Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation (original) (raw)
Cancer cells frequently depend on chromatin regulators to maintain their malignant phenotype. Brg1, an ATPase subunit of SWI/SNF, is known to suppress tumor formation in several cell types. Vakoc and colleagues now show that leukemia cells instead rely on Brg1 to support their oncogenic transcriptional program, which includes Myc as a key target. Brg1 is critical to sustain transcription factor occupancy and enable long-range looping interactions with the Myc promoter. These findings thus implicate enhancer-mediated Myc regulation in leukemia pathogenesis.
Keywords: SWI/SNF, Brg1, leukemia, enhancer, Brd4, Myc
Abstract
Cancer cells frequently depend on chromatin regulatory activities to maintain a malignant phenotype. Here, we show that leukemia cells require the mammalian SWI/SNF chromatin remodeling complex for their survival and aberrant self-renewal potential. While Brg1, an ATPase subunit of SWI/SNF, is known to suppress tumor formation in several cell types, we found that leukemia cells instead rely on Brg1 to support their oncogenic transcriptional program, which includes Myc as one of its key targets. To account for this context-specific function, we identify a cluster of lineage-specific enhancers located 1.7 Mb downstream from Myc that are occupied by SWI/SNF as well as the BET protein Brd4. Brg1 is required at these distal elements to maintain transcription factor occupancy and for long-range chromatin looping interactions with the Myc promoter. Notably, these distal Myc enhancers coincide with a region that is focally amplified in ∼3% of acute myeloid leukemias. Together, these findings define a leukemia maintenance function for SWI/SNF that is linked to enhancer-mediated gene regulation, providing general insights into how cancer cells exploit transcriptional coactivators to maintain oncogenic gene expression programs.
Comprehensive profiling of cancer genomes has revealed that somatic mutation of genes encoding chromatin regulators is a common driver mechanism of tumorigenesis (Garraway and Lander 2013). While the full mechanistic consequences of these mutations remain poorly understood, one expectation is that such events promote acquisition of cancer cell capabilities through alteration of transcriptional programs. Consequently, targeting the chromatin regulatory machinery provides a means of extinguishing oncogenic gene expression programs for therapeutic purposes (Popovic and Licht 2012). In support of this concept, small-molecule-based inhibition of select chromatin regulators has shown efficacy in clinical and preclinical cancer settings (Dawson and Kouzarides 2012). However, a major ongoing challenge remains in identifying and understanding cancer-specific dependencies on chromatin regulatory activities.
ATP-dependent nucleosome remodeling enzymes are a major category of chromatin regulators, of which SWI/SNF (also known as BAF in mammals) is one of the best studied (Hargreaves and Crabtree 2011). First discovered in yeast, SWI/SNF complexes couple ATP hydrolysis to the perturbation of histone:DNA contacts to promote access of transcription factors (TFs) to their cognate DNA elements (Cote et al. 1994). SWI/SNF complexes lack intrinsic DNA sequence specificity; hence, they are typically recruited to genomic sites through physical interactions with sequence-specific TFs (Neely et al. 1999). As such, SWI/SNF functions as a unique coactivator that both stabilizes TF occupancy and facilitates downstream steps in transcriptional activation (Neely et al. 1999). Mammalian SWI/SNF complexes are comprised of ∼11 subunits that are encoded by ∼19 distinct genes, thereby affording diverse combinatorial assemblies with specialized functions (Wu et al. 2009). For example, SWI/SNF contains one of two possible ATPase subunits, Brg1 or Brm, both of which also possess a bromodomain that interacts with acetylated histones (Wang et al. 1996). Despite their similar domain architectures, Brg1 and Brm each interacts with distinct families of TFs to confer unique functional outputs to the complex (Kadam and Emerson 2003). Tissue-specific expression patterns of certain SWI/SNF subunits can also lead to tailoring of subunit configurations for lineage-specific functionalities (Olave et al. 2002).
Individual SWI/SNF subunits are known to perform specialized functions in the hematopoietic system. For example, conditional inactivation of Smarcb1 (encoding BAF47) and Actl6a (encoding BAF53a) in mice leads to severe defects in multilineage hematopoiesis, whereas a mutant allele of Arid1a (encoding BAF250a) leads to hematopoietic stem cell expansion through a non-cell-autonomous mechanism (Roberts et al. 2002; Krosl et al. 2010; Krasteva et al. 2012). Smarca4 (encoding Brg1) mutant mice display defective erythroid and lymphoid differentiation, whereas hematopoietic stem cells, common myeloid progenitors, and mature myeloid populations are maintained at normal levels (Chi et al. 2003; Bultman et al. 2005; Willis et al. 2012; S. Bultman, pers. comm.). SWI/SNF interacts with several hematopoietic TFs (e.g., Runx1 and EKLF) whose functional impairment in SWI/SNF-deficient animals may account for these hematopoietic abnormalities (Kadam and Emerson 2003; Bakshi et al. 2010).
Genomic studies have uncovered a pervasive tumor suppressor role for SWI/SNF complexes, with a frequency of mutation across human cancer being estimated at ∼18%–20% (Kadoch et al. 2013; Shain and Pollack 2013). This includes loss-of-function mutation of genes encoding BAF250a, Brg1, BAF47, and BAF180, which are particularly common in ovarian, lung, rhabdoid, and renal cell cancers, respectively (Kadoch et al. 2013; Shain and Pollack 2013). In leukemias, decreased SWI/SNF subunit expression has been associated with glucocorticoid resistance in acute lymphoblastic leukemia (ALL) (Pottier et al. 2008), and genetic loss of SMARCB1 has been observed in cases of chronic myeloid leukemia (CML) (Grand et al. 1999). However, the functional involvement of SWI/SNF in leukemia progression is currently not well understood. Interestingly, SWI/SNF mutations have not been found as recurrent alterations in large-scale genomic studies of acute myeloid leukemia (AML) (The Cancer Genome Atlas Research Network 2013), raising the possibility that in this particular cancer, SWI/SNF may not act as a tumor suppressor.
We show here instead that Brg1-containing SWI/SNF complexes are critical for the oncogenic transcriptional program of leukemia cells. This includes a direct role for Brg1 in the maintenance of Myc expression, which we link to a cluster of distal enhancer elements at the Myc locus that are lineage-specific and coincide with a region known to be focally amplified in AML. At these enhancers, Brg1 is critical to sustain TF occupancy and enable long-range looping interactions with the Myc promoter. Collectively, our study implicates a role for SWI/SNF in maintaining the epigenetic state of leukemia cells.
Results
Brg1 and other SWI/SNF subunits are required for leukemia cell expansion
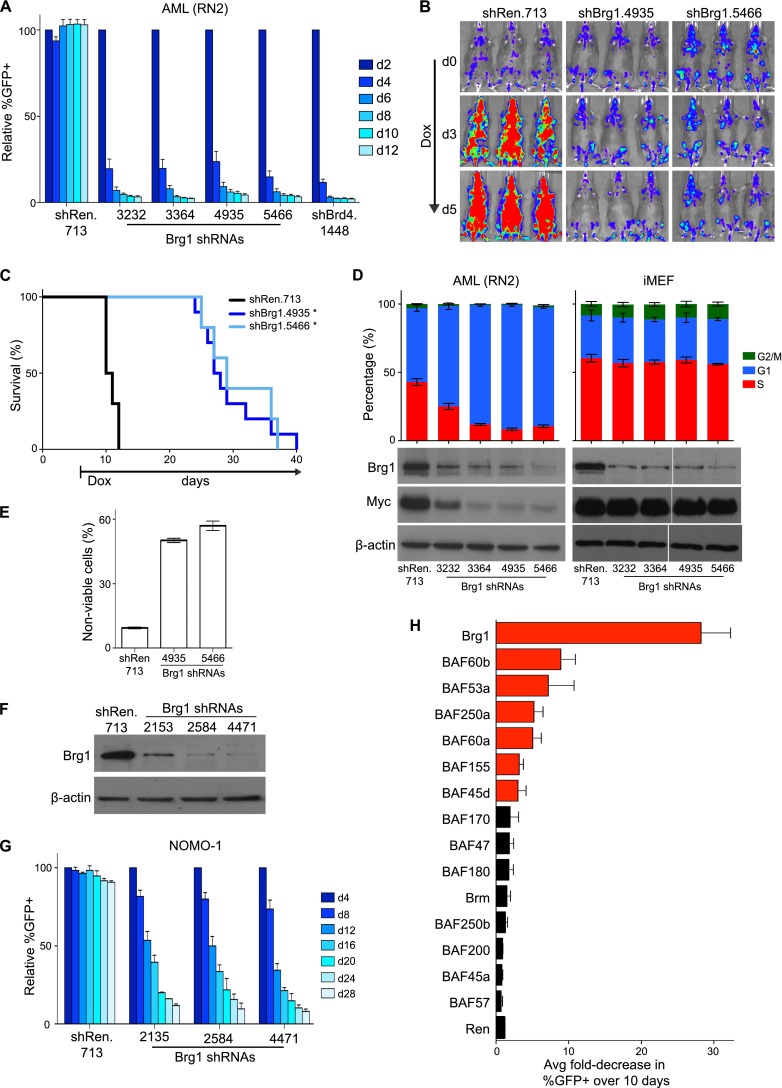
We previously performed an shRNA screen that aimed to identify chromatin regulator dependencies in a mouse model of MLL-AF9/NrasG12D AML (Zuber et al. 2011c). Here we investigated the leukemia maintenance function of Brg1/Smarca4, which scored as a strong hit in this prior screen (Zuber et al. 2011c). We first confirmed the anti-proliferative effects of Brg1 knockdown in leukemia cells using a competition-based assay. Murine MLL-AF9/NrasG12D AML cells (RN2 cell line) transduced with shRNAs targeting Brg1 were rapidly outcompeted by untransduced cells within 6 d in culture—effects nearly as strong as a validated shRNA targeting the BET protein Brd4, which is a known requirement in this model (Fig. 1A; Zuber et al. 2011c). Taking advantage of the Tet-On competence of the RN2 line, we further examined the Brg1 requirement for AML progression under in vivo conditions (Zuber et al. 2011a). Conditional knockdown of Brg1 in established disease led to significant inhibition of disease progression, as measured by bioluminescent imaging of leukemia burden and extension of animal survival (Fig. 1B,C; Supplemental Fig. 1). These findings confirm the Brg1 requirement for disease expansion in a genetically engineered mouse model of MLL-AF9/NrasG12D AML.
Figure 1.
Brg1 and other SWI/SNF subunits are required for AML maintenance. (A) Competition assay to measure the effects of shRNAs on leukemia cell proliferation. The percentage of GFP+/shRNA+ RN2 cells was measured at the indicated time points. Percentages were normalized to day 2 values. shRen.713 is a negative control shRNA targeting Renilla luciferase. n = 3. (B,C) Conditional shRNA experiments performed in vivo. Clonal RN2-TRMPV-Neo lines were transplanted into sublethally irradiated recipient mice followed by administration of doxycycline (dox) at day 6. (B) Bioluminescent imaging of the luciferase+ leukemia burden at the indicated days following dox administration. (C) Kaplan-Meier survival curves. Nine to 10 mice were included in each shRNA group. Statistical significance was calculated using a log-rank test. (*) P < 0.001. (D) Cell cycle and Western blot analysis following dox-induced knockdown of Brg1 in RN2 and iMEF lines transduced with TRMPV-Neo shRNA constructs. shRNA was induced for 48 h with dox followed by BrdU incorporation assays and Western blotting of whole-cell lysates. n = 3. Western blot is a representative of three independent replicates. (E) Measurement of nonviable cells following conditional Brg1 knockdown. shRNA was induced for 72 h with dox. n = 3. (F) Western blotting of Brg1 levels in whole-cell lysates prepared from HeLa cells transduced with the indicated MLP-shRNA constructs. A representative experiment of three independent replicates is shown. (G) Competition assay in NOMO-1 cells (human MLL-AF9+ AML) using the indicated shRNA constructs, as described for A. n = 3. (H) Summary of competition assay data from RN2 cells for 93 independent shRNAs targeting the indicated SWI/SNF subunits. The average fold decrease in GFP percentage over 10 d for four to six independent shRNAs is plotted. Bars are labeled in red if more than two independent shRNAs decreased the percentage of GFP+ more than threefold. All error bars represent SEM.
We next evaluated the specificity of proliferation arrest caused by Brg1 knockdown in various cell backgrounds. Reducing Brg1 levels in RN2 cells led to G1/G0 arrest and cell death, whereas an equivalent knockdown in immortalized murine embryonic fibroblasts (iMEFs) had no significant effect on cell cycle progression or cell survival (Fig. 1D,E; data not shown). We also found that Brg1 shRNAs inhibited proliferation of cells derived from a mouse model of B-cell ALL (B-ALL) initiated by the BCR-ABL oncogene and p19Arf deficiency (Supplemental Fig. 2A; Williams et al. 2006). This indicates that the Brg1 requirement for leukemia proliferation is not restricted to myeloid leukemia or MLL fusion subtypes of disease. In contrast, murine melanoma and breast cancer cell lines proliferated normally despite stable Brg1 knockdown (Supplemental Fig. 2A,B). These findings indicate that the Brg1 requirement for cell proliferation is highly cell context-dependent.
Using shRNAs that are specific to the human gene, we next evaluated the Brg1 requirement in different human leukemia cell lines (Fig. 1F). Proliferation of AML lines harboring endogenous MLL rearrangements (NOMO-1, MV4-11, and MOLM-13) was sensitive to Brg1 knockdown, confirming this subtype of disease as Brg1-dependent (Fig. 1G; Supplemental Fig. 3A). Interestingly, an AML line carrying the AML1-ETO fusion protein (KASUMI-1) was also sensitive to Brg1 knockdown, whereas other AML lines were less affected (HEL and CMK) (Supplemental Fig. 3A). This suggests that Brg1 is required in multiple subtypes of AML but is not a universal requirement in this disease. Proliferation of human T-ALL and B-ALL lines JURKAT and REH, respectively, and the CML blast crisis line K-562 was also inhibited upon Brg1 knockdown (Supplemental Fig. 3A). This overall pattern of sensitivity was distinct from other strong hits in our prior screen (Zuber et al. 2011c), such as SPT16, a subunit of the FACT histone chaperone complex, whose knockdown strongly inhibited proliferation across all human lines examined (Supplemental Fig. 3A,B). These findings suggest that a subset of leukemias require Brg1 to proliferate.
Since Brg1 is a constituent of SWI/SNF, we also evaluated whether other subunits of this complex were required for leukemia proliferation. We designed four to six independent shRNAs targeting each SWI/SNF subunit (93 shRNAs in total), which were evaluated one by one in a competition assay for effects on RN2 cell proliferation. Notably, BAF60c, BAF45b, BAF45c, and BAF53b are not expressed in RN2 cells and were excluded from the screen (Supplemental Fig. 4A). This shRNA screen identified BAF60b, BAF53a, BAF250a, BAF60a, BAF155, and BAF45d as additional requirements for RN2 proliferation based on a criteria of (1) multiple independent shRNAs reducing the GFP+ percentage more than threefold over 10 d in the competition assay and (2) validated on-target knockdown measured by RT-qPCR for growth inhibitory shRNAs (Fig. 1H; Supplemental Fig. 4B,C). Collectively, these results suggest that specific Brg1-containing SWI/SNF complexes are required for leukemia maintenance.
Brg1 promotes self-renewal of leukemia cells by maintaining Myc transcription
Since impaired differentiation is a property of AML, we next considered whether Brg1 regulates the lineage profile of leukemia cells. Following conditional Brg1 knockdown, RN2 cells transitioned from an immature blast morphology to one resembling mature macrophages (Fig. 2A). Consistent with this observation, Brg1 knockdown also decreased the cell surface levels of the leukemia stem cell (LSC) marker ckit and increased the levels of Mac1, a macrophage marker (Fig. 2B,C). We further evaluated the effect on cell differentiation using gene set enrichment analysis (GSEA) of expression microarray data obtained from Brg1-deficient leukemia cells following conditional knockdown using three independent Brg1 shRNAs (Subramanian et al. 2005). This revealed a significant decrease in the expression of genes known to be enriched in LSCs as well as an increase in the expression of macrophage-related genes upon Brg1 knockdown (Fig. 2D,E). Taken together, these findings suggest that Brg1 is required to preserve an undifferentiated cell state in AML.
Figure 2.

Brg1 maintains an undifferentiated cell state in AML. (A) Light microscopy of May-Grunwald/Giemsa-stained RN2 cells transduced with the indicated TRMPV-Neo shRNA constructs. Cells were treated with dox for 3 d. Imaging was performed with a 40× objective. A representative image of three independent biological replicates is shown. (B,C) Flow cytometry analysis of c-kit (B) and Mac-1 (C) surface expression after 96 h of dox treatment. A representative experiment of three biological replicates is shown. (D,E) GSEA of LSC and macrophage development gene sets following Brg1 knockdown. Microarray analysis was performed comparing three independent Ren.713 shRNA RN2 lines with three independent Brg1 shRNA RN2 lines (4935, 3364, and 3232). The dox-inducible TRMPV-Neo vector was used. Dox treatment was for 96 h. (NES) Normalized enrichment score; (FDR),false discovery rate.
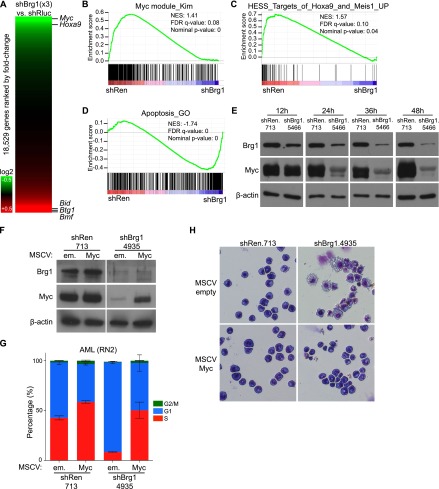
To explain the differentiation phenotype seen in Brg1-deficient AML cells, we examined the expression of known regulators of self-renewal in leukemia using the transcriptome data described above. While expression of several regulators was perturbed upon Brg1 knockdown, Myc was among the most down-regulated genes identified (P < 0.01) (Fig. 3A). Hoxa9, which is a direct target of the MLL-AF9 oncoprotein, was also down-regulated (P < 0.01), albeit to a lesser extent than Myc (Fig. 3A). Gene signatures linked to Myc and Hoxa9 function were globally suppressed following Brg1 knockdown, further confirming an effect on these two pathways (Fig. 3B,C; Supplemental Fig. 5). We noted that the majority of direct MLL-AF9 target genes (Bernt et al. 2011), however, were not down-regulated upon Brg1 knockdown (data not shown), suggesting that MLL-AF9 and Brg1 display overlapping functions at select genes yet are largely distinct from one another in their regulatory activities. We found that Myc expression was also Brg1-dependent in murine BCR-ABL+ B-ALL, whereas Myc levels were unaffected by Brg1 knockdown in iMEFs or melanoma or breast cancer lines. (Fig. 1D; Supplemental Fig. 2B). Thus, the effect on Myc expression correlates with the Brg1 requirement for proliferation across these different lines. We found that Myc expression was also rapidly decreased within the first 24 h of Brg1 knockdown in RN2 cells, suggesting direct regulation (Fig. 3E).
Figure 3.
Brg1 maintains Myc and Hoxa9 expression and represses proapoptotic genes in AML. (A) Heat map of gene expression changes following Brg1 knockdown using microarray data described above. Each protein-coding gene represented on the array is ranked based on fold change in expression, with the position of select genes indicated by horizontal lines. Results are the average of three independent Brg1 shRNAs. (B_–_D) GSEA following Brg1 knockdown, as described in Figure 2. (E) Time-course Western blotting of Myc and Brg1 levels in RN2 cells following dox treatment of TRMPV-Neo RN2 lines with the indicated shRNAs. shRNA expression was triggered by dox for the indicated amount of time before lysate preparation. (F) Western blotting following 48 h of dox-induced Brg1 knockdown performed in AML (RN2) cells that were pretransduced with either MSCV-em (empty) or MSCV-Myc vectors. A representative experiment of three biological replicates is shown. (G) BrdU cell cycle analysis performed on day 3 of dox treatment. n = 3. (H) Imaging of cell morphology of the same cells as described in F. All error bars represent SEM.
Since Brg1 knockdown leads to a differentiation phenotype similar to that observed upon Myc inhibition (Zuber et al. 2011b) and Myc is a potent oncogene in AML (Luo et al. 2005), we evaluated the functional significance of Myc to the Brg1 requirement in leukemia cells. When transcribed from a retroviral promoter, the expression level of Myc was significantly less sensitive to Brg1 knockdown (Fig. 3F), suggesting that Brg1 regulates Myc primarily from its endogenous chromosomal context. Notably, leukemia cells transduced with retroviral Myc constructs were resistant to cell cycle arrest and myeloid differentiation induced by Brg1 knockdown (Fig. 3G,H), indicating that maintenance of endogenous Myc expression is one key function of Brg1 in this disease. _Myc_-transduced RN2 cells still underwent cell death following Brg1 knockdown (data not shown), indicating a Myc-independent function of Brg1 in regulating cell survival. Consistent with this possibility, Brg1 knockdown globally altered the expression of apoptosis regulators (Fig. 3D), with several proapoptotic genes being prominently up-regulated following Brg1 knockdown (e.g., Bid, Btg1, and Bmf; P < 0.01 for each) (Fig. 3A). From this analysis, we conclude that Brg1 maintains a gene expression program that supports leukemia cell expansion, which includes Myc as one of its key downstream targets.
The regulatory functions of Brg1 in RN2 cells are highly similar to the previously described functions of the BET protein Brd4, which also promotes Myc transcription in this leukemia model (Dawson et al. 2011; Zuber et al. 2011c). To evaluate the similarity between Brd4 and Brg1 functions, we first performed a time-course RNA sequencing (RNA-seq) analysis to define a group of 53 genes whose expression is rapidly reduced following exposure to JQ1, a small-molecule inhibitor of BET bromodomains (Supplemental Fig. 6A; Filippakopoulos et al. 2010). Using GSEA, we found that nearly all of these 53 genes were also suppressed following Brg1 knockdown (Supplemental Fig. 6B), suggesting that Brg1 and Brd4 regulate a highly overlapping set of genes in RN2 cells, which includes Myc. Prior proteomic studies found an association between SWI/SNF subunits and BET proteins (Denis et al. 2006; Dawson et al. 2011; Rahman et al. 2011), which we also have confirmed and found to be mediated by the ET domain of Brd4 (Supplemental Fig. 6C–E). However, using ChIP-qPCR, we found that Brg1 and Brd4 occupy chromatin independently of one another at all of the sites we examined (Supplemental Fig. 6F,G), suggesting that, despite their apparent physical association, Brg1 and Brd4 operate in parallel pathways to maintain a common gene regulatory network in this disease. The similar regulatory functions of Brg1 and Brd4 in leukemia might be related to both factors regulating gene expression through common cis elements, as described below.
Brg1 occupies a cluster of lineage-specific enhancers (E1–E5) located 1.7 Mb downstream from Myc
An important question raised by our findings is how Brg1 can promote Myc expression specifically in leukemia but not in other cell types, since both Brg1 and Myc are expressed ubiquitously. To explain this observation, we hypothesized that Brg1 regulates Myc transcription in leukemia cells by occupying cell type-specific regulatory elements. We evaluated this possibility by performing chromatin immunoprecipitation (ChIP) followed by next-generation sequencing (ChIP-seq) analysis of Brg1, histone H3 Lys 4 trimethylation (H3K4me3), and histone H3 Lys 27 acetylation (H3K27ac) in RN2 leukemia cells (Fig. 4A). We also included Brd4, a known regulator of Myc transcription in leukemia cells, in this analysis, since its role at distal regulatory elements was not evaluated in prior studies (Dawson et al. 2011; Mertz et al. 2011; Zuber et al. 2011c). A survey of the extended Myc locus revealed Brg1 and Brd4 occupancy in the vicinity of the Myc promoter and at a prominent cluster of peaks located 1.7 Mb downstream (Fig. 4A). We refer to this cluster of distal elements here as E1–E5 (Fig. 4B). E1–E5 all displayed enrichment for H3K27ac and only low levels of H3K4me3, suggesting that this region may contain active enhancers (Fig. 4A,B; Rada-Iglesias et al. 2011). The strong enrichment of Brd4 occupancy at these sites classifies these elements as a “super-enhancer,” using the criteria of Loven et al. (2013) (Supplemental Fig. 7). Interestingly, the E1–E5 region is closely matched to a previously reported location of recurrent focal amplification seen previously in ∼3% of AML cases (Fig. 4C; Radtke et al. 2009; Kuhn et al. 2012). We observed a trend that The Cancer Genome Atlas (TCGA) tumor samples harboring genomic amplifications 3′ of MYC (not containing MYC itself) tended to have higher MYC mRNA levels (Supplemental Fig. 8). These findings together raised the possibility that E1–E5 are distal enhancer elements that control Myc expression in leukemia cells.
Figure 4.
Intergenic occupancy of Brg1 and Brd4 1.7 Mb downstream from Myc coincides with a region of recurrent focal amplification in AML. (A) ChIP-seq occupancy profiles of Brg1, Brd4, H3K27ac, and H3K4me3 obtained in RN2 cells (in reads per million). Myc is transcribed from left to right in this depiction. Validated transcript models from the mm8 genome assembly are depicted below. (*) Noncoding RNAs. (B) ChIP-seq data in a window of ∼130 kb surrounding the E1–E5 regions. (C) The locations of somatic copy number amplifications identified on human chromosome 8 from prior studies are represented as solid red lines (Radtke et al. 2009; Kuhn et al. 2012). The labels indicate sample IDs used in the prior studies. Locations are depicted on the hg19 genome assembly.
We first examined whether Brg1 occupancy at E1–E5 was lineage-specific and hence could be correlated with the Brg1 requirement for Myc expression. While Brg1 occupied the Myc promoter region in all of the cell types examined, Brg1 occupancy at E1–E5 was found only in leukemia cell lines (RN2, B-ALL, and NOMO-1) (Fig. 5A–E; Supplemental Fig. 9). Indeed, an evaluation of published ChIP-seq data obtained from normal mouse tissues revealed that despite ubiquitous acetylation near the Myc promoter, H3K27ac was absent from the E1–E5 region in most normal mouse tissues (Fig. 5F; Supplemental Fig. 10A; Shen et al. 2012). We detected low-level yet significant H3K27ac enrichment at the E1–E5 elements in normal bone marrow, embryonic day 14.5 (E14.5) fetal livers (which are largely hematopoietic), and mature myeloid cells (bone marrow-derived macrophages), suggesting that E1–E5 is also active in normal hematopoietic cells (Fig. 5F; Supplemental Fig. 10B). Luciferase reporter assays linked to a minimal thymidine kinase promoter revealed that E3 exhibits enhancer activity in K-562 leukemia cells but not in a nonleukemia cell line, HEK293T (Fig. 5G). K-562 was used for this experiment because it was the only leukemia line that we could readily transfect with reporter constructs. K-562 harbors H3K27ac and Brg1 enrichment at its endogenous E3 region but not at E1–E2 or E4–E5 (Supplemental Fig. 11). Notably, we detected occupancy of BAF250a and BAF60a at the E1–E5 regions in RN2 cells, suggesting the presence of a SWI/SNF complex at these sites (Supplemental Fig. 12). These findings together suggest that E1–E5 elements are a cluster of lineage-specific enhancers that are occupied by Brg1-containing SWI/SNF complexes.
Figure 5.
E1–E5 elements are a lineage-specific cluster of enhancers occupied by Brg1. (A_–_E) ChIP-qPCR with Brg1 or control IgG antibodies. PCR primer amplicons are indicated along the _X_-axis. Neg refers to a negative control region in a gene desert region, and −2 kb is relative to the Myc transcription start site. n = 3. A_–_C represent murine cell lines and primer pairs. D and E represent human lines and primer pairs. (F) ChIP-seq profiles of H3K27ac from the indicated normal mouse tissues, obtained from Shen et al. (2012). The AML H3K27ac data were obtained from RN2 cells. (BM) Bone marrow; (BMDM) BM-derived macrophages; (SmInt) small intestine. (G) Enhancer reporter assay. The indicated PGL4.23 constructs were transfected into HEK293T or K562 cells for 48 h followed by measurement of luciferase activity. Results were normalized to the empty vector control. n = 3. (H) ChIP-seq profiles of TF occupancy performed in HPC-7 cells, obtained from Wilson et al. (2010). (I_–_M) ChIP-qPCR with the indicated TFs or control IgG antibodies performed in RN2 cells. PCR primer amplicons are indicated along the _X_-axis. Neg refers to a negative control region in a gene desert region. n = 3. All error bars represent SEM.
Based on the findings described above, we reasoned that enhancer activity at E1–E5 was likely to reflect occupancy of hematopoietic TFs, whose expression and/or function might be deregulated in leukemia cells. By analyzing ChIP-seq data of TF occupancy obtained previously from immortalized HPC-7 hematopoietic stem and progenitor cells (Wilson et al. 2010), we found that seven hematopoietic TFs occupied various combinations of the E1–E5 elements (Erg, Fli1, Gfi1b, Lmo2, Meis1, Pu.1, and Runx1) (Fig. 5H). Since all of these TFs are expressed in RN2 cells (Supplemental Fig. 13), we anticipated that these factors would occupy E1–E5 in leukemia as well. Indeed, we detected occupancy of Lmo2, PU.1, and Erg at various subsets of the E1–E5 enhancers in RN2 using ChIP-qPCR (Fig. 5I–K). Additionally, we found that the hematopoietic TFs Cebpα and Cebpβ, both of which are highly expressed in RN2 cells, also occupied the E1–E5 elements (Fig. 5L,M; Supplemental Fig. 13). Notably, Runx1, Cebpα, and Cebpβ have been shown previously to associate with SWI/SNF (Kowenz-Leutz and Leutz 1999; Pedersen et al. 2001; Bakshi et al. 2010). Furthermore, we found that Flag-tagged PU.1 could immunoprecipitate Brg1 with an efficiency comparable with that of Cebpα (Supplemental Fig. 14), suggesting that multiple TFs at E1–E5 have the capacity to interact with SWI/SNF. These data show that the E1–E5 enhancers are occupied by a set of hematopoietic TFs, of which several have known roles in promoting leukemogenesis (Rosenbauer and Tenen 2007).
We also found sites of Brg1 occupancy in the vicinity of Hoxa9 and at the promoters of Bid, Btg1, and Bmf, suggesting direct regulatory effects at these genes (Supplemental Fig. 15). The presence of Brg1 at the promoters of Bid, Btg1, and Bmf suggests that Brg1 could suppress cell death in leukemia cells through direct transcriptional repression of proapoptotic genes, which is consistent with prior reports that SWI/SNF can repress transcription of specific target genes (e.g., Cd4 in thymocyte progenitors) (Chi et al. 2002). Thus, the direct regulatory effects of Brg1 at multiple genes are likely to account for the entirety of the Brg1 requirement in this disease.
The Myc locus exists in a lineage-specific conformation that juxtaposes the E1–E5 enhancers with the Myc promoter
The E1–E5 enhancers are located nearly 1.7 Mb away from Myc and are actually closer in genomic distance to other flanking genes (Gsdmc, Fam49b, and Asap1) (Fig. 6A). While Gsdmc is not expressed in leukemia cells, Fam49b and Asap1 are expressed but in a manner that is insensitive to Brg1 knockdown (data not shown). In order to establish the regulatory target genes of E1–E5, we performed circular chromosome conformation capture (4C) coupled with high-throughput sequencing (4C-seq) in RN2 cells (Splinter et al. 2012; van de Werken et al. 2012). Using an anchor point fixed near E3, 4C-seq analysis revealed that this enhancer region made preferential contact with Myc as well as several intervening regions (Fig. 6A, top). Strikingly, E3 effectively ignored the nearby Gsdmc, Fam49b, and Asap1 genes, highlighting a selective looping interaction with Myc that occurs over a substantial genomic interval (Fig. 6A, top). The directionality of looping interactions may reflect a spatial constraint imposed by a topological domain whose boundary lies between E1–E5 and the nearby genes (Supplemental Fig. 16; Dixon et al. 2012). We also performed 4C-seq analysis using an anchor point fixed near Myc, which confirmed its preferential association with E1–E5 and only minimal associations with upstream sequences (Fig. 6A, bottom). The E1–E5 enhancers and Myc also make contact with several intervening noncoding regions; however, the low levels of histone acetylation and TF occupancy at these sites suggest that these interactions do not reflect functional enhancer–promoter loops but may instead represent an additional level of higher-order chromatin structure (Supplemental Fig. 17). Chromosome conformation capture (3C) analysis quantified by qPCR also validated an association between MYC and all five enhancers in NOMO-1 cells, whereas in HEK293T, these interactions were undetectable (Fig. 6B). Based on these observations, we conclude that the Myc locus exists in cell type-specific chromatin conformations, which, in leukemia cells, favors contact between the E1–E5 enhancers and the Myc promoter.
Figure 6.
E1–E5 enhancers make contact with Myc in leukemia cells. (A) 4C-seq analysis with the indicated anchor points. The _Y_-axis measures the normalized contact intensities, which plot the relative proximity of various DNA fragments in this region to the anchor-point fragment within the three-dimensional nuclear space. E1–E5 enhancers are at the location indicated. A representative experiment of two independent biological replicates is shown. (B) 3C-qPCR analysis of the interaction frequency of the indicated restriction fragments with an anchor point fixed near the MYC gene. All PCR signals were normalized to digested/religated bacterial artificial chromosome (BAC) templates. The gray boxes highlight the regions containing the E1–E5 enhancer elements. n = 3–5. Error bars represent SEM.
Brg1 facilitates TF occupancy and enhancer–promoter interactions at the Myc locus
Based on the findings described above, we hypothesized that Brg1 regulates Myc transcription in leukemia cells at least in part through its localization at E1–E5. If this is the case, then Brg1 knockdown should influence enhancer–promoter contact and/or the association of other regulatory factors at E1–E5. Using 3C-qPCR in NOMO-1 cells, we observed decreased contact between the E3–E5 enhancers and MYC upon Brg1 knockdown, whereas contact between other adjacent fragments and MYC was less affected (Fig. 7A). To ascertain whether Brg1 knockdown influenced global interactions across the Myc locus, we also performed 4C-seq in Brg1-deficient RN2 cells (Supplemental Fig. 18). This analysis confirmed that Brg1 knockdown led to decreased interactions between Myc and E3–E5 as well as reduced contact between Myc and several other intervening regions (Supplemental Fig. 18). Interestingly, upon Brg1 knockdown, Myc exhibited increased contact with various upstream DNA elements, suggesting that Brg1 is required to not only promote interactions between E3–E5 and Myc but also prevent inappropriate interactions between Myc and upstream elements (Supplemental Fig. 18). Thus, Brg1 is an important factor in leukemia cells for maintaining the fidelity of enhancer–promoter interactions at the Myc locus.
Figure 7.
Brg1 is required for enhancer–promoter proximity and occupancy of hematopoietic TFs at E1–E5. (A) 3C-qPCR analysis of the interaction frequency of the indicated restriction fragments with an anchor point fixed at the MYC gene. All PCR signals were normalized to digested/religated BAC templates. The green boxes highlight the regions containing the E1–E5 elements. The experiment was performed in NOMO-1 following 7 d of dox treatment to induce shRNA expression. n = 3–4. (B_–_I) ChIP-qPCR analysis performed in RN2 cells following conditional Brg1 knockdown. RN2 clones transduced with TRMPV-Neo Brg1 shRNA constructs were treated with dox for 48 h. PCR primer amplicons are indicated along the _X_-axis. Neg refers to a negative control region found at a gene desert region. n = 3. (J) The effect of retroviral overexpression of Brg1 mutants (dominant negatives) on RN2 cell proliferation. Wild-type or mutant Brg1 cDNAs were expressed from an MSCV-IRES-GFP vector. The relative change in GFP percentage over time was used to infer relative proliferation rates. n = 2–3. All error bars represent SEM.
To further define the role of Brg1 at the E1–E5 enhancers, we performed ChIP-qPCR analysis of various enhancer components in Brg1-deficient leukemia cells. Conditional knockdown of Brg1 did not grossly perturb levels of H3K4me1 and H3K27ac at E1–E5, suggesting that enhancer-associated histone modifications were maintained independently of Brg1 (Fig. 7B–D). In contrast, we found that Brg1 knockdown led to marked reductions in the occupancy of several TFs at E1–E5, which included Cebpα, Cebpβ, PU.1, Lmo2, and Erg (Fig. 7E–I). The expression level of these TFs was largely unaffected by Brg1 knockdown, suggesting a direct effect of Brg1 on TF occupancy at these sites (Supplemental Fig. 19). We also detected low levels of RNA produced near E3, whose production was also inhibited upon Brg1 knockdown (Supplemental Fig. 20). Collectively, these results suggest that Brg1 supports TF occupancy, chromatin looping interactions, and the level of noncoding RNA transcription at the E1–E5 enhancers.
The ATPase activity of Brg1 is known to facilitate TF access to DNA by disrupting nucleosome structure (Cote et al. 1994). Since Brg1 knockdown influences TF occupancy at E1–E5, we evaluated the relevance of the Brg1 ATPase activity for leukemia cell proliferation. For this purpose, we employed a dominant-negative approach. We retrovirally transduced leukemia cells with wild-type or various Brg1 mutant-expressing constructs and evaluated their impact on leukemia proliferation (Fig. 7J). While RN2 cells transduced with wild-type Brg1 proliferated normally, an ATPase-defective mutant of Brg1 (K798R) inhibited cell proliferation (Fig. 7J; Khavari et al. 1993). In contrast, the N1506A mutation in the bromodomain pocket of Brg1, which is known to diminish acetyl-histone recognition (Shen et al. 2007), led to minimal effects on proliferation (Fig. 7J). These results suggest that the ATPase activity of Brg1 is required for its leukemia maintenance function.
Discussion
Our study leads to several major conclusions. First, Brg1 and its associated SWI/SNF complex are critical for the proliferation and viability of leukemia cells, a function distinct from its tumor suppressor role described previously in other cancers (Wilson and Roberts 2011). We relate this observation at least in part to a unique role for Brg1 in the maintenance of Myc expression in leukemia cells. To explain this context-dependent regulation, we identified a cluster of lineage-specific enhancers at the Myc locus that loop over a 1.7-Mb distance to contact the Myc promoter. Notably, these lineage-specific enhancers closely align with a region found previously as a site of recurrent focal amplification in AML. At these enhancers, Brg1 is essential to maintain TF occupancy and allow long-range communication with the Myc promoter. Additionally, the ATPase activity of Brg1 is critical for its leukemia maintenance function, presumably by remodeling nucleosome structure to facilitate TF access. Our study implicates SWI/SNF as a novel dependency relevant to multiple subtypes of leukemia, which is linked to a role for this complex in facilitating enhancer-mediated regulation of Myc. SWI/SNF is also likely to play additional roles in leukemia maintenance through repression of proapoptotic genes and activation of Hoxa9 expression.
A key insight from this study is that Myc, despite being broadly expressed in most proliferating cell types, is controlled by highly cell type-specific enhancers, as suggested through comparisons of H3K27ac profiles across different tissues (Supplemental Figs. 10, 16). From this perspective, it would be worthwhile to evaluate whether the multitude of known regulators of Myc transcription (e.g., FBP1 and TCF/LEF) also employ tissue-specific enhancers to optimize Myc expression for lineage-specific purposes. When compared with most other cell lineages, leukemia cells exhibit a highly unusual enhancer configuration at their Myc locus, with the most prominent enhancers being clustered at the extreme boundary of the topological domain and hence at the maximal distance away from Myc. Indeed, most other cell types (such as MEFs) exhibit diffuse localization of nearby enhancers both upstream of and downstream from Myc (Supplemental Fig. 16). It is interesting to note that in leukemia cells, Myc transcription is highly sensitive to targeting general coactivator proteins that occupy the E1–E5 enhancers, such as Brg1 and the BET protein Brd4. We speculate that the unique regulatory configuration at the Myc locus seen in acute leukemia cells might place an elevated demand on activator/coactivator machineries for productive expression. Importantly, the enhancers that we identify here at the wild-type MYC locus are distinct from IgH translocations involving MYC that have been seen previously in multiple myeloma, which may likewise impose a requirement for BET proteins to sustain expression (Delmore et al. 2011). Remarkably, in both situations (IgH enhancers in myeloma and E1–E5 enhancers in leukemia), Brd4 occupancy exists at extremely high levels, a feature recently defined as a “super-enhancer” (Loven et al. 2013). Hence, our findings reinforce the concept that enhancer-mediated regulation is the functionally relevant mechanism for cell type-specific control of MYC expression, with specific enhancer arrangements being differentially sensitive to the targeting of general coactivators.
How might the E1–E5 enhancers become activated in leukemia cells? The assortment of hematopoietic TFs (e.g., Fli1, ERG, and Meis1) that occupy E1–E5 raises several possibilities for how these enhancers are regulated during normal and malignant hematopoiesis. Importantly, the presence of low-level enrichment of H3K27ac at E1–E5 in unfractionated bone marrow suggests that E1–E5 is activated in a subpopulation of normal hematopoietic cells. Nonetheless, the genes encoding Meis1 and its binding partner, Hoxa9, are also direct transcriptional targets of MLL-AF9, suggesting that oncoproteins might indirectly drive E1–E5 hyperactivation by elevating the levels of its constituent TFs. Other TFs that occupy E1–E5 (e.g., ERG) are also known to be overexpressed in AML (Marcucci et al. 2005), which might lead to E1–E5 hyperactivation in other leukemia subtypes. We speculate that elevating the TF content of E1–E5 (and presumably other enhancers) during leukemogenesis could increase the demand for SWI/SNF activity to maintain DNA accessibility at these elements, thereby rendering leukemia cells addicted to SWI/SNF activity for disease maintenance and progression.
An important observation in this study is that previously described focal amplifications at 8q24.21 closely match the position of the E1–E5 Myc enhancers (Radtke et al. 2009; Kuhn et al. 2012). These amplifications are highly focal (80–500 kb), with most lacking any protein-coding genes. The 8q24.21 focal amplifications also overlap with a previously described noncoding transcript called CCDC26, which may possess oncogenic activity (Yin et al. 2006). However, we note that only one of the nine focal amplifications seen in this AML cohort would be expected to contain the full-length CCDC26 transcript (Supplemental Fig. 21), and this RNA does not appear to be conserved in mouse cells (Radtke et al. 2009; Kuhn et al. 2012; data not shown). While several mechanistic possibilities exist for how amplifications at 8q24.21 promote leukemia, our observations raise the possibility that such events expand the enhancer repertoire at the Myc locus, which would drive elevated Myc expression to promote leukemogenesis. Genome editing of E1–E5 duplications/deletions will be required to prove such a model of leukemogenesis and to definitively establish whether Brg1 and Brd4 regulate Myc exclusively through an enhancer mechanism. A knockout of E1–E5 would also clarify whether these enhancers have essential functions during normal hematopoiesis.
Our dominant-negative experiments suggest that targeting the ATPase activity of Brg1, rather than its bromodomain, would be the preferred route to elicit anti-leukemia effects pharmacologically. The ATPase activity of Brg1 is likely required in leukemia cells to sustain TF occupancy at critical enhancers that regulate proto-oncogene expression. While SWI/SNF-deficient mice exhibit a number of phenotypic defects, recent evidence suggests that dual inactivation of Brg1 and Brm leads to only minimal effects on hematopoietic stem and progenitor cells (Willis et al. 2012), suggesting that there may exist a therapeutic window for targeting Brg1 in leukemia. Tumors that are initiated by inactivating mutations of the gene encoding BAF47 are also hypersensitive to Brg1 inhibition, suggesting additional nonleukemia cancer contexts where targeting Brg1 might have therapeutic utility (Wang et al. 2009). Notably, somatic mutations that inactivate the ATPase activity of Brg1 are seen in a variety of cancers (Dykhuizen et al. 2013), highlighting a tumor-protective function for the nucleosome remodeling activity of Brg1 in certain tissues. Therefore, systemic ATPase inhibition of Brg1 would be expected to have both pro- and anti-tumorigenic effects, which would require critical evaluation in preclinical models for the overall safety of such a therapeutic strategy. Nonetheless, given the diversity of SWI/SNF complex assemblies known to exist, it may be possible to avoid the tumorigenic effects of Brg1 inhibition by instead targeting specific SWI/SNF subunits that have cancer maintenance functions while preserving subunit functionalities that are linked with tumor protection.
Materials and methods
shRNA/GFP growth competition assay
Cultures were retrovirally transduced with LMN-shRNA vectors followed by measurement of the GFP percentage at various days post-infection using a Guava Easycyte (Millipore). The rate at which the GFP percentage declines over time was used to infer the relative growth disadvantage conferred by a given shRNA relative to the uninfected cells in the same culture. For human AML cell line experiments, we used the MLS-shRNA vector, which allows a higher retroviral transduction efficiency in these lines. Due to decreased proliferation rates of human AML lines, we also extended the time of GFP measurements to 28 d.
ChIP-seq
Protocols for performing ChIP have been previously described (Boyer et al. 2006). In brief, cells were cross-linked using 1% formaldehyde for 20 min at room temperature. Cells were resuspended, lysed in lysis buffers, and sonicated with a Misonix Sonicator 3000 to solubilize and shear cross-linked DNA. The resulting cell extract was incubated overnight at 4°C with 100 μL of Dynal Protein G magnetic beads that had been preincubated with ∼10 μg of the appropriate antibody. Beads were washed followed by elution from the beads (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% SDS) and heating for 1 h at 65°C with vortexing. Cross-linking was reversed by overnight incubation at 65°C. Following purification of immunoprecipitation DNA, Illumina libraries were prepared essentially as described (http://www.illumina.com/pages.ilmn?ID=203). All ChIP-seq data sets were aligned using Bowtie (version 0.12.2) to build version mm8 of the murine genome. All ChIP-SEQ experiments were performed with two biological replicates, which gave similar results. All sequencing data have been deposited to the Gene Expression Omnibus as SuperSeries GSE52279.
ChIP-qPCR
ChIP-qPCR assays were performed exactly as described (Steger et al. 2008). All results were quantified by qPCR performed using SYBR Green (Applied Biosystems, Inc.) on an ABI 7900HT. Each immunoprecipitation signal was referenced to an input standard curve dilution series (immunoprecipitation/input) to normalize for differences in starting cell number and for primer amplification efficiency.
4C-seq
Preparation of 4C templates was performed as previously described (Splinter et al. 2012). Experimental details are provided in the Supplemental Material.
3C
The 3C assay was performed essentially as described (Jing et al. 2008). Experimental details are provided in the Supplemental Material.
Acknowledgments
We thank Mona Spector for critical insight into genomic copy number data in AML, Wulan Deng for guidance with the 3C-qPCR assay, Charles Lin for super-enhancer analysis, Chris Johns for microarray support, Jerry Pelletier for providing the murine B-ALL cell line, James Bradner and Jun Qi for providing JQ1, Marcus Bosenberg for providing the melanoma cell line, Camila dos Santos for providing the 4T1 breast cancer line, and Anja Hohmann and Anand Bhagwat for feedback on the manuscript. This work was supported by Starr Cancer Consortium grants I4-A430 and I5-A539. Additional funding was provided by the Edward P. Evans Foundation, the Martin Sass Foundation, the F.M. Kirby Foundation, Alex's Lemonade Stand Foundation, the Laurie Strauss Foundation, and the V Foundation. C.R.V. is supported by a Burroughs-Wellcome Fund Career Award and National Institutes of Health grant NCI RO1 CA174793. This work was also supported by Cold Spring Harbor Laboratory National Cancer Institute Cancer Center Support grant CA455087 for developmental funds and shared resource support. D.L.S. is supported by National Institute of General Medical Sciences (NIGMS) 42694 and National Cancer Institute 5P01CA013106. G.A.B is supported by R37DK058044. R.A.Y. is supported by R01 HG002668. R.G.R. was supported by Leukemia and Lymphoma Society Specialized Center of Research (SCOR) grant 7132-08.
Footnotes
Supplemental material is available for this article.
References
- Bakshi R, Hassan MQ, Pratap J, Lian JB, Montecino MA, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS 2010. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J Cell Physiol 225: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Magnuson T 2005. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in β-globin expression and erythroid development. Genes Dev 19: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418: 195–199 [DOI] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR 2003. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19: 169–182 [DOI] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T 2012. Cancer epigenetics: From mechanism to therapy. Cell 150: 12–27 [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE 2006. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res 5: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho YJ, Zhao K, Crabtree GR 2013. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497: 624–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES 2013. Lessons from the cancer genome. Cell 153: 17–37 [DOI] [PubMed] [Google Scholar]
- Grand F, Kulkarni S, Chase A, Goldman JM, Gordon M, Cross NC 1999. Frequent deletion of hSNF5/INI1, a component of the SWI/SNF complex, in chronic myeloid leukemia. Cancer Res 59: 3870–3874 [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR 2011. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389 [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45: 592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366: 170–174 [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A 1999. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell 4: 735–743 [DOI] [PubMed] [Google Scholar]
- Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard MA, Crabtree GR, Lessard JA 2012. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood 120: 4720–4732 [DOI] [PubMed] [Google Scholar]
- Krosl J, Mamo A, Chagraoui J, Wilhelm BT, Girard S, Louis I, Lessard J, Perreault C, Sauvageau G 2010. A mutant allele of the Swi/Snf member BAF250a determines the pool size of fetal liver hemopoietic stem cell populations. Blood 116: 1678–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn MW, Radtke I, Bullinger L, Goorha S, Cheng J, Edelmann J, Gohlke J, Su X, Paschka P, Pounds S, et al. 2012. High-resolution genomic profiling of adult and pediatric core-binding factor acute myeloid leukemia reveals new recurrent genomic alterations. Blood 119: e67–e75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Li Q, O'Neal J, Kreisel F, Le Beau MM, Tomasson MH 2005. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood 106: 2452–2461 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrozek K, Whitman SP, Kolitz JE, Edwards CG, Vardiman JW, Powell BL et al. 2005. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A Cancer and Leukemia Group B study. J Clin Oncol 23: 9234–9242 [DOI] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ 3rd 2011. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci 108: 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell 4: 649–655 [DOI] [PubMed] [Google Scholar]
- Olave I, Wang W, Xue Y, Kuo A, Crabtree GR 2002. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev 16: 2509–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C 2001. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15: 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic R, Licht JD 2012. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov 2: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier N, Yang W, Assem M, Panetta JC, Pei D, Paugh SW, Cheng C, Den Boer ML, Relling MV, Pieters R, et al. 2008. The SWI/SNF chromatin-remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. J Natl Cancer Inst 100: 1792–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J, Ganti R, Cai Z, Goorha S, Pounds SB, et al. 2009. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci 106: 12944–12949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31: 2641–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Leroux MM, Fleming MD, Orkin SH 2002. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2: 415–425 [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG 2007. Transcription factors in myeloid development: Balancing differentiation with transformation. Nat Rev Immunol 7: 105–117 [DOI] [PubMed] [Google Scholar]
- Shain AH, Pollack JR 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 8: e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Xu C, Huang W, Zhang J, Carlson JE, Tu X, Wu J, Shi Y 2007. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 46: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the _cis_-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W 2012. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods 58: 221–230 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28: 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, et al. 2012. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods 9: 969–972 [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 15: 5370–5382 [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, Roberts CW 2009. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res 69: 8094–8101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, Roussel MF, Sherr CJ 2006. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci 103: 6688–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Homeister JW, Rosson GB, Annayev Y, Holley D, Holly SP, Madden VJ, Godfrey V, Parise LV, Bultman SJ 2012. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circ Res 111: e111–e122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492 [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7: 532–544 [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR 2009. Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Rossin A, Clifford JL, Gronemeyer H 2006. Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. Oncogene 25: 3735–3744 [DOI] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW 2011a. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, et al. 2011b. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25: 1628–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011c. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]