Rare chromosomal deletions and duplications increase risk of schizophrenia (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 4.
Published in final edited form as: Nature. 2008 Jul 30;455(7210):237–241. doi: 10.1038/nature07239
Schizophrenia (SCZ) is a severe mental disorder marked by hallucinations, delusions, cognitive deficits and apathy with heritability estimated at 73-90%1. Inheritance patterns are complex and the number and type of genetic variants involved are not understood. Copy number variants (CNVs) have been identified in individual SCZ patients2-7 and also in neurodevelopmental disorders8-11, but large-scale genome-wide surveys have not been performed. We report such a genome-wide survey of rare CNVs in 3,391 patients with SCZ and 3,181 ancestrally-matched controls using high-density microarrays. For CNVs that were observed in less than ∼1% of the sample and greater than 100kb in length, the total burden is increased in SCZ patients compared to controls (_P_=3×10−5; 1.15 fold increase). This effect was more pronounced for rarer, single-occurrence CNVs and for those that involved genes as opposed to those that did not. As expected, deletions were found within the region critical for velo-cardio-facial syndrome (_P_=0.0017, odds ratio (OR) = 21.6), which includes psychotic symptoms in 30% of patients12. Associations with SCZ were also found for large deletions on chromosome 15q13.2 (P = 0.0029, OR = 17.9) and 1q21.1 (P = 0.0076, OR = 6.6). These associations were not previously reported in the literature and remained significant after genome-wide correction. Overall, our results provide strong support for a model of SCZ pathogenesis that includes the effects of multiple rare structural variants, both genome-wide and at specific loci.
The International Schizophrenia Consortium (ISC) was established to promote rapid progress towards the identification of genetic causes underlying SCZ. The ISC is comprised of investigators from the University of Aberdeen, Cardiff University, the University of Edinburgh, the Karolinska Institutet, Massachusetts General Hospital, the University of North Carolina-Chapel Hill, the Queensland Institute of Medical Research, the University of Southern California, the Stanley Center for Psychiatric Research at the Broad Institute of Harvard and MIT, Trinity College Dublin and University College London.
We surveyed single nucleotide polymorphisms (SNPs) and CNVs using the Affymetrix Genome-Wide Human SNP 5.0 and 6.0 Arrays in European SCZ cases and ancestrally-matched controls (Table 1 and Supplementary Information)13. Based on the genome-wide SNP data there was no evidence of major population stratification within each site14 (data not shown). Intensity data from both SNP and CNV probes were used to identify autosomal deletions and duplications, based on a hidden Markov model (HMM)15.
Table 1. Study Sample Characteristics and Genotyping Platforms.
| Sample | Ancestry | Case (N) | Control (N) | Genotyping Platform |
|---|---|---|---|---|
| University of Aberdeen | Scottish | 727 | 694b | 5.0 |
| University College London | British | 547 | n/ac | 5.0 |
| Portuguese Island Collection | Portuguese | 333 | 200b | 5.0 |
| Karolinska Institutet | Swedish | 622 | 437 | 5.0/6.0d |
| Cardiff University | Bulgarian | 479a | 646 | 6.0 |
| Trinity College Dublin | Irish | 280 | 914 | 6.0 |
| University of Edinburgh | Scottish | 403a | 290 | 6.0 |
This study focused on rare but highly penetrant structural variation in schizophrenia, following a natural extension of the classical medical genetic approach. Common CNVs are better identified with different algorithms and better tested for association separately13,15. Considering CNVs present in less than 1% of our total sample, there were 6,753 greater than 100kb that passed sample and CNV quality filtering (see Supplementary Information, Table S1). The median size was 182.1kb (166.3kb for deletions, 194.4kb for duplications), 39% were deletions and the median number per individual was 1. We assessed the impact of rare structural variation on SCZ risk in two ways: first, in terms of an individual's genome-wide burden and second, by searching for specific loci that were significantly associated with disease.
Structural variants have been identified for severe neurodevelopmental disorders9-11,16,17. Since it has been postulated that SCZ might, at least in part, have a developmental etiology18, we posited a role for CNVs in SCZ, as have others2-6. A number of loci have in fact been identified, including variants containing genes with neurodevelopmental roles2-5. However, a critical question is the extent to which this is a general mechanism for producing SCZ in typical clinical populations rather than cases selected for atypical phenotypic features such as very early onset or mental retardation. This motivated our primary hypothesis: that individuals with SCZ have a greater genome-wide burden of CNVs. Considering all CNVs, we observed that cases had a greater average burden than controls (1-sided, empirical _P_= 3×10−5 controlling for array type; Table 2). Controls on average had 0.99 CNVs per person, whereas cases showed a 1.15-fold higher rate.
Table 2. Global CNV burden analysis: event type & frequency.
| CNV burden (number) | CNV burden (gene count) | |||||||
|---|---|---|---|---|---|---|---|---|
| CNV type | Frequency | CNV (N) | P | Case/control ratio | Baseline rate (controls) | P | Case/control ratio | Baseline rate (controls) |
| Deletions & duplications | All | 6753 | 3×10−5 | 1.151 | 0.991 | 2×10−6 | 1.407 | 2.010 |
| Single occurrence | 890 | 5×10−6 | 1.448 | 0.114 | 0.0057 | 1.674 | 0.322 | |
| 2-6 occurrences | 2465 | 0.0013 | 1.163 | 0.353 | 5×10−4 | 1.363 | 0.801 | |
| Deletions only | All | 2652 | 0.11 | 1.077 | 0.402 | 3×10−5 | 1.554 | 0.717 |
| Single occurrence | 470 | 0.011 | 1.287 | 0.064 | 0.005 | 1.774 | 0.115 | |
| 2-6 occurrences | 994 | 0.048 | 1.152 | 0.146 | 0.13 | 1.377 | 0.208 | |
| Duplications only | All | 4101 | 2×10−5 | 1.203 | 0.589 | 1×10−4 | 1.276 | 1.939 |
| Single occurrence | 734 | 8×10−6 | 1.580 | 0.091 | 0.015 | 1.595 | 0.302 | |
| 2-6 occurrences | 1532 | 0.011 | 1.163 | 0.220 | 0.012 | 1.295 | 0.686 |
We next explored this subtle, but highly statistically significant, observation of increased burden. We defined burden in two ways: as the number of CNVs an individual carries (as above), and also as the number of genes spanned by those CNVs. This second metric (the “gene-count”) in fact showed a stronger association with SCZ (1.41-fold increase, empirical _P_=2×10−6) than burden defined simply as the number of CNVs. Characteristics of CNV subgroups studied here are their frequency, type, size, and proximity to a gene (Tables 2 & 3; Table S2). We observed increased burden across multiple independent subgroups of CNVs, a finding that was more pronounced for rarer CNVs and those involving genes. Deletions and duplications also displayed somewhat different profiles: the association of deletions varied more noticeably with respect to CNV size and proximity to a gene, whereas duplications showed a more uniform pattern. Eight hundred ninety CNVs were observed in either a case or a control as a single occurrence. This rarest subset of CNVs would be expected to show enrichment under the model that genetic causes of SCZ are individually unique in some proportion of patients. Indeed, this set of CNVs showed a 1.45-fold increase in cases (empirical _P_=5×10−6). On average, 13.1% of SCZ cases possessed a deletion or duplication observed only once in the sample, in contrast to 10.4% of controls. Under a model in which very rare (occurring in under 1/1000 individuals) inherited or recurrently de novo events increase risk, we would expect to observe a greater overall burden in SCZ. Although our study was statistically under-powered to identify the actual loci involved, such variants could in theory be mapped in extremely large samples. In this intermediate group, we observed 2,465 CNVs occurring between 2 and 6 times in the total sample, for which there was an increased burden, both for number of CNVs (empirical P = 0.0013) and gene-count (empirical P = 5×10−4).
Table 3. Global CNV burden analysis: event type & size.
| CNV burden (number) | CNV burden (gene count) | |||||||
|---|---|---|---|---|---|---|---|---|
| CNV Type | Size Range (kb) | CNV (N) | P | Case/control ratio | Baseline rate (controls) | P | Case/control ratio | Baseline rate (controls) |
| Deletions & duplications | 100 – 200 | 3725 | 0.0017 | 1.145 | 0.55 | 8×10−6 | 1.346 | 0.73 |
| 200 – 500 | 2156 | 0.028 | 1.106 | 0.32 | 0.0088 | 1.249 | 0.66 | |
| ≥500 | 872 | 0.0013 | 1.315 | 0.12 | 8×10−4 | 1.788 | 0.62 | |
| Deletions only | 100 – 200 | 1612 | 0.54 | 1.016 | 0.25 | 0.28 | 1.069 | 0.26 |
| 200 – 500 | 755 | 0.39 | 1.041 | 0.12 | 0.059 | 1.274 | 0.14 | |
| ≥500 | 285 | 3×10−4 | 1.671 | 0.03 | 2×10−5 | 3.567 | 0.14 | |
| Duplications only | 100 – 200 | 2113 | 1×10−4 | 1.256 | 0.30 | 2×10−6 | 1.500 | 0.47 |
| 200 – 500 | 1401 | 0.017 | 1.143 | 0.20 | 0.026 | 1.243 | 0.52 | |
| ≥500 | 587 | 0.11 | 1.174 | 0.09 | 0.17 | 1.285 | 0.48 |
Because several known genomic disorders of the nervous system result from large CNVs, that are often many hundreds of kilobases11, we additionally stratified by size of event (Table 3). Of deletions, only larger (>500kb) variants were enriched (empirical _P_=3×10−4) despite being the least frequent set of CNVs (N=285), displaying a 3.57-fold increase in gene-count between cases and controls (empirical P = 2×10−5). In contrast, shorter duplications showed a stronger association with disease than longer duplications, albeit with a smaller fold increase than deletions (Table 3).
In general, the gene-count definition of CNV burden yielded stronger results, particularly for deletions (gene-count P = 3×10−5 versus number P = 0.11; Table 2). In fact, dividing all CNVs into two sets, of those that intersect at least one gene and those that do not, we saw an increased burden only in the number of “genic” CNVs (P = 5×10−6; Table S2) and not for non-genic CNVs (P = 0.16). There was a similar trend for CNVs seen 2-6 times when comparing enrichment in genic and non-genic CNVs (P = 7×10−4 and 0.19) but not single-occurrence CNVs (_P_=6×10−4 and 6×10−4). These results may reflect biological distinctions, although they may to some extent also reflect variable performance in CNV detection for different classes of variant. We conducted a set of analyses to rule out several sources of bias and confounding in the primary genome-wide burden analysis (Tables S3, S4, S5 & S6). While, in general, low specificity and sensitivity decrease power, of concern here is potential measurement error that systematically varied between cases and controls, leading to spurious results. In this respect, an obvious concern is that both Affymetrix 5.0 and 6.0 arrays were used; as a consequence, we performed all analyses controlling for array type. As described in the Supplementary Information, the primary result was also robust to the following. First, in addition to array type, we controlled for sample collection site, genotyping plate and average probe variance. Second, sensitivity analyses showed that no single sample collection site accounted for the observations. Third, we restricted analysis to the most homogeneous 90% of the sample with respect to intra-individual probe variance. Fourth, if case/control differences in CNV burden were purely due to unmeasured confounders, we would not expect an enriched gene-count after controlling for the overall extent and rate of CNVs. Of note, however, is that after controlling for overall (genic and non-genic) CNV burden there remained a significantly enriched gene-count burden in SCZ patients.
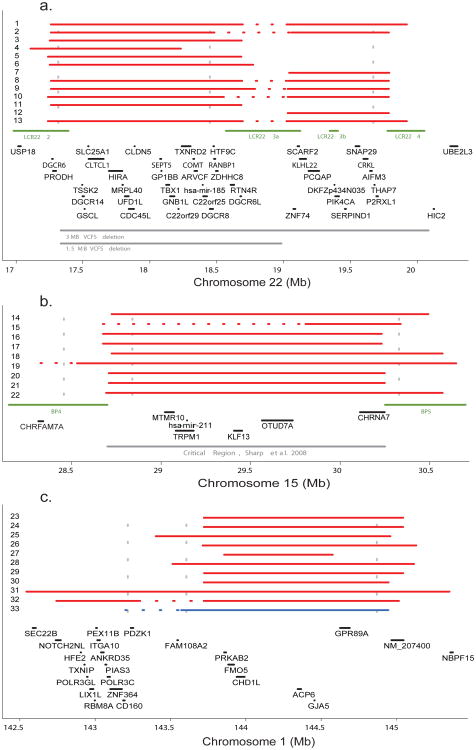
Our large sample size further enabled us to search for specific CNV regions associated with SCZ. One locus previously reported to increase risk for SCZ is 22q11.2 (17-21Mb), at which hemizygosity occurs in 1:4000 live births12. These deletions produce a range of clinically heterogeneous phenotypes, including velo-cardio facial syndrome (VCFS) and DiGeorge syndrome that together are known as 22q11.2 deletion syndrome (22q11.2DS)12; approximately 30% of carriers develop psychosis12. Previous studies estimated the frequency of 22q11.2 deletions to be 0.6%-1.0% in SCZ cases although many of these studies had technically incomplete characterization of this region19. Thus we expected to find examples of 22q11.2 deletions in our sample of 6,572 individuals. The most common form of 22q11.2DS is a 3Mb loss (∼90% frequency), although a nested 1.5Mb deletion is also observed (∼7%) along with infrequent (∼3%) atypical deletions20. We identified 13 large deletions (>500 kb) in SCZ cases within this interval and none in controls (Table S7). Of these, six were consistent with the larger deletion, five with the shorter deletion, and two were atypical. The 11 samples with typical deletions defined an interval with the strongest association (empirical P = 0.0017; genome-wide corrected P = 0.0046; odds ratio = 21.6) (Figure 1A). Controlling for sample collection site or genotyping plate instead of array type did not change the results (Table S10). The two other atypical deletions in this region overlap the distal end of the 3Mb variant. Deletion events within the region were confirmed in all 13 patients by quantitative polymerase chain reaction with three individual assays (qPCR, Tables S11 & S12 and Figure S1). Our findings provide additional evidence that hemizygosity in 22q11.2 is a rare but powerful risk factor for SCZ.
Figure 1. Regions with excess large deletions in cases.
a)The positions of CNVs >500kb across chromosome 22q11.2 region. Case deletions: red lines. Horizontal dashed sections: qPCR or visual inspection of array intensity data suggest an extended deletion. Green lines: locations of low-copy repeats (LCR22-2-LCR22-4). Gray lines: recurrent ∼3MB and ∼1.5MB VCFS deletions. qPCR primers marked by vertical dashed lines. b) Chromosome 15q13.2 region, as above, except for the locations of breakpoint regions (BP4, BP5) (green) and the MR critical region previously defined17. c) Chromosome 1q21.1: as above, except single deletion identified in a control subject is marked by blue line.
The larger 22q11.2 deletion harbors 43 genes (Table S8). Despite the efforts of many groups, the psychiatric symptoms observed in 22q11DS have not been ascribed to reduced copy number of any individual gene12. Variants within catechol-O-methyltransferase(COMT), an enzyme responsible for degrading catecholamines including dopamine, have been implicated in a wide variety of phenotypes, but with inconsistent results12.
Removing the 13 22q11.2DS individuals, we observed a further 271 deletions >500 kb (175 in cases and 110 in controls). Two additional regions (15q13.2 and 1q21.1) were identified that harbor a significant excess of deletions in SCZ cases after correction for multiple testing (Figure 1B and 1C; Table S7 for case descriptions). On chromosome 15 (28 – 31Mb) there were deletions in 9 cases and 0 controls (empirical P = 0.0029; genome-wide corrected P = 0.046; odds ratio = 17.9). On chromosome 1 (142.5-145.5Mb) there were 10 deletions in cases and 1 in controls (empirical P = 0.0076; genome-wide corrected P = 0.046; odds ratio = 6.6). All 20 large deletions at 15q13.2 and 1q21.1 were validated by one or more qPCR reactions (Tables S11 & S12 and Figure S1). The multiple test correction factors were small as a consequence of restricting our attention to this small class of rare variants. We did not observe any regions with a corrected P < 0.05 for either duplications or smaller (<500kb) deletions. Also of note, the primary CNV burden tests remained significant after removing individuals with a deletion at one of these three loci (number _P_ = 1 × 10−4 and gene-count _P_ = 3 × 10−5); for >500kb deletions specifically, the burden test remained significant for number (P = 0.02) but not for gene-count (P = 0.11).
The large deletions on chromosome 15q13.2 have not been previously associated with SCZ. This region does not include the nearby critical region for Prader-Willi/Angelman syndrome21 but is consistent with the critical region defined by recurrent deletion in cases of mental retardation (MR) with seizures recently reported17. Furthermore, our estimated breakpoints fall within the segmental duplications reported (BP4 and BP5). In the present study, evidence consistent with mildly-impaired cognition was seen in five of the nine patients with deletions and one individual also had a history of epilepsy (Table S7). This broad region has been the focus of previous genetic studies in SCZ. The alpha 7 subunit of the nicotinic acetylcholine receptor gene (CHRNA7) is a candidate based on an initial identification from linkage analysis of the P50 auditory evoked potential deficits observed in SCZ patients22,23 though there are no prior reports of large deletions associated with SCZ.
The deleted region on 1q21.1 is consistent with a previously reported de novo deletion in a patient with MR and seizures17 and two patients with autism (one de novo and one inherited)10. In the present study, three cases had mild cognitive abnormalities and one had a history of epilepsy (Table S7). The region contains 26 known genes, the majority of which are expressed in the brain (Table S8) and has previous reports of linkage24-26 but no prior reports of CNVs associated with SCZ.
Regions of highly homologous segmental duplication (SD) flank the deletions we report at 22q11.2, 15q13.2, and 1q21.1. A prominent mechanism for CNV genesis is non-allelic homologous recombination (NAHR) mediated by SDs, resulting in deletions and reciprocal duplications of the interval between SDs16,27. Neurodevelopmental and psychiatric syndromes have been associated with deletions and duplications flanked by SDs, many of which occur de novo10,11,17. Segmental duplications and NAHR mediate CNVs at 22q11.228 and may be involved in the genesis of CNVs at 15q13.214, and 1q21.1, though other mechanisms may be involved29. While this work was under review, Walsh et al.2 reported a higher frequency of CNVs in cases (15%, 22 in 150 SCZ patients) versus controls (5%). Among the 21 autosomal case CNVs they identified, we observed overlapping control CNVs at seven loci (for example, DLG2 and PTPRM, Table S9), illustrating that large sample numbers are needed to conclude that any one particular CNV or implicated gene can cause SCZ. Our global burden analysis demonstrated that, in aggregate, single-occurrence and very rare (under ∼1/1000) CNVs have increased rates in SCZ cases compared to controls, in line with Walsh et al. This suggests that at least some proportion of these rare CNVs seen in cases but not controls are likely SCZ risk factors, although like Walsh et al, we are unable to distinguish exactly which. Some examples of possible risk CNVs that were observed multiple times only in cases include deletions at 12p11.23 (4 cases) and 16p12.2-12.1 (6 cases). These deletions were >500kb, flanked by SDs and span several brain-expressed genes. In addition, duplications in two genes relevant to neural development and growth (NOTCH1 and p21-activated kinase 7, PAK7) were found in 5 and 6 cases respectively and 0 controls. Furthermore, we identified CNVs at two recently reported loci, NRXN1 and CNTNAP23,5 (Figure S2).
The etiology of SCZ has been vigorously debated. We now have strong and replicated2 evidence that individuals with SCZ have a greater burden of structural variation across their genomes. Our data show that CNVs in at least three loci act as strong risk factors for SCZ in a minority of individuals. Thus we can now posit that some cases of SCZ are “genomic disorders”16 although we do not yet know whether the risk is specific for SCZ as opposed to a more general risk factor for neuropsychiatric or central nervous system illness.
Exactly how a subtle, 1.15-fold increase in CNV burden translates mechanistically into illness in a given patient is currently unknown. We also do not know whether common genetic variants of more subtle effect are components of the etiology of SCZ, an empirical question that we and others are addressing. Similarly, we do not know how environmental risk or protective factors might act in concert with specific CNVs or with overall burden of CNVs.
A critically important goal will be to determine the full clinical and phenotypic spectrum in carriers of these deletions. Our data provide preliminary evidence of a variable phenotype in SCZ patients who would otherwise be regarded as clinically typical. Examining the role of these variants in related psychotic disorders, such as bipolar disorder, is imperative. Further work explicating the epidemiology and mechanism of these variants in SCZ may ultimately lead to a role for them in genetic counseling and understanding disease biology.
Methods Summary
Cases satisfied DSM-IV or ICD-10 criteria for SCZ and were broadly representative of clinical cases in contact with psychiatric services. DNA was extracted from whole blood, with approval from institutional review boards. CNVs were identified using the Birdseye package15 and analysed using PLINK v1.0314. See the Supplementary Information for detail. A list of all CNVs passing quality control is available at http://pngu.mgh.harvard.edu/isc/
Supplementary Material
Supplementary Data
Acknowledgments
We would like to thank the patients and families who have contributed their time and DNA to these studies. We would also like to thank David Altshuler and members of the Medical and Population Genetics group at the Broad Institute of Harvard and MIT for valuable discussion.
The Stanley Center for Psychiatric Research at the Broad Institute: The Sylvan C Herman Foundation (EMS); the Stanley Medical Research Institute (EMS)
Cardiff University: The Cardiff group was supported by an MRC (UK) Programme Grant and NIMH (USA) (CONTE: 2 P50 MH066392-05A1)
Karolinska Institutet: Swedish Council for Working Life and Social Research FO 184/2000; 2001-2368
Massachusetts General Hospital: Stanley Medical Research Institute (PS); MH071681 (PS); Narsad Young Investigator Award (SP)
Queensland Institute of Medical Research: Supported by the Australian National Health and Medical Research Council.
Trinity College Dublin: The Trinity College Dublin research was supported by Science Foundation Ireland, the Health Research Board (Ireland), the Stanley Medical Research Institute and the Wellcome Trust. Irish controls were supplied by Dr. Joe McPartlin from the Trinity College Biobank.
University of Aberdeen: This work was part funded by GSK and Generation Scotland, Genetics Health Initiative.
University College London: The University College London clinical and control samples were collected with support from the Neuroscience Research Charitable Trust, the Camden and Islington Mental Health and Social Care Trust, East London and City Mental Heath Trust, The West Berkshire NHS Trust, The West London Mental Health Trust, Oxfordshire & Buckinghamshire Mental Health Partnership NHS Trust, South Essex Partnership NHS Foundation Trust, Gloucestershire Partnership NHS Foundation Trust. Mersey Care NHS Trust, Hampshire Partnership NHS Trust and North East London Mental Health Trust.
University of Edinburgh: The collection of the Edinburgh cohort was supported by grants from The Wellcome Trust, London and the Chief Scientist Office of the Scottish Executive.
University of North Carolina, Chapel Hill: Supported by MH074027, MH077139, and MH080403; the Sylvan C Herman Foundation (PFS); the Stanley Medical Research Institute (PFS)
University of Southern California: This work would not have been possible without the collaboration of our patients and their families. We would also like to acknowledge the support of the National Institute of Mental Health and the Department of Veterans Affairs.
Author Contributions
Manuscript Preparation: Jennifer L Stone, Michael C O'Donovan, Hugh Gurling, George K Kirov, Douglas HR Blackwood, Aiden Corvin, Nick J Craddock, Michael Gill, Christina M Hultman, Paul Lichtenstein, Andrew McQuillin, Carlos N Pato, Douglas M Ruderfer, Michael J Owen, David St. Clair, Patrick Sullivan, Pamela Sklar, Shaun M Purcell
Data Analysis: Jennifer L Stone, Douglas M Ruderfer, Joshua Korn, George K Kirov, Stuart MacGregor, Andrew McQuillin, Derek W Morris, Colm T O'Dushlaine, Mark J Daly, Peter M Visscher, Peter A Holmans, Michael C O'Donovan, Patrick Sullivan, Pamela Sklar, Shaun M Purcell
Management Committee: Hugh Gurling, Aiden Corvin, Douglas HR Blackwood, Nick J Craddock, Michael Gill, Christina M Hultman, George K Kirov, Paul Lichtenstein, Andrew McQuillin, Michael C O'Donovan, Michael J Owen, Carlos N Pato, Shaun M Purcell, Edward M Scolnick, David St. Clair, Jennifer L Stone, Patrick Sullivan, Pamela Sklar
International SCZ Consortium Sites
Stanley Center for Psychiatric Research and Broad Institute of MIT and Harvard: Shaun M Purcell1-4, Jennifer L Stone1-4, Kimberly Chambert2,3, Douglas M Ruderfer1-4, Joshua Korn3,4, Steve A McCarroll3,4, Casey Gates3, Stacey B Gabriel3, Scott Mahon3, Kristen Ardlie3, Mark J Daly2,3,4, Edward M Scolnick2,3, Pamela Sklar1-4
Cardiff University: Michael C O'Donovan5, George K Kirov5, Nick J Craddock5, Peter A Holmans5, Nigel M Williams5, Lucy Georgieva5, Ivan Nikolov5, N Norton5, H Williams5, Draga Toncheva6, Vihra Milanova7, Michael J Owen5
Karolinska Institutet/University of North Carolina at Chapel Hill Christina M Hultman8.9, Paul Lichtenstein8, Emma F Thelander8, Patrick Sullivan10
Massachusetts General Hospital: Jennifer L. Stone1-4, Douglas M. Ruderfer1-4, Joshua Korn3,4, Steve A McCarroll3,4, Mark Daly2,3,4, Shaun M Purcell1-4, Pamela Sklar1-4
Trinity College Dublin: Derek W Morris11, Colm T O'Dushlaine11, Elaine Kenny11, John L Waddington12, Michael Gill11, Aiden Corvin11
University College London: Andrew McQuillin13, Khalid Choudhury13, Susmita Datta13, Jonathan Pimm13, Srinivasa Thirumalai14, Vinay Puri13, Robert Krasucki13, Jacob Lawrence13, Digby Quested15, Nicholas Bass13, David Curtis16, Hugh Gurling13
University of Aberdeen: Caroline Crombie17, Gillian Fraser17, Noelle Kwan18, Nicholas Walker19, David St. Clair18
University of Edinburgh: Douglas HR Blackwood20, Walter J Muir21, Kevin McGhee21, Alan W Maclean21, Margaret Van Beck20
Queensland Institute of Medical Research: Peter M Visscher22, Stuart Macgregor22
University of Southern California: Michele T. Pato23, Helena Medeiros23, Frank Middleton24, Celia Carvalho23, Christopher Morley24, Ayman Fanous23,25-27, David Conti23, James Knowles23, Carlos Paz Ferreira28, Antonio Macedo29, M. Helena Azevedo29, Carlos N. Pato23
- Department of Psychiatry, Massachusetts General Hospital and Harvard Medical School, 185 Cambridge St, Boston, MA 02114, USA
- Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge MA 02142 USA
- Broad Institute of Harvard and MIT, Cambridge, MA, 02142 USA
- Center for Human Genetic Research, Massachusetts General Hospital, 185 Cambridge St, Boston MA 02114 USA
- School of Medicine, Department of Psychological Medicine, School of Medicine, Cardiff University, Cardiff, C144XN UK
- Department Medical Genetics, University Hospital Maichin Dom, Sofia, Bulgaria
- Department of Psychiatry, First Psychiatric Clinic, Alexander University Hospital, Sofia, Bulgaria
- Medical Epidemiology and Biostatistics, Karolinska Institutet, Sweden
- Department of Neuroscience, Psychiatry, Ulleråker, Uppsala University, Sweden
- Departments of Genetics, Psychiatry, & Epidemiology, University of North Carolina at Chapel Hill, USA
- Neuropsychiatric Genetics Research Group, Dept. of Psychiatry and Institute of Molecular Medicine, Trinity College Dublin, Ireland
- Molecular & Cellular Therapeutics and RCSI Research Institute, Royal College of Surgeons in Ireland
- University College London: Molecular Psychiatry Laboratory, Department of Mental Health Sciences, University College London Medical School, Windeyer Institute of Medical Sciences, 46 Cleveland Street, London, W1T 4JF, UK
- West Berkshire NHS Trust, 25 Erleigh Road, Reading, RG3 5LR, UK
- West London Mental Health Trust, Hammersmith & Fulham Mental Health Unit and St Bernard's Hospital, London, W6 8RF, UK; Current address, Department of Psychiatry, University of Oxford, Warneford Hospital, Headington, Oxford, OX3 7JX, UK
- Queen Mary College, University of London and East London and City Mental Health Trust, Royal London Hospital, Whitechapel, London, E1 1BB, UK
- Department of Mental Health, University of Aberdeen, AB252ZD UK
- Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen AB252ZD, UK
- Ravenscraig Hospital, Inverkip Road, Greenock, PA16 9HA, UK
- Division of Psychiatry, School of Molecular and Clinical Medicine, University of Edinburgh, Edinburgh UK
- Division of Psychiatry, School of Molecular and Clinical Medicine, University of Edinburgh, Edinburgh UK
- Queensland Institute of Medical Research, Brisbane, Australia
- Center for Genomic Psychiatry, University of Southern California, Los Angeles, California, USA
- State University of New York- Upstate Medical University, Syracuse, New York, USA
- Washington VA Medical Center
- Department of Psychiatry, Georgetown University
- Department of Psychiatry, Virginia Commonwealth University
- Department of Psychiatry, Sao Miguel, Azores, Portugal
- Department of Psychiatry University of Coimbra, Portugal
Footnotes
The authors report no competing financial interests statement.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Walsh T, et al. Rare Structural Variants Disrupt Multiple Genes in Neurodevelopmental Pathways in Schizophrenia. Science. 2008 doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 3.Kirov G, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–65. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 4.Flomen RH, et al. Association study of CHRFAM7A copy number and 2 bp deletion polymorphisms with schizophrenia and bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141:571–5. doi: 10.1002/ajmg.b.30306. [DOI] [PubMed] [Google Scholar]
- 5.Friedman JI, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–6. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 6.Wilson GM, et al. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet. 2006;15:743–9. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]
- 7.Moon HJ, et al. Identification of DNA copy-number aberrations by array-comparative genomic hybridization in patients with schizophrenia. Biochem Biophys Res Commun. 2006;344:531–9. doi: 10.1016/j.bbrc.2006.03.156. [DOI] [PubMed] [Google Scholar]
- 8.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 11.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–21. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 12.Williams NM, O'Donovan MC, Owen MJ. Chromosome 22 deletion syndrome and schizophrenia. Int Rev Neurobiol. 2006;73:1–27. doi: 10.1016/S0074-7742(06)73001-X. [DOI] [PubMed] [Google Scholar]
- 13.McCarroll S, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, deBakker PW, Maller J, Kirby A, Elliott AE, Parkin M, Hubbell E, Webster T, Mei R, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones K, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet, in press. 2008 doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korn J, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Nizzari MM, Gabriel SB, Purcell SM, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms, and rare CNVs. Nat Genet, in press. 2008 doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–22. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 17.Sharp AJ, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–8. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross CA, Pearlson GD. Schizophrenia, the heteromodal association neocortex and development: potential for a neurogenetic approach. Trends Neurosci. 1996;19:171–6. doi: 10.1016/s0166-2236(96)10022-9. [DOI] [PubMed] [Google Scholar]
- 19.Karayiorgou M, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–6. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaikh TH, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 21.Butler MG, Fischer W, Kibiryeva N, Bittel DC. Array comparative genomic hybridization (aCGH) analysis in Prader-Willi syndrome. Am J Med Genet A. 2008 doi: 10.1002/ajmg.a.32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman R, et al. Linkage of a neurophysiological deficit in schizophrenia to a chromosome 15 locus. Proc Natl Acad Sci U S A. 1997;94:587–92. doi: 10.1073/pnas.94.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, et al. Evidence for linkage disequilibrium between the alpha 7-nicotinic receptor gene (CHRNA7) locus and schizophrenia in Azorean families. Am J Med Genet. 2001;105:669–74. doi: 10.1002/ajmg.1549. [DOI] [PubMed] [Google Scholar]
- 24.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–82. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurling HM, et al. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet. 2001;68:661–73. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwu HG, Liu CM, Fann CS, Ou-Yang WC, Lee SF. Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry. 2003;8:445–52. doi: 10.1038/sj.mp.4001235. [DOI] [PubMed] [Google Scholar]
- 27.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 28.Shaikh TH, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–91. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data