Molecular architecture of the prolate head of bacteriophage T4 (original) (raw)
Abstract
The head of bacteriophage T4 is a prolate icosahedron with one unique portal vertex to which the phage tail is attached. The three-dimensional structure of mature bacteriophage T4 head has been determined to 22-Å resolution by using cryo-electron microscopy. The T4 capsid has a hexagonal surface lattice characterized by the triangulation numbers _T_end = 13 laevo for the icosahedral caps and _T_mid = 20 for the midsection. Hexamers of the major capsid protein gene product (gp)23* and pentamers of the vertex protein gp24*, as well as the outer surface proteins highly antigenic outer capsid protein (hoc) and small outer capsid protein (soc), are clearly evident in the reconstruction. The size and shape of the gp23* hexamers are similar to the major capsid protein organization of bacteriophage HK97. The binding sites and shape of the hoc and soc proteins have been established by analysis of the _soc_– and _hoc_–_soc_– T4 structures.
Bacteriophage T4 is a large, tailed, double-stranded DNA (dsDNA) virus (family Myoviridae) that uses Escherichia coli as a host. The mature T4 virion, which contains ≈50 different proteins, consists of a prolate capsid, 172-kbp genomic DNA, and a tail with a contractile sheath terminating in a base plate to which are attached six long tail fibers. The architecture and the molecular composition of the T4 head, tail, and fibers have been characterized extensively by using a variety of techniques (1–3) leading to a structural model (4). Recent studies of the T4 components by cryo-electron microscopy (cryo-EM) and x-ray crystallography extended the structural knowledge to higher resolution (5).
T4 has one of the most complex structures of any virus that has been studied. There are >2,000 protein molecules of at least five different gene products (gps) in the head alone. The molecular mass of the DNA-filled head is 194 MDa and of the capsid alone is 82 MDa (6). The T4 head assembly proceeds via a number of intermediate stages. First, a DNA-free precursor, or prohead, is assembled that is processed proteolytically. Next, the genomic DNA is packaged into the prohead in a process that requires ATP energy (1). The prohead assembly is initiated by the portal protein gp20. The prohead contains an internal core made up of the major core protein, gp22, the minor core proteins, gp_alt_, a serine-type protease, gp21, and other internal proteins (1). The major capsid protein, gp23, is assembled around the scaffolding core together with the minor capsid protein, gp24. After completion of prohead assembly, the inactive gp21 enzyme is converted to the active protease, which cleaves the scaffold proteins into small peptides. A 65-residue-long amino-terminal “Δ-piece” is also cleaved from the 56-kDa gp23 molecule, thus yielding 48.7-kDa gp23* (1)∥. In addition, a 2.2-kDa amino-terminal piece of the 48.4-kDa gp24 is cleaved, giving rise to gp24* during head maturation. Most of the small peptides produced by the gp21 protease are expelled from the prohead, thus providing space necessary to accommodate the genomic DNA.
After cleavage of the gp23 Δ-piece, the near-hexagonal capsid lattice expands, thus increasing the capsid volume by ≈50% (7). Two proteins, highly antigenic outer capsid protein (hoc, 39.1 kDa) and small outer capsid protein (soc, 9.7 kDa), are attached to the capsid surface (8–10). Whereas the soc protein helps to stabilize the capsid against extremes of pH and temperature, hoc only has a marginal effect on head stability (11, 12). However, both proteins are dispensable for the head morphogenesis and phage infection.
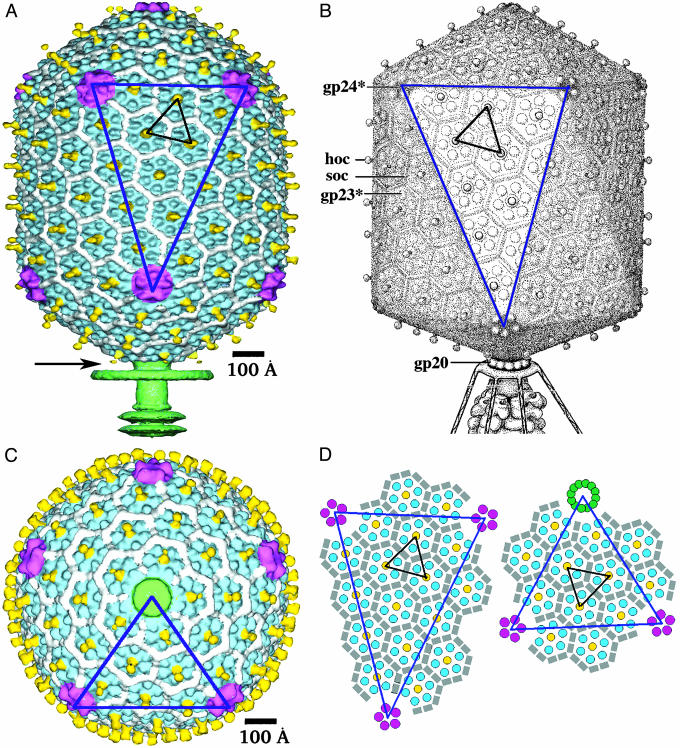
The mature T4 head is a prolate icosahedron elongated along a fivefold axis (7, 9, 13, 14). The surface of the prolate icosahedron is composed of two end caps, each made of five equilateral triangular facets and connected by an elongated midsection made of 10 triangular facets (Fig. 1 and Appendix). The facets of the T4 capsid are composed of gp23*. The 11 vertices are occupied by pentamers of gp24*, whereas the 12th vertex is a special portal for DNA packaging, tail attachment, and DNA exit. The portal vertex protein, gp20 (61 kDa), assembles as a dodecamer often called the “connector” (15).
Fig. 1.
Structure of the bacteriophage T4 head (_T_end = 13 laevo, _T_mid = 20, _h_1 = 3, _k_1 = 1, _h_2 = 4, and _k_2 = 2) compared with the previously proposed model (_T_end = 13 laevo, _T_mid = 21, _h_1 = 3, _k_1 = 1, _h_2 = 3, and _k_2 = 3) (4, 9). The facet triangles are shown in blue and the basic triangles (see Fig. 4) are shown in black as appropriate. (A) Shaded surface representation of the cryo-EM reconstruction viewed perpendicular to the fivefold axis. gp23* is shown in blue, gp24* is in magenta, soc is in white, hoc is in yellow, and the tail is in green. (B) Model of the previously proposed T4 head structure (adapted from ref. 4). (C) View of the reconstruction along the fivefold axis with the portal vertex toward the observer; the tail has been cut away at the level shown by the black arrow in A. Proteins are colored as described for A. (D, Left) Schematic representation of the distribution of proteins in the elongated midsection facet. (D, Right) Schematic representation of an end-cap facet. Proteins are colored as described for A except the soc molecules are shown as gray rectangles. A and C were prepared with the help of the programs dino (www.dino3d.org) and povray (www.povray.org).
The organization of gp23* into a surface lattice on the prolate head can be described by two triangulation numbers: _T_end and _T_mid (the latter is often called Q; ref. 16 and Appendix). The icosahedral end caps were shown to conform to the triangulation number _T_end = 13 laevo (7, 17). This number, but not the hand, was later confirmed by three-dimensional cryo-EM reconstructions of isometric T4 capsids (18, 19). The value of _T_mid for the T4 capsid had been determined previously to be 21 (9).
Here, we present the three-dimensional structure of the T4 prolate head of the mature virus and of the urea-treated virus in which the tail has contracted by using cryo-EM and image-reconstruction techniques. We show that, in both cases, _T_mid is equal to 20. We also report the structure of the head when soc or hoc and soc are missing, and hence we were able to determine the shapes of the hoc and soc proteins and their binding interfaces.
Methods
Phage Particle Preparation. Concentrated crude lysate of wild-type bacteriophage T4D was prepared on E. coli BE/1 cells by using a glucose-salts medium, M9A (7, 20). Phage particles were purified by sucrose gradient centrifugation as described in ref. 17. The _soc_– and _hoc_–_soc_– mutant phages (V. B. Rao collection) were grown on E. coli P301 and purified by sucrose gradient centrifugation by using the basic procedure described (7, 21).
Particles with contracted tails were prepared as follows. A 1-ml sample of the T4 phage, with a titer of ≈1011 plaque-forming units per ml, was diluted 10-fold by using 3 M urea, buffered with 50 mM Tris·HCl at pH 8.0 complemented with 1 mM MgCl2, and incubated for 2 h at 4°C. DNase I was added to the sample to a final concentration of 30 μg/ml to eliminate residual phage DNA. The sample was further 3-fold diluted and centrifuged for 1 h at 75,000 × g and 4°C. The pellet was dissolved slowly in 100–200 μl of water.
EM. Low-dose cryo-EM was performed as described by Baker et al. (22). Images of frozen-hydrated samples were recorded on Kodak film by using a Philips (Eindhoven, The Netherlands) CM300 FEG microscope at a magnification of ×47,000 and an irradiation dose of ≈29 electrons per Å2 using a defocus distance of 1–3 μm. The images were digitized with a ZI scanner. Particles were picked from the micrographs by using the program robem (http://bilbo.bio.purdue.edu/∼workshop/help_robem).
Results
The Three-Dimensional Reconstruction. The analysis of the 3 M urea-treated sample was the first to be completed successfully. The three-dimensional image reconstructions were performed with the program spider (23) using the projection-matching technique to orient the particles. The initial model was a prolate icosahedron with flat facets of uniform density, represented on a grid with a 12-Å pixel size. The dimensions of the phage facets and the relative orientation of the caps were calculated assuming _T_end = 13 laevo and _T_mid = 21 symmetry, given the overall dimensions of the phage head (9). Several cycles of image reconstruction were performed using five- and twofold-symmetry averaging. The resultant map had a smooth surface and did not show a recognizable hexagonal surface lattice.
A more complex model was subsequently constructed as a map with a 6-Å pixel size. The end caps of the model were derived from a reconstruction of T4 isometric particles (19). The cylindrical midsection was based on the reconstruction that was initiated from the original mask model. This reconstruction was performed by imposing five- and twofold-symmetry averaging as before. The resultant map had featureless end caps, suggesting that the relative orientation of the end caps in the model might be incorrect. Therefore, the model was remade assuming _T_end = 13 laevo and _T_mid = 20 symmetry (_T_mid = 21 and _T_mid = 20 produce similarly shaped prolate particles, although the end caps are related by roughly opposite rotations about the unique fivefold axis). This reconstruction showed, even after only one cycle of averaging, a hexameric surface lattice on the end caps and small protrusions corresponding to hoc molecules on the smooth midsection facets. After several additional cycles using fivefold and twofold averaging, a lattice of hexameric capsomers became evident on the midsection surface. At this stage, a cylinder of constant density was added to the model to simulate a part of the phage tail. The iterative reconstruction procedure then was completed by using fivefold averaging only.
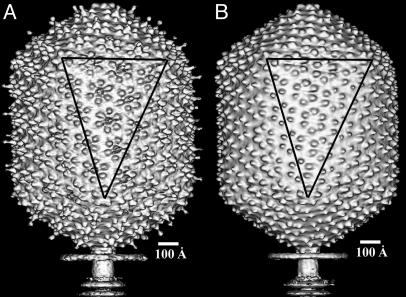
The reconstruction of the native T4 particle with an extended tail (Fig. 1 A and C) and the _soc_– heads (Fig. 2_A_) were made by using the urea-treated sample as an initial model. To reconstruct the _hoc_–_soc_– head (Fig. 2_B_), the structure of the _soc_– head (Fig. 2 A) was used as a model. The final reconstructions were corrected with the contrast transfer function (ref. 24 and Table 1). The limit of resolution varies roughly linearly with the number of particles, suggesting that increasing the number of particles would be beneficial to obtain improved results.
Fig. 2.
Shaded surface representations of the _soc_– (A) and _hoc_–_soc_– (B) reconstructions. A facet triangle is shown.
Table 1. Number of particles used in the reconstructions and the final resolution (Fourier shell correlation cutoff = 0.3).
| Sample | No. of particles | Resolution, Å | Figure |
|---|---|---|---|
| T4 with extended tail | 5,140 | 22.5 | 1 A and C |
| T4 with contracted tail | 1,757 | 24 | Not shown |
| T4 _soc_- | 763 | 32 | 2_A_ |
| T4 _hoc_-_soc_- | 3,156 | 25 | 2_B_ |
Overall Architecture of the T4 Prolate Head. The T4 head is an elongated icosahedron with rounded edges (Fig. 1 A and C), with a length of 1,195 Å measured along the fivefold axis and a width of 860 Å measured along a pseudo twofold axis passing perpendicular to the unique fivefold axis. The capsid surface is covered with a complex pattern of protrusions, attributed to the gp23*, gp24*, hoc, and soc proteins (Fig. 1). The major capsid protein gp23* forms a hexagonal lattice conforming to the triangulation numbers _T_end = 13 and _T_mid = 20 (_h_1 = 3, _k_1 = 1, _h_2 = 4, and _k_2 = 2; see Appendix), with a separation of 140 Å between hexamer centers. Each gp23* hexamer has six small protrusions separated by 45 Å from each other. Although the structure of the end caps is closely similar to the structure of the T4 isometric capsid (18, 19), the organization of the hexagonal lattice in the midsection facets is different from that reported previously (refs. 4 and 9; Fig. 1 A and B). The total number of gp23* monomers forming the capsid, _N_gp23*, is given by
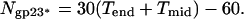 |
[1] |
|---|
The first term in Eq. 1 corresponds to Eq. 6 (Appendix), and the last term accounts for the 11 pentameric vertices of the capsid that are occupied by gp24* pentamers and one vertex that is occupied by the gp20 connector. Thus, the total number of gp23* monomers in the T4 capsid is 930.
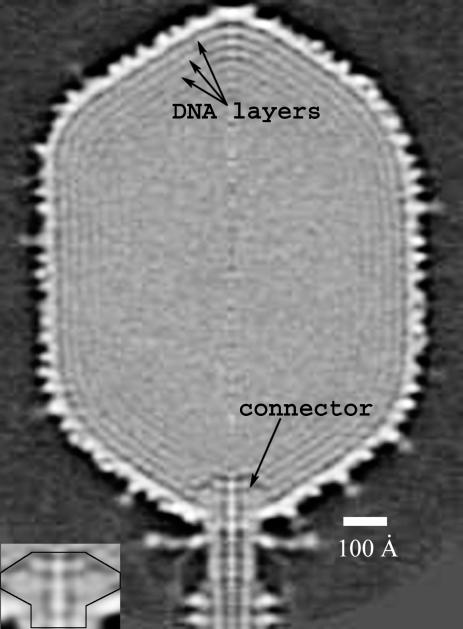
The protrusions corresponding to the gp24* monomers in the 11 pentagonal vertices are larger and more exposed (separated by 48 Å from each other) than the protrusions of the gp23* hexamers. The distance between the center of the gp24* pentamer and the nearest gp23* hexamer is 127 Å. The 12th portal vertex, to which the tail is attached, is occupied by a connector formed by a dodecamer of gp20. Because the reconstruction assumed fivefold symmetry, the actual structures of the sixfold-symmetric tail and twelvefold-symmetric connector were not resolved. Nevertheless, the reconstruction shows a part of the tail with a collar and two of the disks belonging to the contractile sheath (Fig. 1 A). At the portal vertex, there is density corresponding to the gp20 connector (Fig. 3). The mushroom shape of this density is analogous to that of the bacteriophages SPP1 (25) and φ29 (26) connector assemblies.
Fig. 3.
The central section of the reconstructed cryo-EM density map viewed from a direction perpendicular to the fivefold axis. The concentric layers beneath the outer capsid shell are attributed to the densely packaged dsDNA. The spacing between successive layers is ≈25 Å. The high-density region in the capsid interior next to the portal vertex is attributed to the gp20 connector (see the enlargement in Inset).
The most protruding features on the capsid surface are the hoc molecules, which extend ≈60 Å away from the shell surface. One hoc molecule (9) is attached to the center of each gp23* hexamer (18, 19), hence the total number of hoc monomers is given by
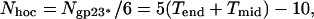 |
[2] |
|---|
and is equal to 155.
On the capsid surface, the soc molecules form a nearly continuous mesh that encircles the gp23* hexamers. As also seen in the structure of the isometric capsid (18, 19), soc molecules bind between two gp23* subunits but not between gp23* and gp24*. Therefore, the soc molecules do not bind around pentamers of gp24*. The reconstruction shows that soc molecules are also absent around the portal vertex, between gp23* and gp20 (Fig. 1_C_), confirming the finding that one molecule of soc interacts with two molecules of gp23* (18). Therefore, the total number of soc molecules is given by
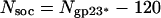 |
[3] |
|---|
and is equal to 810.
The Structure of the Encapsidated DNA. The density distribution in the interior of the head consists of concentric layers with alternating high and low density, separated by 25 Å, just inside the capsid shell (Fig. 3). These layers can be attributed to densely packed dsDNA. At least six DNA layers can be detected and are arranged like a Russian doll. Eight layers of DNA with a similar spacing were observed in the cryo-EM reconstructions of the isometric T4 head (19).
Comparison of the soc– and hoc–soc– T4 Heads. The lack of hoc and soc proteins does not cause any rearrangement of the gp23* capsomer lattice (Fig. 2). The reconstructions do not show depressions on the capsid surface in the places at which hoc and soc bind. Hence, the hoc and soc molecules have no substantial portion buried in the gp23* lattice. The soc monomer is a rod-like molecule with a length of ≈40 Å and diameter of ≈20 Å, which is a slight modification of earlier results (18). The protrusions corresponding to the hoc molecule have the shape of a dumbbell with a globular head (≈25 Å in diameter), a constricted neck, and a base (≈32 Å in diameter) that is bound to the center of each gp23* hexamer (Fig. 1 A and C).
Absolute Hand and Relative Head–Tail Orientation. To determine the chirality of the head reconstruction, three-dimensional models of the whole phage were created, consisting of the head, the contracted sheath, baseplate, and approximately two thirds of the tail tube (P.G.L., P.R.C., V. A. Kostyuchenko, V.V.M., and M.G.R., unpublished data). In these models, the tail and head were related by 0°, 4°, and 8° rotations, thus uniformly covering the 12°-interval of the five- to sixfold-symmetry mismatch between the head and the tail. The hand of the head was determined based on the hand of the contracted sheath and the baseplate. The latter had been established by fitting the cryo-EM density with the known x-ray structures of the baseplate proteins gp8 (27), gp9 (28), and gp11 (29). Initially, each urea-treated phage particle was oriented by using only the head part, assuming its fivefold symmetry, leaving unknown which of five possible orientations of the head matched the current asymmetric model. To solve this problem, the whole phage particle (head + tail) was used to differentiate the five possible orientations. The reconstructions calculated by using the models with the dextro head chirality had a lattice of recognizable hexamers on the capsid surface but had a smooth pattern for the tail part. However, the reconstructions calculated by using the models with laevo head chirality had not only a lattice of hexamers on the head but also the anticipated helical pattern for the tail-sheath subunits. Thus, the hand of the head is laevo, consistent with results obtained from metal-shadowed phages (17). The three reconstructions, calculated starting with the laevo model density distributions, were all of similar quality, making it impossible to establish the relative head–tail rotational relationship to better than 6°. However, by applying sixfold averaging to the tail component during the reconstruction procedure, it was possible to differentiate between the three starting models, thus determining the unique relationship between the fivefold-symmetric head and sixfold-symmetric tail to an accuracy of ≈2°.
Stability of the T4 Head Treated with 3 M Urea. After attachment of phage to the host cell surface, the tail sheath contracts. This process can be mimicked in vitro, without DNA leaving the capsid, by 3 M urea treatment (30). There were no detectable differences between the native (Fig. 1 A) and urea-treated capsids in their lattice organization, capsomer shape, and orientation of gp23*, gp24*, and the hoc and soc proteins. The presence of hoc and soc in the urea-treated samples demonstrates the stability of their interaction with the gp23* shell.
Discussion
Four different reconstructions were made with virus propagated in two laboratories (Moscow and Washington, DC). These reconstructions each had identical hexameric lattices with _T_end = 13 and _T_mid = 20, demonstrating that the prolate head of wild-type strain T4D (described by Doermann et al. in ref. 21) phage has no polymorphic variants. Earlier results had depended primarily on length-to-width measurements of the prolate head as well as the stoichiometry of the capsid proteins. These quantities are rather similar for _T_mid = 20 or 21, resulting in an ambiguous conclusion (9). Using freeze-etching EM, Lane and Eiserling (31) visualized T4 particles with _T_mid = 20, but they attributed this unexpected result to a mutation in the major capsid protein. The revised geometry of the head now permits the exact determination of the number of protein subunits forming the T4 mature capsid.
Kellenberger (20, 32) proposed a template and also a vernier mechanism that might determine the size and shape of the T4 prolate head based on the interactions of the internal scaffolding proteins or “core” with the capsid shell. In the template model, the structure of the assembled scaffolding core is assumed to account for the shape of the prohead. In contrast, the vernier model assumes a vernier-type matching between the assembly of the core and shell of the growing prohead such that when the core and shell are in register, the elongation is terminated. Current data suggest that the primary determinant of the prohead shape is its scaffolding core, and it is likely that the core and shell grow concurrently (1). The three-dimensional structure of the T4 head as described here should be helpful in determining the mechanism that controls the formation of the geometry in the prolate head. Similar mechanisms might be expected in other tailed phages.
Layers of dsDNA have been observed in a variety of other viruses including adenovirus (33) and tailed phages (34, 35). Furthermore, layers of dsRNA have been observed in reoviruses (36). The separation of 25 Å between the DNA layers in the T4 head agrees with that found in isometric heads of T4 (19) and in phage λ DNA toroids condensed in vitro (37). Hud and Downing (37) have suggested that such a layered structure is the consequence of hexagonal packing of the DNA strands separated by ≈29 Å. A geometrical estimation assuming hexagonal packing for the DNA strands throughout the head shows that essentially all of the volume of the phage head is required to accommodate the genomic DNA supporting the “head-full” mechanism of DNA packaging (1). The presence of the connector does not indent the DNA layers inward (Fig. 3) but probably causes the DNA to wind around the connector.
The three-dimensional fold of polypeptides is far more conserved than their primary amino acid sequence. Thus, proteins with similar folds frequently have little, if any, recognizable sequence similarity. The number of unique folds of protein domains thus is limited (38, 39), although the actual number cannot be defined without first making an arbitrary definition of what degree of difference is required to define a new fold and what defines a domain. The capsid structure of viruses seems to be subject to an exceptional degree of conservation. The large majority of known virus capsid structures are based on the jelly-roll fold (40–42). However, there are exceptions such as the capsid structure of the MS2 phage (43), alphaviruses (44), and the tailed bacteriophage HK97 (45). Indeed, the HK97 structure has been identified in some other tailed phage capsids (46). Whereas the jelly-roll motif creates hexagonal surface lattices with pseudo (actually trimeric) repeating units separated by ≈75 Å, the HK97 motif creates hexagonal surface lattices of true hexamers separated by ≈132 Å. The similarity of major capsid protein molecular weight and separation between hexameric motifs makes it likely that most or all tailed phages would have major capsid proteins that are based on the HK97 structure (Table 2). The molecular mass of T4 gp23* is ≈1.6 times larger than that of the coat protein of HK97, which also is consistent with a slight increase in the hexamer separation in T4 compared with HK97. Presumably, the additional amino acids make loop insertions around the periphery of the folding motif. Similar loop insertions occur, for instance, in the jelly-roll fold found in parvoviruses (47) when compared with picornaviruses (40). Although the HK97 monomers are linked in the manner of chain mail (45), the structure of P22 bacteriophage uses the HK97 motif without forming chain-mail links (46). Superposition of the HK97 hexamer onto a typical hexamer of T4 shows an acceptable visual fit to the available resolution map. None of these considerations establish with certainty that the fold of gp23* is similar to HK97, but they do show that it is possible. Considering other structural similarities in the structural components of tailed phages, such as the connector dodecamer in T4, φ29 (26), and SPP1 (25) or the scaffolding protein in T4 (48), P22 (49), and φ29 (50), it would seem probable that the structural proteins of most tailed phages might have had a common evolutionary origin.
Table 2. Comparison of the molecular masses of capsid proteins and distances between the capsomers in tailed dsDNA bacteriophages.
| Phage | Molecular mass of capsid protein, kDa | Distance between capsomers, Å |
|---|---|---|
| φ29 | 50 | 142 |
| P22 | 47 | ≈132 |
| T4 | 49 | 140 |
| HK97 | 31 | 132 |
Acknowledgments
We are grateful for helpful discussions with Victor Kostyuchenko and Marc Morais. We thank Cheryl Towell and Sharon Wilder for help with preparation of the manuscript. This work was supported by National Science Foundation Grant MCB-9986266 (to M.G.R.), Human Frontier Science Program Grant RGP-28/2003 (to M.G.R., V.V.M., and Fumio Arisaka), Howard Hughes Medical Institute Grant 55000324 (to V.V.M.), and the Keck Foundation for the CM300 Philips electron microscope (Grant 1419991440 to M.G.R.).
Appendix: Quasisymmetry in Prolate Icosahedral Heads
An icosahedron can be considered as having 10 equilateral triangular facets on the two opposing end sections and 10 equilateral triangular facets in the midsection. The vertex of each facet is on a fivefold axis. For a prolate head elongated along a fivefold axis, the facets of the midsection are extended.
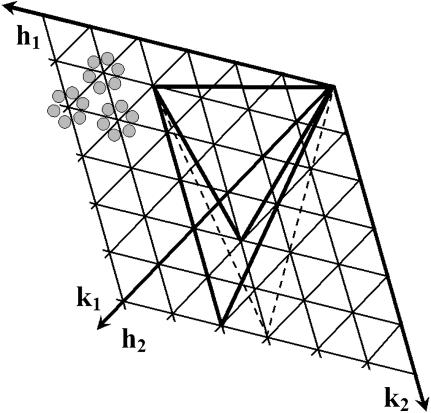
If there are >60 asymmetric units in the icosahedron, then the asymmetric units that are related by quasisymmetry (51) lie on a hexagonal surface lattice defined by the h and k axial lattice directions. A “basic” triangle then can be defined as an equilateral triangle joining the centers of adjacent hexagonal units. Thus, the basic triangle will contain three asymmetric units (Fig. 4).
Fig. 4.
Surface lattice organization for a prolate bacteriophage T4 head. Small triangles represent the “basic” triangles. Each such triangle contains three asymmetric subunits schematically shown in gray (top left). The large equilateral triangle corresponds to an end-cap facet. It is defined by one vertex at the origin and another vertex at (_h_1 = 3, _k_1 = 1) and corresponds to a _T_end = 13 laevo lattice. The large dashed and solid elongated triangles describe the midsection facets. One vertex of these triangles is at the origin, another is at the point (_h_1 = 3, _k_1 = 1), and the third is at (_h_2, _k_2). The dashed triangle corresponds to the midsection structure of the previously proposed bacteriophage T4 model (4, 9), whereas the solid triangle corresponds to the cryo-EM reconstruction reported here.
The triangulation number T specifies the number of basic triangles in a facet (Fig. 4), which is equal to the number of quasiasymmetric units per icosahedral asymmetric unit. Each of the 10 equilateral facets forming the icosahedral end caps are defined by the vector joining the origin to the point (_h_1, _k_1). _T_end is the number of basic triangles in each facet of the end caps and is given by the equation (51)
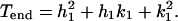 |
[4] |
|---|
For bacteriophage T4, _h_1 = 3 and _k_1 = 1, thus giving a _T_end = 13 laevo lattice (7, 17). The midsection triangles, which have unequal sides, are defined (Fig. 4) by the two vectors from the origin to the points (_h_1, _k_1) and (_h_2, _k_2). The triangulation _T_mid number, which determines the number of basic triangles in each midsection facet, is expressed as (16)
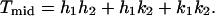 |
[5] |
|---|
The _T_mid number is related to the often-used Q number by _T_mid = fQ, where f is an integer (16).
The surface of the head is formed by 10 end-section triangles and 10 midsection triangles, making the total number of basic triangles in the head equal to 10(_T_end + _T_mid), and the total number of protein subunits, N, is given by
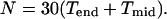 |
[6] |
|---|
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: cryo-EM, cryo-electron microscopy; dsDNA, double-stranded DNA; gp, gene product; hoc, highly antigenic outer capsid protein; soc, small outer capsid protein.
Footnotes
∥
According to phage genetics usage, gp_X_* signifies the product of maturation produced by the cleavage of gp_X_ to gp_X_*.
References
- 1.Black, L. W., Showe, M. K. & Steven, A. C. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 218–258.
- 2.Coombs, D. H. & Arisaka, F. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 259–281.
- 3.Wood, W. B., Eiserling, F. A. & Crowther, R. A. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 282–290.
- 4.Eiserling, F. A. & Black, L. W. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 209–212.
- 5.Leiman, P. G., Kanamaru, S., Mesyanzhinov, V. V., Arisaka, F. & Rossmann, M. G. (2003) Cell. Mol. Life Sci. 60**,** 2356–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baschong, W., Baschong-Preschianotto, C., Engel, A., Kellenberger, E., Lustig, A., Reichelt, R., Zulauf, M. & Aebi, U. (1991) J. Struct. Biol. 106**,** 93–101. [DOI] [PubMed] [Google Scholar]
- 7.Aebi, U., Bijlenga, R., van der Broek, J., van der Broek, R., Eiserling, F., Kellenberger, C., Kellenberger, E., Mesyanzhinov, V., Müller, L., Showe, M., et al. (1974) J. Supramol. Struct. 2**,** 253–275. [DOI] [PubMed] [Google Scholar]
- 8.Aebi, U., van Driel, R., Bijlenga, R. K., ten Heggeler, B., van den Broek, R., Steven, A. C. & Smith, P. R. (1977) J. Mol. Biol. 110**,** 687–698. [DOI] [PubMed] [Google Scholar]
- 9.Baschong, W., Aebi, U., Baschong-Preschianotto, C., Dubochet, J., Landmann, L., Kellenberger, E. & Wurtz, M. (1988) J. Ultrastruct. Mol. Struct. Res. 99**,** 189–202. [DOI] [PubMed] [Google Scholar]
- 10.Ishii, T. & Yanagida, M. (1977) J. Mol. Biol. 109**,** 487–514. [DOI] [PubMed] [Google Scholar]
- 11.Ishii, T., Yamaguchi, Y. & Yanagida, M. (1978) J. Mol. Biol. 120**,** 533–544. [DOI] [PubMed] [Google Scholar]
- 12.Steven, A. C., Greenstone, H. L., Booy, F. P., Black, L. W. & Ross, P. D. (1992) J. Mol. Biol. 228**,** 870–884. [DOI] [PubMed] [Google Scholar]
- 13.Moody, M. F. (1965) Virology 26**,** 567–576. [DOI] [PubMed] [Google Scholar]
- 14.Boy de la Tour, E., Bolle, A. & Kellenberger, E. (1965) Pathol. Microbiol. (Basel) 28**,** 229–234. [PubMed] [Google Scholar]
- 15.Driedonks, R. A., Engel, A., tenHeggeler, B. & van Driel, R. (1981) J. Mol. Biol. 152**,** 641–662. [DOI] [PubMed] [Google Scholar]
- 16.Moody, M. F. (1999) J. Mol. Biol. 293**,** 401–433. [DOI] [PubMed] [Google Scholar]
- 17.Aebi, U., Bijlenga, R., tenHeggeler, B., Kistler, J., Steven, A. C. & Smith, P. R. (1976) J. Supramol. Struct. 5**,** 475–495. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki, K., Trus, B. L., Wingfield, P. T., Cheng, N., Campusano, G., Rao, V. B. & Steven, A. C. (2000) Virology 271**,** 321–333. [DOI] [PubMed] [Google Scholar]
- 19.Olson, N. H., Gingery, M., Eiserling, F. A. & Baker, T. S. (2001) Virology 279**,** 385–391. [DOI] [PubMed] [Google Scholar]
- 20.Kellenberger, E. (1968) Virology 34**,** 549–561. [DOI] [PubMed] [Google Scholar]
- 21.Doermann, A. H., Eiserling, F. A. & Boehner, L. (1973) J. Virol. 12**,** 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker, T. S., Olson, N. H. & Fuller, S. D. (1999) Microbiol. Mol. Biol. Rev. 63**,** 862–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. & Leith, A. (1996) J. Struct. Biol. 116**,** 190–199. [DOI] [PubMed] [Google Scholar]
- 24.Frank, J. & Penczek, P. (1995) Optik 98**,** 125–129. [Google Scholar]
- 25.Orlova, E. V., Gowen, B., Droge, A., Stiege, A., Weise, F., Lurz, R., van Heel, M. & Tavares, P. (2003) EMBO J. 22**,** 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson, A. A., Tao, Y., Leiman, P. G., Badasso, M. O., He, Y., Jardine, P. J., Olson, N. H., Morais, M. C., Grimes, S., Anderson, D. L., et al. (2000) Nature 408**,** 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leiman, P. G., Shneider, M. M., Kostyuchenko, V. A., Chipman, P. R., Mesyanzhinov, V. V. & Rossmann, M. G. (2003) J. Mol. Biol. 328**,** 821–833. [DOI] [PubMed] [Google Scholar]
- 28.Kostyuchenko, V. A., Navruzbekov, G. A., Kurochkina, L. P., Strelkov, S. V., Mesyanzhinov, V. V. & Rossmann, M. G. (1999) Structure (London) 7**,** 1213–1222. [DOI] [PubMed] [Google Scholar]
- 29.Leiman, P. G., Kostyuchenko, V. A., Shneider, M. M., Kurochkina, L. P., Mesyanzhinov, V. V. & Rossmann, M. G. (2000) J. Mol. Biol. 301**,** 975–985. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg, E., Grinius, L. & Letellier, L. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 347–356.
- 31.Lane, T. & Eiserling, F. (1990) J. Struct. Biol. 104**,** 9–23. [DOI] [PubMed] [Google Scholar]
- 32.Kellenberger, E. (1972) in Polymerization in Biological Systems. Ciba Foundation Symposium 7, eds. Wolstenholme, G. E. Q. & O'Connor, M. (Churchill, London), pp. 189–206.
- 33.Stewart, P. L., Burnett, R. M., Cyrklaff, M. & Fuller, S. D. (1991) Cell 67**,** 145–154. [DOI] [PubMed] [Google Scholar]
- 34.Lepault, J., Dubochet, J., Baschong, W. & Kellenberger, E. (1987) EMBO J. 6**,** 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerritelli, M. E., Cheng, N., Rosenberg, A. H., McPherson, C. E., Booy, F. P. & Steven, A. C. (1997) Cell 91**,** 271–280. [DOI] [PubMed] [Google Scholar]
- 36.Gouet, P., Diprose, J. M., Grimes, J. M., Malby, R., Burroughs, J. N., Zientara, S., Stuart, D. I. & Mertens, P. P. C. (1999) Cell 97**,** 481–490. [DOI] [PubMed] [Google Scholar]
- 37.Hud, N. V. & Downing, K. H. (2001) Proc. Natl. Acad. Sci. USA 98**,** 14925–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossmann, M. G., Moras, D. & Olsen, K. W. (1974) Nature 250**,** 194–199. [DOI] [PubMed] [Google Scholar]
- 39.Jones, D. & Thornton, J. (1993) J. Comput. Aided Mol. Des. 7**,** 439–456. [DOI] [PubMed] [Google Scholar]
- 40.Rossmann, M. G. & Johnson, J. E. (1989) Annu. Rev. Biochem. 58**,** 533–573. [DOI] [PubMed] [Google Scholar]
- 41.Harrison, S. C. (2001) Curr. Opin. Struct. Biol. 11**,** 195–199. [DOI] [PubMed] [Google Scholar]
- 42.Nandhagopal, N., Simpson, A. A., Gurnon, J. R., Yan, X., Baker, T. S., Graves, M. V., Van Etten, J. L. & Rossmann, M. G. (2002) Proc. Natl. Acad. Sci. USA 99**,** 14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golmohammadi, R., Valegård, K., Fridborg, K. & Liljas, L. (1993) J. Mol. Biol. 234**,** 620–639. [DOI] [PubMed] [Google Scholar]
- 44.Choi, H. K., Tong, L., Minor, W., Dumas, P., Boege, U., Rossmann, M. G. & Wengler, G. (1991) Nature 354**,** 37–43. [DOI] [PubMed] [Google Scholar]
- 45.Wikoff, W. R., Liljas, L., Duda, R. L., Tsuruta, H., Hendrix, R. W. & Johnson, J. E. (2000) Science 289**,** 2129–2133. [DOI] [PubMed] [Google Scholar]
- 46.Jiang, W., Li, Z., Zhang, Z., Baker, M. L., Prevelige, P. E., Jr., & Chiu, W. (2003) Nat. Struct. Biol. 10**,** 131–135. [DOI] [PubMed] [Google Scholar]
- 47.Tsao, J., Chapman, M. S., Agbandje, M., Keller, W., Smith, K., Wu, H., Luo, M., Smith, T. J., Rossmann, M. G., Compans, R. W. & Parrish, C. R. (1991) Science 251**,** 1456–1464. [DOI] [PubMed] [Google Scholar]
- 48.Mesyanzhinov, V. V., Sobolev, B. N., Marusich, E. I., Prilipov, A. G. & Efimov, V. P. (1990) J. Struct. Biol. 104**,** 24–31. [DOI] [PubMed] [Google Scholar]
- 49.Tuma, R., Parker, M. H., Weigele, P., Sampson, L., Sun, Y., Krishna, N. R., Casjens, S., Thomas, G. J., Jr., & Prevelige, P. E., Jr. (1998) J. Mol. Biol. 281**,** 81–94. [DOI] [PubMed] [Google Scholar]
- 50.Morais, M. C., Kanamaru, S., Badasso, M. O., Koti, J. S., Owen, B. A., McMurray, C. T., Anderson, D. L. & Rossmann, M. G. (2003) Nat. Struct. Biol. 10**,** 572–576. [DOI] [PubMed] [Google Scholar]
- 51.Caspar, D. L. D. & Klug, A. (1962) Cold Spring Harbor Symp. Quant. Biol. 27**,** 1–24. [DOI] [PubMed] [Google Scholar]