An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription (original) (raw)
Significance
The transcription factor Aire controls an unusual mode of transcriptional regulation, important to establish immune tolerance to self, which allows the ectopic expression in the thymic epithelium of RNA transcripts normally restricted to defined tissues. Through a genome-scale RNAi, we identify 51 functional partners of Aire, reaffirming a role for Aire in unleashing stalled transcription, interestingly through involvement of RNA processing factors.
Keywords: autoimmunity, negative selection, hnRNP
Abstract
Aire induces the expression of a large set of autoantigen genes in the thymus, driving immunological tolerance in maturing T cells. To determine the full spectrum of molecular mechanisms underlying the Aire transactivation function, we screened an AIRE-dependent gene-expression system with a genome-scale lentiviral shRNA library, targeting factors associated with chromatin architecture/function, transcription, and mRNA processing. Fifty-one functional allies were identified, with a preponderance of factors that impact transcriptional elongation compared with initiation, in particular members of the positive transcription elongation factor b (P-TEFb) involved in the release of “paused” RNA polymerases (CCNT2 and HEXIM1); mRNA processing and polyadenylation factors were also highlighted (HNRNPL/F, SFRS1, SFRS3, and CLP1). Aire’s functional allies were validated on transfected and endogenous target genes, including the generation of lentigenic knockdown (KD) mice. We uncovered the effect of the splicing factor Hnrnpl on Aire-induced transcription. Transcripts sensitive to the P-TEFb inhibitor flavopiridol were reduced by Hnrnpl knockdown in thymic epithelial cells, independently of their dependence on Aire, therefore indicating a general effect of Hnrnpl on RNA elongation. This conclusion was substantiated by demonstration of HNRNPL interactions with P-TEFb components (CDK9, CCNT2, HEXIM1, and the small 7SK RNA). Aire-containing complexes include 7SK RNA, the latter interaction disrupted by HNRNPL knockdown, suggesting that HNRNPL may partake in delivering inactive P-TEFb to Aire. Thus, these results indicate that mRNA processing factors cooperate with Aire to release stalled polymerases and to activate ectopic expression of autoantigen genes in the thymus.
The transcription factor Aire plays a very specific role in the management of immunological tolerance, promoting the ectopic expression, in medullary epithelial cells (MECs) of the thymus, of a wide array of peripheral-tissue antigens (PTA) (1). Differentiating T cells are thus exposed to the self-antigens that they will encounter later on, and potentially self-reactive cells are deleted or deviated to regulatory phenotypes as a result. _Aire_-deficient mice and human patients develop autoantibodies and multiorgan autoimmune infiltration, similarly to human patients with a mutation at the AIRE gene locus (reviewed in ref. 2).
Aire is an unusual transcriptional regulator. It impacts thousands of PTA-encoding genes whose expression is very differently controlled in parenchymal tissues (3), does not have a clear DNA binding motif, adjusts its targets according to the cell in which it is expressed (4), and takes as cues nonspecific marks of inactive chromatin such as unmethylated H3K4 (5, 6). Indeed, Aire-interacting proteins include factors involved in chromatin structure/modification, transcriptional elongation, and pre-mRNA processing (7). Several lines of investigation have shown that Aire primarily impacts the elongation steps of transcription, in particular by releasing promoter-bound polymerases that remain paused after abortive initiation (8, 9). Correspondingly, Aire has been shown to interact with subunits of the key controller of polymerase release, the positive transcription elongation factor b (P-TEFb) (8, 10, 11). P-TEFb is recruited to stalled initiation complexes in many systems of regulated transcription. It phosphorylates Ser2 on the C-terminal domain (CTD) of Pol-II and the elongation inhibitors DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF), dislodging NELF and converting DSIF into an activator (12–14). P-TEFb itself exists in an inactive form when bound to the 7SK small nuclear RNA or to HEXIM1 (15, 16).
To better understand the molecular mechanisms that Aire exploits to promote ectopic gene expression, and to complement the biochemical interaction analyses (7), we undertook a systematic large-scale RNAi screen to identify Aire’s transcriptional allies. The results confirmed the importance of factors involved in transcriptional elongation and in RNA processing and identified several previously unrecognized elements of the Aire pathway, which we validated in RNAi knockdown (KD) lentigenic mice. We also identified physical interaction and functional involvement of RNA splicing regulators, in particular Hnrnpl, in elongation control, helping to integrate the diverse classes of Aire-associated proteins previously uncovered.
Results
Identification of AIRE’s Functional Allies by a High-Throughput RNAi Lentivirus Screen.
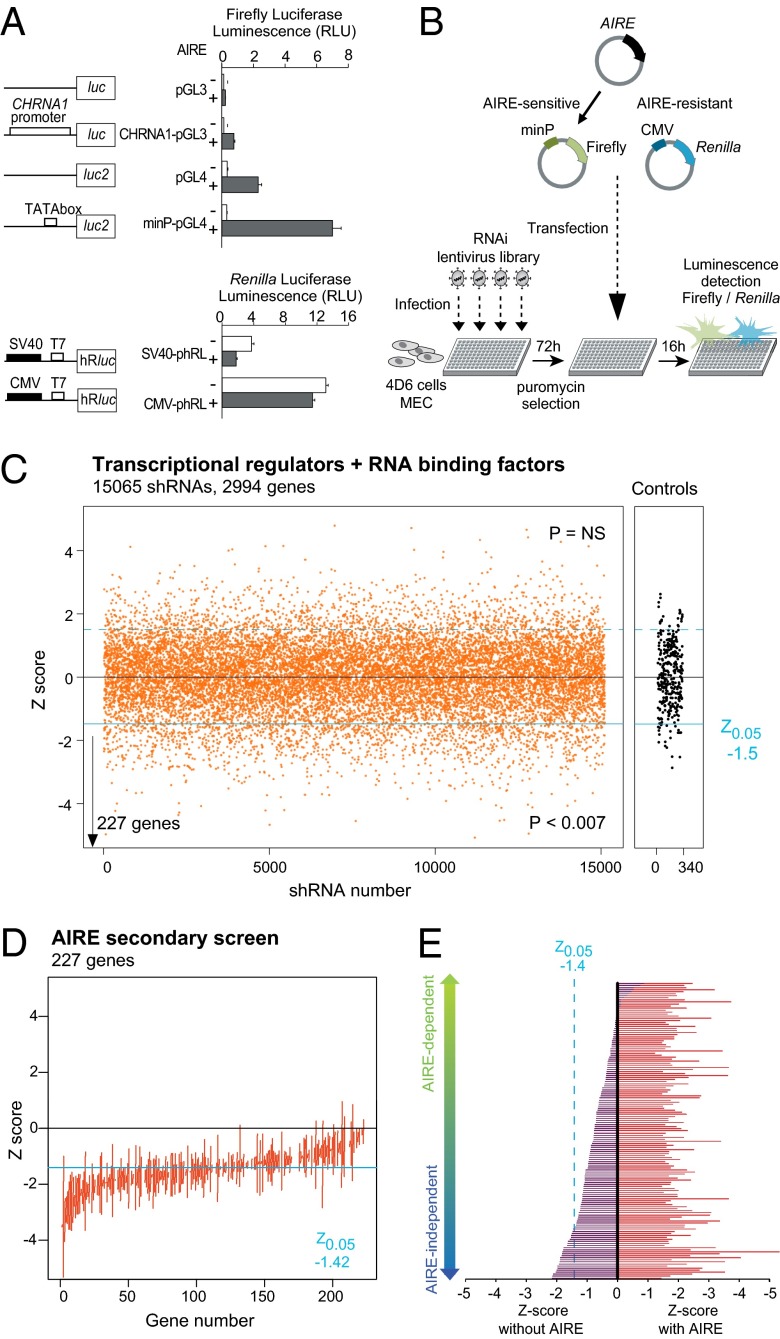
To perform the intended RNAi screen for Aire allies, we needed to develop a robust assay for Aire-induced gene expression, with a strong signal of Aire activity, minimal noise to avoid masking true signals, and amenable to high throughput. It was impossible to use primary MECs, given their very low yield from a mouse thymus, so we opted for transient transfection of an AIRE-expression construct into the AIRE-negative human MEC line 4D6 (17). An assay of AIRE-induced transcription based on a paired chemiluminescence readout (firefly/Renilla luciferase) was implemented. Expanding from our earlier experience (18), we tested a panel of luciferase reporter plasmids cotransfected with AIRE into 4D6 cells and found that AIRE’s impact was highest for plasmids with largely inactive minimal promoters (human CHRNA1, or an isolated TATA box; Fig. 1_A_). In contrast, AIRE did not increase luciferase expression driven by strong eukaryotic promoters (SV40 or CMV).
Fig. 1.
Luciferase-based RNAi screening for AIRE transactivation partners. (A) Effect of AIRE expression in human MEC 4D6 cells transfected with firefly or Renilla luciferase reporter vectors, driven by no, minimal, or full promoters. Data are average ± SEM of four transfections. (B) Schematic of the screening strategy. The 4D6 cells were plated in 384-well plates, infected with lentivirus-containing shRNAs, selected for puromycin resistance, and transfected with AIRE, minP-pGL4, and CMV-phRL. (C) Primary screen summary. Z scores for >15,065 shRNAs targeting 2,994 genes or irrelevant controls (gray); negative scores indicate a decrease of AIRE transactivation for the given shRNA. (D) Replication screen on the two or more “hit shRNAs” targeting the 227 genes identified in the primary screen. The vertical line indicates the range of Z scores observed. (E) Screen for AIRE dependence performed on hit shRNAs targeting the 77 genes most robustly confirmed in the replication screen; values on the Left represent the effect of each shRNA when the AIRE-expression vector is replaced by an empty plasmid. There is a spectrum of “AIRE dependence,” but genes listed below as AIRE independent are those for which the effect in the absence of AIRE meets the Z0.05 criterion.
To validate the specificity of the assay, we verified that transactivation of minP-luciferase luminescence by AIRE did correspond to increased reporter mRNA (Fig. S1_A_), and that Aire expression constructs bearing deletions of certain of the Aire functional domains were indeed impaired in luciferase induction (Fig. S1_B_). The screen strategy is outlined in Fig. 1_B_: infection of 4D6 cells by a lentivirus library in the LKO.1 vector encoding short hairpin RNAs (shRNAs) under the U6 promoter (one virus per well in 384-well plates); selection of transduced cells by puromycin treatment; cotransfection with the AIRE-expressing construct, an AIRE-dependent (minimal promoter driven) firefly luciferase reporter and an AIRE-insensitive (CMV promoter driven) Renilla luciferase reporter; and quantitation of luminescence from the two reporters. Renilla luciferase provided an internal control for shRNAs that would generically affect transcription or cell viability and served as a normalization for transfection efficiency, which varied with the density of puromycin-surviving cells and thus with the titer of each lentivirus (Fig. S1_C_). The primary screen targeted 2,994 human genes (15,065 nontoxic shRNAs) including basal transcription factors, transcription activators, chromatin modifiers, and RNA-binding proteins. The libraries included 5 different shRNAs for each gene, 55% of which provoke a >70% reduction of their target mRNA, and each of the 384-well screening plates included 40 wells with irrelevant controls or empty shRNAs for a robust estimate of assay noise and plate- or batch variance. Renilla and firefly luminescence levels for each plate were fit with a locally smoothed regression, the residuals from the fit for each data point reflecting the effect of the corresponding shRNA on AIRE activity. To estimate the significance of these effects over the whole screen, a null distribution of the firefly/Renilla residuals was determined for a set of 340 control shRNAs, and a Z score was computed for each test shRNA. As illustrated in Fig. 1_C_, 1,315/15,065 hairpins met a Z-score criterion of P < 0.05, indicating that the screen was indeed identifying shRNAs with a negative impact on AIRE-induced gene expression (P < 0.007, Fisher’s exact), many inhibitory targets being identified by 2 or more shRNAs (Fig. S1_D_). The screen also identified shRNAs with a positive impact on AIRE activity, but with a distribution similar to that of control shRNAs. To conservatively flag robust candidates in this primary screen and to avoid false positives due to assay noise and off-target effects, we selected those genes identified by 2 shRNAs passing the Z-score criterion, or one passing if the aggregate of the 5 shRNAs for that gene showed a significantly skewed distribution in a Student t test. Applying this rule, we identified 227 candidate genes (∼9.2% hit rate) whose knockdown diminished AIRE-activated transcription (Table S1). We then performed a secondary replication screen for these 227 genes (using only those shRNAs that scored in the primary screen). The Z scores showed an excellent replication of the primary screen, confirming the reliability of the high-throughput system (Fig. 1_D_).
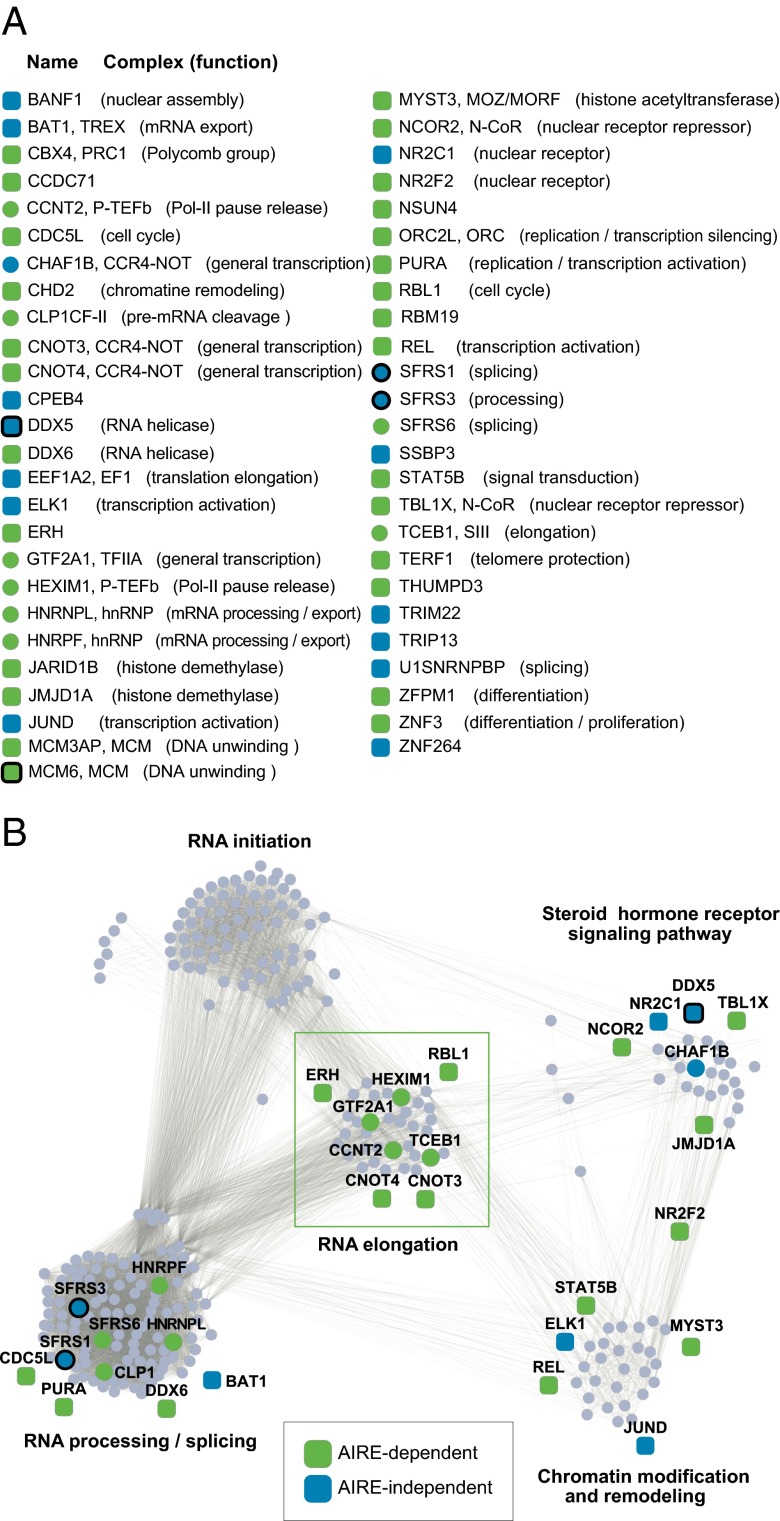
It was theoretically possible that these cofactors influenced AIRE-activated transcription only when AIRE was present, or that they modified the transcriptional structure on which AIRE docks before its recruitment, in which case one might also expect an impact of the knockdown in the absence of AIRE. Thus, we repeated the analysis of shRNAs for the 77 AIRE “allies” identified, but without AIRE. Although the effects of the shRNAs were never as strong as in the presence of AIRE, two types of results were observed (Fig. 1_E_): some shRNAs lost all effect in the absence of AIRE, whereas others did have marked activity even without AIRE, presumably acting on a baseline state, which AIRE amplified. To further increase the likelihood that the candidates were not false positives, we selected genes with significant expression in 4D6 cells, based on microarray analysis (Table S1), and also dropped from further consideration genes whose knockdown resulted in sequestration of Aire in the cytoplasm (Fig. S1_E_ and Table S1). This RNAi screen ultimately led to the identification of 51 genes most likely involved in the Aire-transactivation function; comfortingly, these molecules included several Aire-interaction factors highlighted in our previous screen (7), e.g., DDX5, SFRS1, SFRS3, and MCM6 (Fig. 2_A_).
Fig. 2.
AIRE’s functional allies. (A) List of the 51 AIRE allies most robustly identified, the molecular complex to which they belong, and their currently ascribed molecular function. (B) AIRE functional allies were positioned on a transcription-related network where the function of each cluster is represented by Gene Ontology terms (RNA elongation, Pcorr = 3.10−33; RNA initiation, Pcorr = 1.10−31; RNA processing/splicing, Pcorr = 1.10−200; steroid signaling pathway, Pcorr = 4.10−10; and chromatin modification and remodeling, Pcorr = 4.10−11). AIRE allies that belong to a cluster are shown as circles, those with predicted interactions with members of each cluster as squares. A thick line denotes factors found to physically interact with Aire in our earlier Mass Spec screen.
AIRE’s Functional Allies Are Primarily Involved in RNA Elongation.
We then investigated the nature of the 51 confirmed candidates, categorizing the molecular steps of transcription in which they are involved. The STRING database (19), which groups and scores all known molecular and genetic interactions between gene products, was used to generate a network of transcription-related genes. The network was clustered based on the frequency and validity of the interactions by the MCODE algorithm, and the inferred biological function of each cluster was defined by a frequency of Gene Ontology identifiers. Although there were distinct cross-relationships between clusters, as expected, this process grouped distinct clusters predominantly involved in specific functions such as transcriptional initiation, elongation, or pre-mRNA processing (Fig. S2). We mapped onto this framework 38 AIRE allies (Fig. 2_B_; 13 of the 51 were unlisted in the STRING database). Remarkably, 8 of them, all of which belonged to the AIRE-dependent group, mapped to the “elongation” cluster, whereas none mapped to the “initiation” cluster (Fisher’s exact P = 0.0001). A high proportion of AIRE allies also mapped to the “processing/splicing,” “chromatin modification” and ”nuclear receptor signaling pathway” clusters. Thus, the functional screen dovetailed with the physical association screen in showing that AIRE impacts several steps in the formation of mature transcripts, and some of the factors were hits in both screens (symbols outlined by thick lines in Fig. 2 A and B).
Validation of AIRE’s Functional Allies.
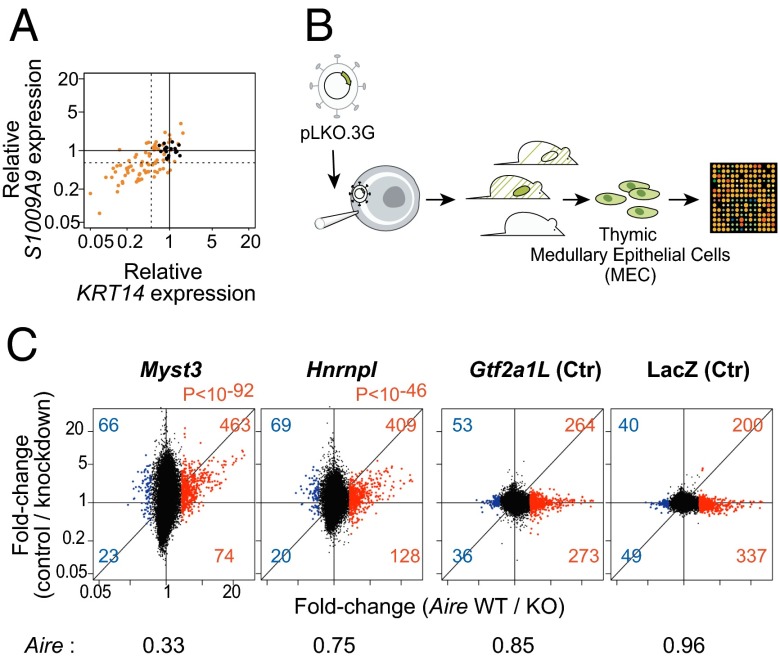
The reporter system used in our screen, although well-suited for a high-throughput process, was rather artificial, so we felt it important to confirm the validity of the candidates by evaluating their influence on AIRE’s induction of its normal endogenous targets. After performing the same lentiviral shRNA infection and subsequent AIRE transfection as above, we measured the expression of two of AIRE’s endogenous targets in 4D6 cells, KRT14 and S100A9 (6). Knockdown of the candidates had a clear dampening effect on their induction by AIRE, with a good correlation between the reduction in KRT14 or S100A9 mRNA and the magnitude of the shRNA’s effect in the luciferase assay (Fig. 3_A_).
Fig. 3.
Validation of AIRE’s functional allies. (A) Effect of hit shRNAs on AIRE transactivation of two AIRE targets in 4D6 cells (KRT14 and S100A9); orange dots are for test shRNAs, black dots for control shRNAs. (B) Schematic of the lentigenic knockdown strategy. Each selected shRNA was transferred to PLKO.3G, packaged to >109 viral titer for microinfection of fertilized oocytes under the zona pellucid. Resulting offspring were selected for activity of the lentigene in >10% of somatic cells, and GFP+ MECs were sorted and profiled. (C) MEC gene expression profiles for mice with knockdown expression of Myst3 and Hnrnpl, or Aire-irrelevant controls Gtf2a1L and LacZ. Control shRNA lentigenes targeting RFP were used as reference, and the results are shown as a FoldChange relative to RFP controls (y axis) vs. Aire’s transcriptional footprint (x axis; from a comparison of _Aire_-KO and WT MECs). Myst3 and Hnrnpl KD effects on Aire-induced genes are shown in red. Aire expression values from microarray profiling of the knockdown lentigenics are shown normalized to those of the RFP control.
To address the functional relevance of these candidates in the natural mouse thymus, we first selected a set of 10 genes for which we could identify shRNAs that effectively reduced AIRE-activated transcription in the 1C6 mouse thymic epithelial cell line (Fig. S3_A_ and Table S1). High-titer lentiviruses containing U6-driven shRNAs, and the phosphoglycerate kinase-GFP as a marker of activity, were used to microinfect fertilized oocytes, which were reimplanted into pseudopregnant females (Fig. 3_B_). Of the 15 hairpins tested, 2 pups with lentigene expression and target knockdown >50% were obtained for only Myst3 and Hnrnpl (Table S2). No pups, or only pups with complete extinction of the lentigene, or with little to no effect on the target transcript, were obtained for shRNAs targeting most of our candidates, likely reflecting a lethal effect or competitive disadvantage of the knockdown (success rates for control lentigenics were far greater). GFP+ MECs were sorted from these mice, gene-expression profiles were generated, and the changes induced by each knockdown (relative to averages from three control lentigenics) were plotted in relation to Aire’s transcriptional impact (Fig. 3_C_). As indicated by the vertical shift of transcripts with an _Aire_-WT/KO ratio >2 (Fig. 3_C_, red dots), a clear relationship was observed with knockdowns for Myst3 and Hnrnpl, as the vast majority of genes activated by Aire were underexpressed in the corresponding knockdowns (although not all transcripts sensitive to Myst3 and Hnrnpl were Aire responsive). This deviation was highly significant (χ2 P < 10−46 overall); the significance of the effects on individual genes was also estimated by computing their probabilities based on the variance observed among control lentigenics (Fig. S3_B_). In contrast, no deviation in the expression of Aire-responsive genes was observed in MECs expressing shRNAs targeting the Gt2a1l or LacZ control genes. In addition the expression of Aire itself proved to be impaired in the Myst3 but not in the Hnrnpl knockdown (Fig. 3_C_). To confirm the specificity of the effects observed in the Hnrnpl knockdown, we compared the changes to those reported by Huang et al. (20) after knockdown of HNRNPL in HeLa cells; the results showed a clear reciprocal enrichment of the genes impacted by Hnrnpl (Fig. S3_C_).
In addition, we were able to test one conventional knockout for a factor identified in the screen, Stat5 (21). Gene-expression profiling of MECs from Stat5a/b double-deficient mice and matched controls revealed a moderate but significant impact of Stat5 on Aire-dependent transcripts (Fig. S3_D_).
An Hnrnpl Effect on Transcriptional Elongation.
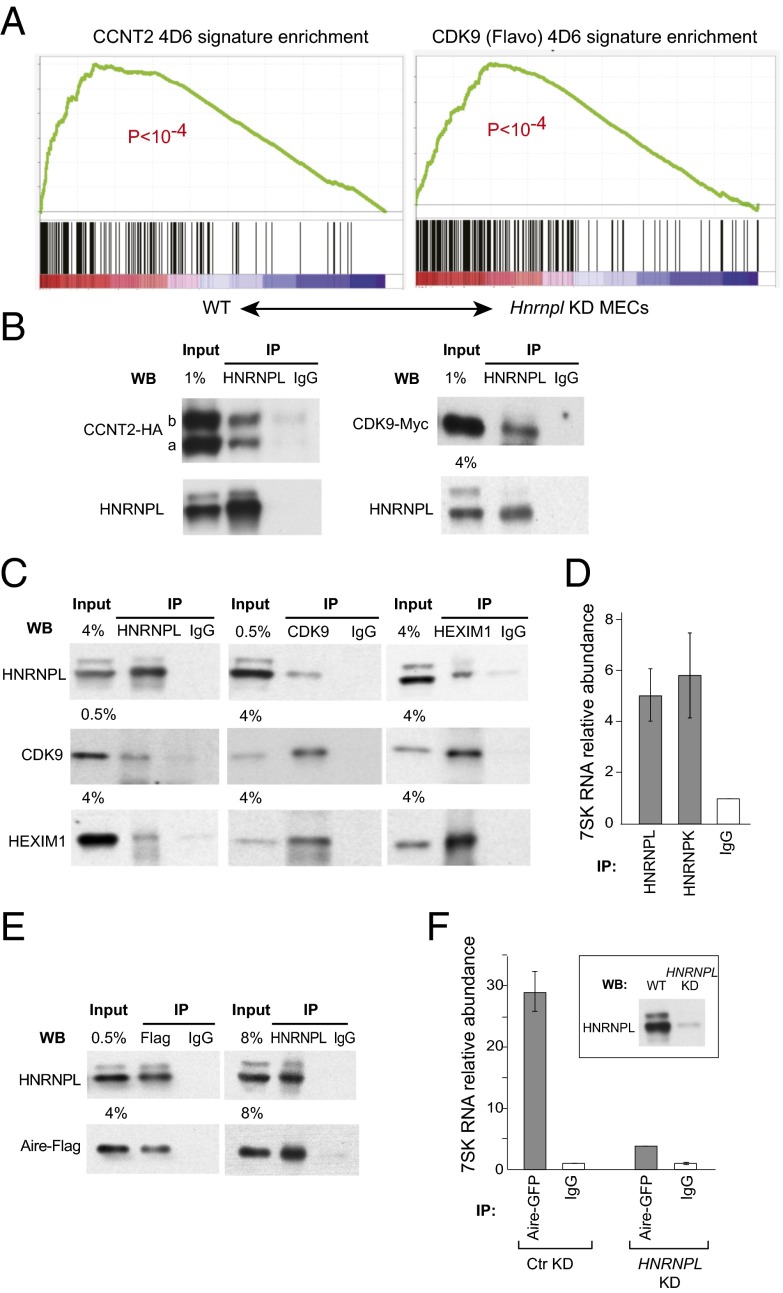
HNRNPL is an RNA processing factor from the heterogeneous nuclear ribonucleoprotein (hnRNP) family, with a well-defined role in alternative splicing and in recognition of 3′ UTR motifs (22–25), and it was intriguing that it had such a broad impact on Aire’s in vivo footprint. Other members of the hnRNP family, like HNRNPK, impact on transcriptional elongation through interactions with P-TEFb, via binding to the small 7SK RNA (26). We hypothesized that HNRNPL has similar properties and interacts with P-TEFb and its subunits CCNT2 and CDK9 (CCNT2 being a validated hit in our screen; Fig. S3_A_). We observed that there was, among the transcripts affected by Hnrnpl knockdown in MECs, a strong overlap with the genes controlled by CCNT2 (as determined by profiling of CCNT2 knockdown 4D6 cells) and by CDK9 (determined by inhibition of CDK9 by Flavopiridol) (Fig. 4_A_). In addition, we could show an interaction between endogenous HNRNPL and tagged versions of CDK9 and CCNT2 in transduced HEK293 cells (7) (Fig. 4_B_). Binding of HNRNPL to the endogeneous P-TEFb complex was also found by reciprocal coimmunopreciptations between HNRNPL and CDK9 (Fig. 4_C_).
Fig. 4.
Cooperation of Hnrnpl with the inactive P-TEFb and Aire. (A) Gene Set Enrichment Analysis in the WT vs. Hnrnpl KD ranked expression dataset from primary MECs, of the genes impacted by Flavopiridol or CCNT2 knockdown in 4D6 cells. (B) Coimmunoprecipitation of HNRNPL and Myc-tagged CDK9 or HA-tagged CCNT2 in HEK293 cells. CCNT2 is expressed as two isoforms, CCNT2a and CCNT2b. (C) Reciprocal coimmunoprecipitations of endogenous HNRNPL, CDK9, and HEXIM1 in HEK293s. (D) RT-PCR quantitation of 7SK RNA in HNRNPL and HNRNPK immunoprecipitates, relative to control IgG. Average ± SEM of three experiments. (E) Reciprocal coimmunoprecipitation of HNRNPL with the Flag-tagged Aire. (F) Quantitation of 7SK RNA bound to Aire complex, as affected by HNRNPL knockdown, in stably transduced Aire-GFP 4D6 cells.
P-TEFb is inhibited by binding to the 7SK small nuclear RNA (15, 16) and/or to HEXIM1, which also interacts with the small 7SK RNA on the inactive P-TEFb. The deinhibition of P-TEFb at the transcriptional start sites (TSS) can be regulated by various factors, notably c-Myc, BRD4, or SFRS2, the latter a serine/arginine-rich (SR) splicing factor, reflecting the functional link between the splicing machinery and transcriptional elongation (27–29). We asked whether HNRNPL also interacted with P-TEFb inhibitory elements. Reciprocal coimmunoprecipitation was found between HNRNPL and HEXIM1 (Fig. 4_C_). In addition, 7SK RNA was enriched in HNRNPL immunoprecipitates (Fig. 4_D_) and in HNRNPK precipitates as expected (30). The amount of 7SK associated with HNRNPL increased upon transcription blockade, as it does for HNRNPK (30).
If Hnrnpl is a necessary Aire ally, and if it interacts with transcriptional control components that are also important for Aire’s activity, how do Aire and Hnrnpl interact? Reciprocal coimmunoprecipitations showed a physical interaction between HNRNPL and Aire in HEK293 cells (Fig. 4_E_). Importantly, we found that anti-Aire coprecipitated a readily detectable amount of 7SK RNA, but that this was decreased when HNRNPL was knocked down (Fig. 4_F_). Thus, HNRNPL plays an important role in the interaction of Aire with 7SK-containing complexes. That Aire is functionally involved with inactive P-TEFb is consistent with the deleterious effect of HEXIM1 knockdown on AIRE-induced expression in the screen.
Discussion
Aire-dependent ectopic gene expression in MECs operates on an array of genes that are normally controlled by a wide variety of transcription factors in diverse tissues, thus posing an unusual challenge to gene regulation. To identify the cast of molecules that cooperate with Aire to perform this function, we conducted an RNAi screen that combined a high-throughput in vitro component with validation in lentigenic mice. The results further connect the activity of Aire with elongation control, and to lifting of the paused polymerases, and help bridge the relation between splicing- and elongation-control factors.
The disparity in the number of initiation and elongation factors identified in our shRNA screen was quite striking. In hindsight, this is perhaps not surprising, because Aire is specifically effective on reporters with enhancer-less minimal promoters, or on promoters for which key initiation factors are absent. Our earlier work had proposed a potential role for Aire in boosting elongation, through the recruitment of the DNA-PK/TOP2a/FACT “eviction” complex, thought to facilitate transcription by relieving torsional constraints during Pol-II progression (7). Oven et al. (8) had also shown a preferential effect of Aire on elongation, through interaction with P-TEFb, as had our detailed analysis of Aire-regulated transcripts in MECs (9). To ensure the specificity of the factors identified in the RNAi screen, we cotransfected an AIRE-insensitive reporter, which ruled out factors with generic effects on all transcription. Thus, some ubiquitously active factors that are also important for AIRE activity may have been missed, and this may explain why some important factors of transcriptional elongation, such as CDK9 or CCNT1, did not score as hits.
If Aire and its allies partake in pause release, how is this accomplished at the biochemical level? Two allies identified in the screen are at the center of Pol-II release. CCNT2, a subunit of P-TEFb, was highlighted in the screen, and was found to interact with Aire. The elongation inhibitor HEXIM1 was also a hit in the screen, two shRNA hairpins strongly decreasing AIRE’s activity on both the transfected reporter and endogenous targets. This result may seem paradoxical at first, as HEXIM1 actually inhibits P-TEFb by sequestering it within the nucleoplasm, and Oven et al. (8) reported the opposite effect: activation of Aire activity by transient RNAi repression of Hexim1 in 1C6 cells versus stable RNAi in our screen, a difference which may be related to different limiting factors in the two systems. A requirement for HEXIM1 is compatible with a model in which AIRE, like the TAT protein, targets for activation the inactive P-TEFb, bound to 7SK RNA and HEXIM1 (31). Increasing the pool of free active P-TEFb by knocking down HEXIM1 would lead to a relative decrease in AIRE’s activity. The necessity for AIRE to recruit the inactive 7SK RNA-associated P-TEFb complex to activate transcription is consistent with the fact that the 7SK RNA coprecipitates with Aire, as do CDK9 and HEXIM1.
A number of mRNA processing factors were identified in our screen, therefore confirming the involvement of the splicing machinery in the Aire transactivation function, as first demonstrated in a proteomic interaction screen for Aire’s partners (7). It is well established that tight connections between splicing and transcription exist, in particular with the release of Pol-II pausing (27–29). Of the 10 splicing factors that were validated in our screen, two were also identified in the Aire interactome, the two SR-proteins SFRS1 and SFRS3. Relevant to a role for RNA processing factors in transcriptional elongation, SFRS1 and SFRS2 were reported to recruit the inactive 7SK-associated P-TEFb complex to the polymerase and to promote the dissociation of 7SK RNA from P-TEFb (29). The role of SFRS1 in recruiting and activating the inactive P-TEFb complex is consistent with the notion that 7SK RNA-associated P-TEFb is the substrate of the Aire-dependent machinery to activate paused polymerases. A similar explanation may well hold for HNRNPL, whose function was validated in MECs of lentigenic mice. HNRNPL is primarily known for its major known role in alternative splicing and in the recognition of 3′ UTR motifs (22, 24, 25). It has also been shown to interact with the Mediator complex, therefore coupling mRNA processing to transcription (20). Here, analyses of the expression data from Hnrnpl knockdown mice revealed that Hnrnpl preferentially stimulates the expression of genes (some of which are Aire insensitive) whose transcription is also affected by inhibition of CDK9 (flavopiridol inhibition), i.e., genes under dominant elongation control. The interaction of HNRNPL and P-TEFb was confirmed biochemically, including to the 7SK inhibitory RNA (in keeping with the demonstration that several hnRNPs can trap 7SK RNA and alter the balance of active to inactive P-TEFb) (30, 32).
A key to conceptually integrating the interplay between Aire and P-TEFb factors and inhibitors may lie in the observation (Fig. 4_F_) that Aire interacts with 7SK-containing complexes, and that this interaction requires HNRNPL, leading to the following speculative scenario: Aire binds to 7SK- and HEXIM1-containing inactive P-TEFb complexes at the position of stalled polymerases, which it helps activate in concert with other factors, and perhaps in combination with short promoter-associated transcripts, by analogy to HIV TAT and to the model of Ji et al. (29). HNRNPL might help recycle P-TEFb from the the site of transcription termination where hnRNP factors were shown to facilitate pre-mRNA 3′ end processing (33) also facilitated by CLP1, a cleavage and polyadenylation factor, which was also a hit in the RNAi screen.
Thus, and although much work remains to elucidate the interplay between Aire’s allies in enabling ectopic transcription, the set of characters that has been identified here should be instrumental in piecing the puzzle together.
Experimental Procedures
Full methods are detailed in SI Experimental Procedures.
Luciferase-Based RNA-Interference Screening.
The screen was performed using the lentiviral library prepared, titered,and arrayed as described (34) as schematized in Fig. 1_B_, in 384-well plates and in duplicate, using automated pipetting robotics.
Lentigenic Mouse Generation.
Selected shRNAs from the mouse screen were transferred from pLKO.1 to pLKO.3G with GFP as a reporter gene. Lentivirus stocks were generated in 293-FT cells, concentrated by ultracentrifugation to very high titer (>109 infectious unit/mL) for microinfection into fertilized oocytes (20–40 per construct) (35). Pools of 30 transduced oocytes were reimplanted into pseudopregnant females. Newborns with GFP expression were selected and the shRNA present in each pup was identified by PCR amplification and sequencing (Table S3).
Network Modeling.
A transcription-related network was generated using the STRING database (http://string-db.org/). This search was seeded, blind to AIRE cooperants, by introducing factors known to be important in various transcriptional processes, secondarily incorporating all other factors with which these seeds interact. Within this network, proteins with a high density of interaction were clustered and the biological function of these clusters identified by a Gene Ontology analysis.
Gene Expression Profiling.
Total RNA, isolated from sorted MECs of 4-wk-old individual mice were profiled as described on Affymetrix Mouse Gene ST1.0 arrays.
Coimmunoprecipitations.
HEK293 cells were transfected with CCNT2, CDK9, and Aire expression vectors. Nuclear extracts were prepared after 48 h, protein complexes captured with specific antibodies on magnetic beads (ActiveMotif) followed by immunoblotting. For 7SK RNA quantitation, the immunoprecipitated RNA was extracted by adding TRIzol directly on the beads and retrotranscribed with a specific RT primer.
Supplementary Material
Supporting Information
Acknowledgments
We thank Drs. J. Ihle for mice; N. Fujikado, J. Lopes, and S. Silver for materials or advice; H. Le for help with the screen; and S. Davis and H. Paik for computation. This work was supported by National Institutes of Health DK060027 and AI088204 (to C.B. and D.M.), ANR-2011-CHEX-001-R12004KK and PCIG9-GA-2011-294212 (to M.G.), National Institute of Allergy and Infectious Diseases U01AI074575 (to N.H.). M.G. received fellowships from the Campbell and Hall Charity Fund, the Harold Whitworth Pierce Charitable Trust, the Philippe Foundation, and “la Fondation pour la Recherche Médicale.”
Footnotes
The authors declare no conflict of interest.
References
- 1.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 2.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 3.Venanzi ES, Melamed R, Mathis D, Benoist C. The variable immunological self: Genetic variation and nongenetic noise in Aire-regulated transcription. Proc Natl Acad Sci USA. 2008;105(41):15860–15865. doi: 10.1073/pnas.0808070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerau-de-Arellano M, Mathis D, Benoist C. Transcriptional impact of Aire varies with cell type. Proc Natl Acad Sci USA. 2008;105(37):14011–14016. doi: 10.1073/pnas.0806616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9(4):370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh AS, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci USA. 2008;105(41):15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell. 2010;140(1):123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Oven I, et al. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27(24):8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giraud M, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci USA. 2012;109(2):535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Žumer K, Plemenitaš A, Saksela K, Peterlin BM. Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription. Nucleic Acids Res. 2011;39(18):7908–7919. doi: 10.1093/nar/gkr527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S, Bansal K, Lopes J, Benoist C, Mathis D. Aire’s plant homeodomain(PHD)-2 is critical for induction of immunological tolerance. Proc Natl Acad Sci USA. 2013;110(5):1833–1838. doi: 10.1073/pnas.1222023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20(9):2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujinaga K, et al. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24(2):787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada T, et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21(2):227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 16.Michels AA, et al. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23(14):4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández E, et al. Establishment and characterization of cloned human thymic epithelial cell lines. Analysis of adhesion molecule expression and cytokine production. Blood. 1994;83(11):3245–3254. [PubMed] [Google Scholar]
- 18.Giraud M, et al. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448(7156):934–937. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 19.Jensen LJ, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45(4):459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teglund S, et al. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93(5):841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 22.Oberdoerffer S, et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321(5889):686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z, et al. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity. 2008;29(6):863–875. doi: 10.1016/j.immuni.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003;10(1):33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 25.Ergun A, et al. ImmGen Consortium Differential splicing across immune system lineages. Proc Natl Acad Sci USA. 2013;110(35):14324–14329. doi: 10.1073/pnas.1311839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13(6):868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15(8):819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji X, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153(4):855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol. 2007;27(20):6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17(7):815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Herreweghe E, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26(15):3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millevoi S, et al. A physical and functional link between splicing factors promotes pre-mRNA 3′ end processing. Nucleic Acids Res. 2009;37(14):4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Stockton J, Mathis D, Benoist C. Modeling CTLA4-linked autoimmunity with RNA interference in mice. Proc Natl Acad Sci USA. 2006;103(44):16400–16405. doi: 10.1073/pnas.0607854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information