scholar]
- 16.Jameson, J. L., R. C. Jaffe, P. J. Deutsch, C. Albanese, and J. F. Habener. 1988. The gonadotropin alpha-gene contains multiple protein binding domains that interact to modulate basal and cAMP-responsive transcription. J. Biol. Chem. 263**:**9879-9886. [PubMed] [Google Scholar]
- 17.Kuo, C. T., and J. M. Leiden. 1999. Transcriptional regulation of T lymphocyte development and function. Annu. Rev. Immunol. 17**:**149-187. [DOI] [PubMed] [Google Scholar]
- 18.Lai, M. M., H. R. Luo, P. E. Burnett, J. J. Hong, and S. H. Snyder. 2000. The calcineurin-binding protein Cain is a negative regulator of synaptic vesicle endocytosis. J. Biol. Chem. 275**:**34017-34020. [DOI] [PubMed] [Google Scholar]
- 19.Lamarre-Vincent, N., and L. C. Hsieh-Wilson. 2003. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J. Am. Chem. Soc. 125**:**6612-6613. [DOI] [PubMed] [Google Scholar]
- 20.Lenglet, S., E. Louiset, C. Delarue, H. Vaudry, and V. Contesse. 2002. Activation of 5-HT7 receptor in rat glomerulosa cells is associated with an increase in adenylyl cyclase activity and calcium influx through T-type calcium channels. Endocrinology 143**:**1748-1760. [DOI] [PubMed] [Google Scholar]
- 21.Liu, F. C., and A. M. Graybiel. 1996. Spatiotemporal dynamics of CREB phosphorylation: transient versus sustained phosphorylation in the developing striatum. Neuron 17**:**1133-1144. [DOI] [PubMed] [Google Scholar]
- 22.Lonze, B. E., and D. D. Ginty. 2002. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35**:**605-623. [DOI] [PubMed] [Google Scholar]
- 23.Luscombe, N. M., T. E. Royce, P. Bertone. N. Echols, C. E. Horak, J. T. Chang, M. Snyder, and M. Gerstein. 2003. ExpressYourself: a modular platform for processing and visualizing microarray data. Nucleic Acids Res. 31**:**3477-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao, D., E. A. Warner, S. A. Gurwitch, and D. R. Dowd. 1998. Differential regulation and transcriptional control of immediate early gene expression in forskolin-treated WEH17.2 thymoma cells. Mol. Endocrinol. 12**:**492-503. [DOI] [PubMed] [Google Scholar]
- 25.Martone, R., G. Euskirchen, P. Bertone, S. Hartman, T. E. Royce, N. M. Luscombe, J. L. Rinn, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. Distribution of NF-κB binding sites across human chromosome 22. Proc. Natl. Acad. Sci. USA 100**:**12247-12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2**:**599-609. [DOI] [PubMed] [Google Scholar]
- 27.Newman, J. R. S., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300**:**2097-2101. [DOI] [PubMed] [Google Scholar]
- 28.Parker, D., U. S. Jhala, I. Radhakrishnan, M. B. Yaffe, C. Reyes, A. I. Shulman, L. C. Cantley, P. E. Wright, and M. Montminy. 1998. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell 2**:**353-359. [DOI] [PubMed] [Google Scholar]
- 29.Powell, J. D., C. G. Lerner, G. R. Ewoldt, and R. H. Schwartz. 1999. The −180 site of the IL-2 promoter is the target of CREB/CREM binding in T-cell anergy. J. Immunol. 163**:**6631-6639. [PubMed] [Google Scholar]
- 30.Quackenbush, J. 2002. Microarray data normalization and transformation. Nat. Genet. Suppl. 32**:**496-501. [DOI] [PubMed] [Google Scholar]
- 31.Rinn, J. L., G. Euskirchen, P. Bertone, R. Martone, N. M. Luscombe, S. Hartman, P. M. Harrison, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. The transcriptional activity of human chromosome 22. Genes Dev. 17**:**529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassone-Corsi, P. 1995. Transcription factors responsive to cAMP. Annu. Rev. Cell Dev. Biol. 11**:**355-377. [DOI] [PubMed] [Google Scholar]
- 33.Shieh, P. B., S. C. Hu, K. Bobb, T. Timmusk, and A. Ghosh. 1998. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron 20**:**727-740. [DOI] [PubMed] [Google Scholar]
- 34.Song, H., and M. M. Poo. 2001. The cell biology of neuronal navigation. Nat. Cell Biol. 3**:**E81-E88. [DOI] [PubMed] [Google Scholar]
- 35.Sun, P., H. Enslen, P. S. Myung, and R. A. Maurer. 1994. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 8**:**2527-2539. [DOI] [PubMed] [Google Scholar]
- 36.Tabuchi, A., H. Sakaya, T. Kisukeda, H. Fushiki, and M. Tsuda. 2002. Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J. Biol. Chem. 277**:**3592-35931. [DOI] [PubMed] [Google Scholar]
- 37.Tan, K. M. L., S. L. Chan, K. O. Tan, and V. C. Yu. 2001. The Caenorhabditis elegans sex-determining protein FEM-2 and its human homologue, hFEM-2, are Ca2+/calmodulin-dependent protein kinase phosphatases that promote apoptosis. J. Biol. Chem. 276**:**44193-44202. [DOI] [PubMed] [Google Scholar]
- 38.Tao, X., S. Finkbeiner, D. B. Arnold, A. J. Shaywitz, and M. E. Greenberg. 1998. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20**:**709-726. [DOI] [PubMed] [Google Scholar]
- 39.Vicini, E., and M. Conti. 1997. Characterization of an intronic promoter of a cyclic adenosine 3′,5′-monophosphate cAMP-specific phosphodiesterase gene that confers hormone and cAMP inducibility. Mol. Endocrinol. 11**:**839-850. [DOI] [PubMed] [Google Scholar]
- 40.Vinson, C., M. Myakishev, A. Acharya, A. A. Mir, J. R. Moll, and M. Bonovich. 2002. Classification of human b-Zip proteins based on dimerization properties. Mol. Cell. Biol. 22**:**6321-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waeber, G., N. Thompson, T. Chautard, M. Steinman, P. Nicod, F. P. Pralong, T. Calandra, and R. C. Gaillard. 1998. Transcriptional activation of the macrophage migration-inhibitory factor gene by the corticotropin-releasing factor is mediated by the cyclic adenosine 3′,5′-monophosphate responsive element-binding protein CREB in pituitary cells. Mol. Endocrinol. 12**:**698-705. [DOI] [PubMed] [Google Scholar]
- 42.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitations. Methods 26**:**37-47. [DOI] [PubMed] [Google Scholar]
- 43.Young, M., and M. J. McPhaul. 1998. A steroidogenic factor-1-binding site and cyclic adenosine 3′,5′-monophosphate response element-like elements are required for the activity of the rat aromatase promoter in rat Leydig tumor cell lines. Endocrinology 139**:**5082-5093. [DOI] [PubMed] [Google Scholar]