The Cell Biology of the Endocytic System from an Evolutionary Perspective (original) (raw)
Abstract
Evolutionary cell biology can afford an interdisciplinary comparative view that gives insights into both the functioning of modern cells and the origins of cellular systems, including the endocytic organelles. Here, we explore several recent evolutionary cell biology studies, highlighting investigations into the origin and diversity of endocytic systems in eukaryotes. Beginning with a brief overview of the eukaryote tree of life, we show how understanding the endocytic machinery in a select, but diverse, array of organisms provides insights into endocytic system origins and predicts the likely configuration in the last eukaryotic common ancestor (LECA). Next, we consider three examples in which a comparative approach yielded insight into the function of modern cellular systems. First, using ESCRT-0 as an example, we show how comparative cell biology can discover both lineage-specific novelties (ESCRT-0) as well as previously ignored ancient proteins (Tom1), likely of both evolutionary and functional importance. Second, we highlight the power of comparative cell biology for discovery of previously ignored but potentially ancient complexes (AP5). Finally, using examples from ciliates and trypanosomes, we show that not all organisms possess canonical endocytic pathways, but instead likely evolved lineage-specific mechanisms. Drawing from these case studies, we conclude that a comparative approach is a powerful strategy for advancing knowledge about the general mechanisms and functions of endocytic systems.
Comparing the endocytic machinery of diverse organisms has provided insights into the origins and evolution of endocytic systems, as well as basic functions of modern cells.
The endomembrane system mediates transport of lipids, proteins, and other molecules to the various locations in the eukaryotic cell. It also underlies the interactions with the extracellular environment, presenting material at the cell surface as well as secreting and internalizing material. In modern cells, these latter aspects are important for signal transduction, surface remodeling, and nutrient acquisition. Just as these abilities are crucial to modern cells, they were likely equally important for the very first eukaryotes as they underwent speciation from prokaryotic-like ancestors via niche competition in the ancient world (Cavalier-Smith 2002). Understanding the events and biological processes involved in the evolution of the membrane-trafficking system in general, and the endocytic system in particular, gives us insights into landmark events in our cellular past.
Evolutionary insight about cellular phenomenon is derived from two basic types of comparative study: from molecular cell biological analyses of increasingly tractable model organisms across the diversity of eukaryotes, and by computational analyses of genomic information (i.e., the genes encoding the membrane-trafficking machinery). Whereas the information gathered from taking this comparative, or evolutionary cell biology, approach (Brodsky et al. 2012) is valuable for evolutionary content, these same analyses are potentially highly valuable in understanding basic cell biology, a benefit that is perhaps less obvious and hence less appreciated. In this article, we frame what has been learned about the evolution of the endocytic system, in the dual context of what it tells us about ancient cells together with what it can tell us about modern ones. We begin with a brief introduction to eukaryotic diversity and the evolution of the membrane-trafficking system. We then delve into the evolution of specific endocytic factors to illustrate the ways in which cell biologists of all stripes can benefit from the emerging field of evolutionary cell biology.
EUKARYOTIC DIVERSITY AND THE EVOLUTION OF THE EUKARYOTIC MEMBRANE-TRAFFICKING SYSTEM
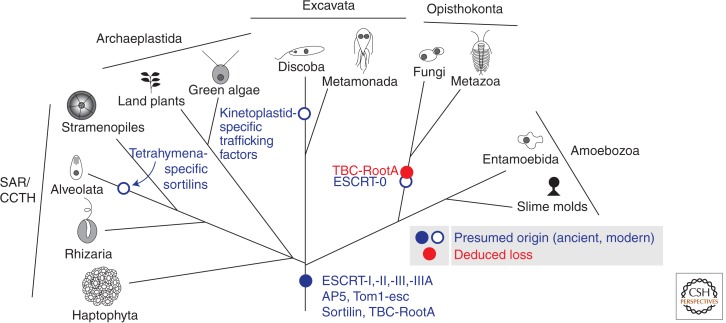
The possession of a true endocytic system is likely restricted to and ubiquitous in the eukaryote domain (Fig. 1). This domain can be divided into six taxonomic supergroups (Walker et al. 2011; Adl et al. 2012). The Opisthokonta comprise animals, fungi, and their unicellular relatives. The Amoebozoa include slime molds, for example, Dictyostelium discoideum, and many familiar amoebae, including the pathogen Acanthamoeba castellanii and the archetypal amoebozoan Amoeba proteus. The Amoebozoa and Opisthokonta are clearly sister to one another and likely form a megaclade, recently renamed Amorphea (Adl et al. 2012). The Archaeplastida include land plants and the red and green algae. The SAR supergroup is composed of Stramenopiles, Alveolates, and the understudied Rhizaria. Stramenopiles include brown algae (e.g., kelp), oomycetes (e.g., the blight causing the Irish potato famine, Phytophthora infestans), and diatoms. The Alveolates include ciliates (e.g., Paramecium and Tetrahymena), dinoflagellates (protists that cause red tides), and the parasitic apicomplexans (e.g., the malaria parasite, Plasmodium falciparum and Toxoplasma gondii). The Rhizaria include diverse organisms such as Radiolaria, Foraminifera, and Cercozoa. The CCTH supergroup includes cryptophytes, centrohelids, telonemids, and the oceanographically important haptophytes. The Archaeplastida and SAR supergroups were postulated to be sister to one another, and along with lineages within the CCTH supergroup, to form a megaclade termed Diaphoratickes (Adl et al. 2012), but new analyses have brought the unity of CCTH and the respective relationships of the component taxa to the rest of the Diaphoratickes into question (Burki et al. 2012). Finally, the supergroup Excavata lies between the megaclades Amorphea and Diaphoratickes (Hampl et al. 2009), although the monophyly of this supergroup is also currently debated (Zhao et al. 2012). This group includes several protist taxa including the free-living Naegleria gruberi as well as some familiar parasites including trypanosomes (the causative agents of African sleeping sickness and Chagas disease), Giardia intestinalis, and the sexually transmitted pathogen Trichomonas vaginalis. The relationship between these groups, the placement of cell biologically interesting orphan taxa (e.g., Collodictyon), and the placement of the eukaryotic root are all questions of intense interest and importance, because accurate systematics is a bedrock of biology.
Figure 1.
Endocytic proteins as distributed across the eukaryotic supergroups. Some factors, such as the ESCRT-I, -II, -III, and -III-associated proteins, are found in most or all the lineages and thus deduced as being present in the ancestor of eukaryotes (filled blue circle). Other factors, such as ESCRT-0 or the trypanosome-clathrin machinery, are found in a narrowly distributed range of lineages and are thus deduced as arising more recently (open circles). Because the exact relationship of the CCTH and SAR lineages is currently unresolved, these are grouped as a single supergroup. (Blue) Gains of proteins; (red) losses. Although AP5, Tom1-esc, and sortilin all have more complicated distributions, to illustrate ancient factors lost in opisthokonts, only the loss of TBC-rootA is shown. For the full details of the evolutionary history of these proteins, see Herman et al. (2011), Hirst et al. (2011), and Koumandou et al. (2011).
GENERALIZABILITY OF THE ENDOCYTIC SYSTEM
Regardless of the precise relationship of the eukaryotic supergroups, examination of the membrane-trafficking machinery of diverse organisms and comparisons with the better studied model systems of animals, fungi, and plants have provided insights relevant to both modern and deduced ancient cells. The basic principle is that the evolutionary pattern of conservation (Fig. 1) can indicate if a protein or protein complex is ancient (found across the diversity of eukaryotes) or derived more recently (found in only a restricted subset).
Such analyses indicate that most of the structural features and basic processes of the membrane-trafficking system are present in diverse eukaryotes (Roger 1999; Lee et al. 2000). This includes organelles such as the endoplasmic reticulum (ER), the Golgi, peroxisomes, lysosomes, early and late endosomes, and multivesicular bodies, as well as processes such as phagocytosis, endocytosis, and membrane recycling. Microscopic examination of some organisms, like Giardia, suggested that they lacked major aspects of the membrane trafficking, and particularly the Golgi complex, and, were at one point thought to be members of deeply diverging lineages retaining primitive cellular states (Cavalier-Smith 1987). However, the discovery of highly reduced Golgi-derived organelles and genes encoding Golgi-related proteins in these organisms (Mowbrey and Dacks 2009 and references therein) solidified the model that the Golgi complex and many other features of the endomembrane system are, indeed, ancient features and were present in the last eukaryotic common ancestor (LECA).
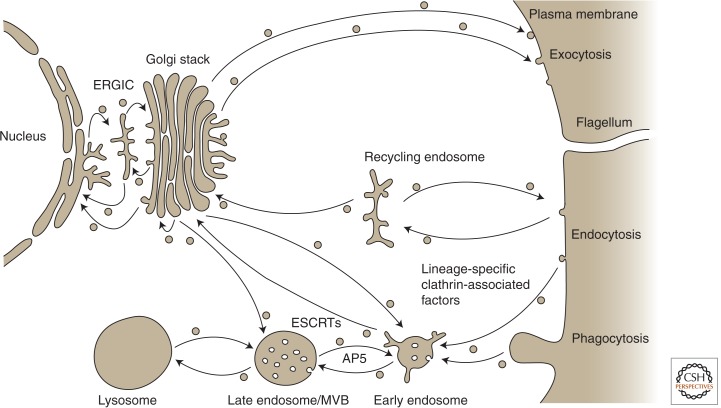
The endocytic system encompasses many aspects of the endomembrane system, including processes involving plasma membrane uptake, sorting of molecules at the endosomes, and either recycling back to the plasma membrane or entry into the degradative/lysosomal pathway (Fig. 2). Although similar processes are observed in diverse organisms, their homology at the molecular level is not always clear; that is, are these activities really using proteins retained from an ancient common set or is the apparent conservation actually an example of convergence?
Figure 2.
Endomembrane organelles and vesicle trafficking pathways. The general locations of endocytic factors discussed in detail (ESCRTs, the adaptin 5 complex, and clathrin complexes) are shown on a simplified cartoon of the endomembrane organelles in the eukaryotic cell.
Comparative genomics analysis suggests that much of the molecular machinery involved in conferring specificity and function to membrane trafficking is evolutionarily ancient. The known major protein families involved in endocytic pathways were all present in the LECA. Specifically, clathrin, epsinR, eps15R, AP180, AP1 to 5, Fab1, Vps34, Syntaxins, SNAREs, SM proteins, ArfGAPs, TBC Rab-GAPs, most components of the HOPS, GARP, ESCRT and retromer complexes, and several classes of GTPases, including Rabs, Arfs, Sar1, and Arls, are present in organisms across the eukaryote lineage (Koumandou et al. 2013, and references therein). From these studies, the LECA appears to have had an endocytic system of similar complexity to that of many extant taxa and that may have exceeded the complexity of some important modern organisms, including Saccharomyces cerevisiae and Trypanosoma brucei. Conversely, these studies also uncovered proteins that, although important to familiar model organisms, are lineage-specific novelties. For example, epsin, eps15, Dab2, GGA, the stonins, and caveolin (Field et al. 2007) all have distributions restricted to either the metazoa or opisthokonts.
This work also suggested some of the evolutionary mechanisms by which the endomembrane system grew in complexity between the first eukaryotic common ancestor to LECA. The organelle paralogy hypothesis (OPH) proposes that the increase in complexity was caused by iterative gene duplications, followed by sequence divergence and neofunctionalization in multiple interacting proteins encoding organelle identity and pathway specificity (Dacks and Field 2007; Dacks et al. 2008). The hypothesis is based largely on phylogenetic patterns of paralogous gene families of such organelle-specific trafficking factors (Rabs, Syntaxins, adaptins, Arf-GAPs). A key analysis (Dacks et al. 2008) showed that, whereas most of the gene duplications giving rise to these paralogs predate the LECA (Boehm and Bonifacino 2001; Pereira-Leal and Seabra 2001; Dacks and Doolittle 2002, 2004; Elias et al. 2012; Schlacht et al. 2013), several distinct lineage-specific expansions of endocytic proteins occurred after the primary eukaryote radiation into the major supergroups. For example, further specialization of early endosomal compartments is modulated by multiple isoforms of Rab5 in humans and trypanosomes. However, it was shown that the Rab5 isoforms in these diverse organisms are paralogs (homologous owing to lineage-specific gene duplications) and not orthologs (homologous owing to vertical inheritance), and the Rab5 expansions occurred independently in each lineage (Field et al. 1998; Dacks et al. 2008). A similar situation was found for the anterograde endocytic SNARE proteins Vam3/Pep12 vs Syntaxins 7 and 13, as well as the (β1 and β2) subunits of the adaptin complexes (Dacks et al. 2008). This implies parallel evolution of the endocytic machinery along a trajectory of expansion following the OPH model. Further validation of the OPH has come from in silico simulations that used the basic precepts of the model (gene duplication and organelle identity) as inputs. These simulations showed increased complexity, in the sense that additional distinct, but related, organelles emerged over time (Ramadas and Thattai 2013).
Additionally, the model also predicts that resolution between organelle-specific paralogs should reveal the order in which organelles evolved. Recent analyses of Rab GTPases have determined a likely set of Rabs present in the LECA to number more than 20 (Diekmann et al. 2011; Elias et al. 2012). Phylogenetic studies (Elias et al. 2012) have furthermore allowed for the identification of two well-supported clades of Rab GTPases: one corresponding to likely primordial endocytic Rabs (Rab5, 20, 21, 22, 24, and 50), the second to primordial exocytic Rabs (Rab1, 2, 4, 8, 11, 14, and 18). This is fully consistent with the OPH proposal that all endocytic Rabs originated from an ancestral endocytic Rab, whereas all exocytic Rabs originated from an ancestral exocytic Rab. It also sets up a model for the order of organelle/pathway origins against which to test resolved phylogenies of additional trafficking components.
EVOLUTIONARY CELL BIOLOGY AND ENDOCYTOSIS
Thus far, by examining the distribution of endomembrane components, we have illustrated that a common set of core membrane-trafficking machinery was likely present in the LECA. Next, we show that coupling the examination of these distributions with direct empirical analysis has yielded several unexpected findings. These include (1) ways in which our models of endocytosis actually might not fit with eukaryotes in general (i.e., the identification of lineage-specific adaptations in opisthokonts; (2) endocytic proteins in eukaryotes, including mammalian cells, identified by their ancient nature that have turned out to be more important than initially thought; and (3) identification of endocytic proteins unique to non-opisthokont lineages, representing possible novel adaptations in organisms of ecological or medical importance.
LINEAGE-SPECIFIC ADAPTATIONS IN OPISTHOKONTS
The model systems of animals and fungi have provided extensive and valuable information, but it is commonly assumed that this is applicable to eukaryotic cells in general. Although this is often true, there are many exceptions. A good example is found within a very well-known set of proteins, the endosomal complexes required for transport or ESCRTs (Conibear 2002; Henne et al. 2013).
Multivesicular bodies (MVBs) are late endosomal compartments that contain intralumenal vesicles (ILVs) made up of proteins and lipids often destined for degradation by lysosomes (Fig. 2). ILVs are produced by the invagination of endosomal membranes by the sequential action of the ESCRT system. According to information derived from mammalian cells and yeast (Henne et al. 2011), ubiquitylated proteins and PI3P in the endosomal membrane first bind proteins of the ESCRT-0 subcomplex, which directs them to late endosomes and initiates sorting into ILVs by recruiting the ESCRT-I subcomplex. Next, ESCRT-I works in conjunction with ESCRT-II subcomplex components to form a stable vesicle neck within the lumen of the endosome. ESCRT-III proteins bind ESCRT-II and are the best candidates for contributing to membrane pinching and, along with ESCRT-III-associated (e.g., Vps4) components, are responsible for ILV membrane scission. Several electron microscopy studies (Yang et al. 2004; Walker et al. 2006; Haas et al. 2007; Hurley 2008) have shown the existence of MVB-like organelles in diverse organisms, but until recently it was unknown if these organelles also used the same underlying molecular machinery.
Molecular evolutionary surveys of diverse eukaryotes for homologs of the various ESCRT components showed that ESCRT-I, -II, -III, and -III-associated are found in several organisms from each of the eukaryote supergroups (Field et al. 2007; Leung et al. 2008; Herman et al. 2011). The presence of the ESCRTs in all supergroups suggests that at least some features of ESCRT biology are therefore generalizable. To verify inferences drawn from their bioinformatic analyses, Leung et al. (2008) investigated the localization and endocytic involvement of Vps23 (Tsg101 in mammals) and Vps28, both ESCRT-I components, in the divergent excavate T. brucei. ESCRT-I localization (Fig. 3) and function is similar in T. brucei, and these proteins locate to late endosomal compartments, as has previously been reported in opisthokont model systems. The presence of MVBs in trypanosomes has been challenged recently, as has the role of Vps23 in turnover of ubiquitylated proteins (Silverman et al. 2013); this underscores the need for detailed analysis in divergent organisms because even when mapping apparently conserved functions, subtle differences in experimental approach or interpretation can lead to differing conclusions; in trypanosomes, MVB-like structures are rare, albeit with sufficient examples to indicate that such a compartment is most likely present. Additional studies in plants (Scheuring et al. 2012), Dictyostelium (Blanc et al. 2009), and Plasmodium (Yang et al. 2004) have confirmed the involvement of ESCRT proteins in the function of the MVB in diverse eukaryotes.
Figure 3.
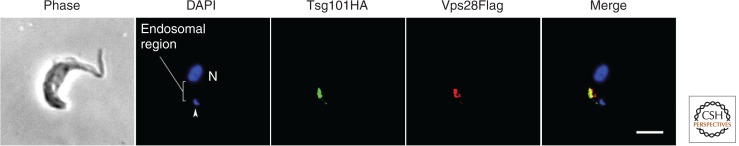
Localization of components of the ESCRT machinery in trypanosomes. Two components of the ESCRT-I system were localized using ectopic expression of epitope-tagged versions of the relevant proteins. Tsg101/Vps23 (green) and Vps28 (red) are shown, together with the location of the nucleus (N) and kinetoplast (arrowhead), visualized with DAPI (blue). The region of the cell that contains essentially all of the endocytic apparatus is highlighted in the DAPI panel, and is clearly where both Tsg101 and Vps28 are localized. The absence of complete concordance between the two ESCRT factors suggests possible discrete targeting/functional differentiation of late endosome/MVB membranes in trypanosomes. Note that in trypanosomes there is a potential second Tsg101 paralog, which has not so far been investigated. Scale bar, 2 μm. (For more details, see Leung et al. 2008 and Silverman et al. 2013.)
However, although the basic involvement of ESCRT machinery in MVB function is generalizable, the complete failure to identify components of ESCRT-0 from all organisms sampled outside the Opisthokonta (Leung et al. 2008; Herman et al. 2011) suggested that another mechanism must exist in eukaryotes for binding ubiquitylated cargo and recruiting ESCRT-I to endosomes. The Tom1 (Target of Myb protein 1) family of proteins comprises candidates that may perform this function. For example, a study using D. discoideum implicated DdTom1 as a potential component of an alternative ESCRT-0 complex (Blanc et al. 2009). This result is consistent with reports in metazoa showing that Tom1 family member Tom1L1 is recruited to MVBs and interacts with the ESCRT component Tsg101 (Puertollano 2005). By a comparative genomic and phylogenetic analysis, it was shown that, whereas ESCRT-0 is lacking from all eukaryote lineages outside Opisthokonta, organisms from across eukaryotic diversity were found to encode at least one Tom1 family homolog (Herman et al. 2011). These findings indicate that Tom1 is an ancient protein, whereas the ESCRT-0 components are not, implying that a Tom1-based mode of ESCRT function may be the more general eukaryotic mechanism. Of course, additional functional analyses in diverse model organisms will be required to confirm and further characterize the role of Tom1 family proteins in sorting ubiquitylated cargo to MVBs.
ANCIENT AND UNDERAPPRECIATED PROTEINS
The principle behind the prediction that Tom1 might be playing a role in ESCRT function is based on the premise that, if a protein has been retained for more than ∼1.5 billion years of evolution spanning eukaryotic history (Yoon et al. 2004), it is likely functionally important. Nowhere is this principle better illustrated than in the recent discovery of the fifth adaptin complex.
The adaptin (AP) complexes comprise a group of evolutionarily related heterotetramers that are involved in various stages of membrane trafficking (for review, see Robinson 2004). All AP complexes consist of two large (γ/δ/α/ε) subunits plus a medium (µ) and a small (σ) subunit. It had been held, until recently, that there were only four such AP complexes, each with a unique cellular localization and function. The AP components are derived from a set of gene duplications that gave rise to the subunits specific to each complex and furthermore link the adaptins to the COPI complex of the early secretory system (Schledzewski et al. 1999). Importantly, with the exception of the β1 and β2 subunits discussed above, these gene duplications, by comparative genomics and phylogenetic evidence, occurred before the LECA (Boehm and Bonifacino 2001; Field et al. 2007; Dacks et al. 2008). This implies that the adaptins are both ancient and conserved features of the eukaryotic endocytic system. In addition to the de facto APµ subunits, there are several proteins in the human genome that contain µ-homology domains (Boehm and Bonifacino 2001). Until recently, these proteins were poorly explored and thought to possibly represent adaptations arising out of the increased complexity of metazoans, despite their identification in Arabidopsis thaliana, which alternatively suggested a more general role (Boehm and Bonifacino 2001).
The presence of one of these µ-homology proteins (MUDENG) in the genome of the free-living excavate Naegleria gruberi (Fritz-Laylin et al. 2010) implied an ancient and potentially important functional role and prompted more intense study. A yeast two-hybrid screen identified an uncharacterized human protein, DKFZp761E198, with homology and predicted secondary structural similarity to the adaptin β subunits, as one of the potential MUDENG interactors (Hirst et al. 2011). In a separate high-throughput screen of proteins associated with the human disease hereditary spastic paraplegia (Słabicki et al. 2010), DKFZp761E198 interacted with four additional human proteins: SPG11, SPG15, C20orf29, and KIAA0415. The latter two showed distant homology and predicted structural similarity to the γ/δ/α/ε proteins, respectively (Hirst et al. 2011). Tagged versions of the candidate σ5 were shown to have a similar localization as the µ5 and β5 subunits, as did SPG11 and SPG15 (Hirst et al. 2011, 2013). RNAi knockdowns of any of the four AP5 subunits or SPG11 and SPG15 resulted in similar phenotypes (Hirst et al. 2011, 2013). Recently, it has been shown that all six proteins can be coimmunoprecipitated in a complex with 1:1:1:1:1:1 stoichiometry (Hirst et al. 2013). Furthermore, SPG11 and SPG15 possess α-solenoid domains, with a β-propellor domain preceding the α-solenoid, similar to the clathrin and COPI/COPII membrane-deformation subcomplexes (Hirst et al. 2013). Together these data are strongly suggestive that AP5 is a heterotetrameric adaptor complex, similar to the other APs, with SPG11 and 15 forming a membrane-deformation coat. Colocalization of tagged versions of the AP5 proteins with LAMP1 (Hirst et al. 2011, 2013) suggests that AP5 may participate in the formation or regulation of late endosomes, a novel pathway for AP complex participation (Fig. 2).
The initial discovery of the protein µ5 in N. gruberi, humans, and plants suggested that the AP5 complex is much more widespread than originally imagined, but how widespread? Comparative genomics and confirmation by phylogenetics (Hirst et al. 2011) allowed for the identification of AP5 components plus SPG11 and SPG15 in at least two organisms in every eukaryote supergroup including D. discoideum, N. gruberi, A. thaliana, and T. gondii, suggesting that AP5 was likely present in the LECA. At the same time, absence of AP5 and SPG11/SPG15 from many diverse taxa suggests that, although the AP5 complex is ancient, it has been lost independently numerous times.
In this case, a broad distribution in eukaryotes highlighted a protein complex with a function in animal cells, prompting experiments that give a first glimpse into the cell biology underlying at least two human diseases (Hirst et al. 2012). However, there might be proteins absent in animals but present in the remaining diversity of eukaryotes. These could be ancient factors inherited from the eukaryotic ancestor but lost in our particular lineage. Such factors not only allow us to glimpse the complexity of our cellular past and how it has evolved, but also they provide opportunities to better understand and potentially manipulate the biology of parasites, crops, or ecologically important organisms. Although not necessarily endocytic proteins, examples of membrane-trafficking proteins with precisely this distribution (Fig. 1) have recently been reported. Analysis of both Rab proteins and their TBC-GAPs identified several uncharacterized Rab and Rab-GAP homologs that are ancient but without homologs in humans (Elias et al. 2012; Gabernet-Castello et al. 2013). Finally, an analysis of the ArfGAP protein family (Schlacht et al. 2013) expanded the number of subfamilies from 10 to 11, with the new subfamily (ArfGAPC2) being absent from humans but present in diverse eukaryotic lineages, including plants and important agricultural pathogens like oomycetes.
LINEAGE-SPECIFIC ENDOCYTIC FACTORS IN NON-OPISTHOKONTS
One of the limitations to a comparative understanding of endocytosis is experimental sampling bias, and this bias is rather pronounced. The most important biological model systems are mammalian cells and S. cerevisiae, with minor parts being played, up until now, by a few other organisms. The dominance of molecular cell biology by opisthokont taxa has the consequence of asymmetry, such that it is comparatively easy to find novel opisthokont components, for example, ESCRT-0, but substantially more demanding to uncover unique factors or systems in the remaining supergroups, or even to pin down the precise mechanisms by which homologous components might operate. This requires careful molecular cell biology without the benefits of easily available resources such as antibodies or genetics. Nonetheless, this difficult work is not only possible, but can yield exciting insights.
Some “unique” endocytic factors may actually represent novel paralogs of protein families. These paralogs can be identified by using similar methods to those that identified the ancient proteins described above (like ArfGAPC2), which are absent from opisthokonts. However, without molecular cell biological characterization, an exact functional context is lacking. For example, sortilins are, as their name implies, proteins with an intimate involvement in protein sorting. They are widely conserved as a class (Koumandou et al. 2011), but there is significant variation in their domain organization across the eukaryotes, and they participate in many trafficking steps. One class is involved in regulated secretion in mammalian cells and has been implicated in diabetes (Bogan 2012). In Tetrahymena thermophila, sortilins also contribute to the regulated exocytosis performed by mucocysts, specialized exocytic structures (Briguglio et al. 2013). However, it is significant that in both Homo sapiens and T. thermophila the sortilins involved in regulated secretion are not direct orthologs, but distinct paralogs, conceptually similar to the lineage-specific Rab5 expansions discussed above. This, then, is a fine example of coalescing function, albeit not a full convergent evolution. Interestingly, similar findings have also been reported for the role of dynamin in endocytosis in the ciliate model and supported by homology searching (Elde et al. 2005; Field et al. 2007). However, in the absence of well-sampled and resolved phylogenies of the greater dynamin family, as well as a highly detailed and orthogonal dissection of the endocytic pathways of organisms where dynamin is suspected to not play a role, for example, in trypanosomes (Morgan et al. 2004), the evolution of the dynamin family and its association with endocytosis remains an exciting area to be better explored.
Other endocytic factors might truly lack any form of homologs in opisthokonts, or may lie beyond detection by current bioinformatic methods. Such a crucial example comes from T. brucei. Here there is a well-characterized, single mode of clathrin-dependent endocytosis (Field and Carrington 2009), as well as clear divergence from most organisms in that the African trypanosome lacks AP2 (Manna et al. 2013), a major clathrin-interaction hub that organizes multiple protein complexes and binds to cargo. In the absence of AP2 it is questionable if the mechanisms that subtend clathrin endocytosis are conserved, and direct investigation of this process by proteomics of clathrin-interacting proteins reveals a substantial cohort of trypanosome-specific proteins (Adung’a et al. 2013). These trypanosome-specific factors likely operate within the system together with clearly conserved components.
What both of these studies highlight, by rather distinct approaches, is that what superficially may appear to be an example of conservation of function, that is, sortilin-mediated regulated secretion or clathrin-mediated endocytosis, may, in fact, be a rather distinct process when considered from an evolutionary context and at the level of molecular mechanism. This may indicate some constraints on how these various trafficking steps can operate, as well as suggest that aspects of cellular physiology may be even more divergent between supergroups than we have assumed or imagined.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
We began this article with the question of whether endocytic processes are truly homologous or are, instead, an example of convergence. As our examples attest, the answer is that both scenarios are in operation. A major portion of the endocytic machinery is common to eukaryotes, passed down from the LECA more than a billion years ago. However, there are subtleties and innovations that have occurred in diverse lineages, including the ones on which we often base our understanding of cell biology.
Each species has a unique evolutionary history and has thus become uniquely adapted to its particular ecological niche. Thus, generalizations between even relatively closely related organisms should be taken with caution. To fully understand the diversity of life, we must develop a cohort of more diverse model organisms. The establishment of an organism as a model requires the coalescence of many things, and which can become self-reinforcing as resources are accumulated, a sort of natural selection in itself. However, we should be open to the possibility that not all of the models have been optimally chosen in terms of the biological questions that can be addressed, but additional considerations, such as disease impact, are major contributors behind their development. It is also possible that certain organisms that have served extremely well can effectively become exhausted, and that the uncovering of novel biology requires a move into additional systems to reinvigorate discovery. Of course, it is impractical to study every cellular phenomenon in every organism, but our current popular models only span, at most, one-sixth of the full diversity of eukaryotes (see Fig. 1). Fortunately, some less-familiar organisms (e.g., the ciliate T. thermophila, the apicomplexan T. gondii, the slime mold D. discoideum, the kinetoplastid T. brucei, and the plant A. thaliana) have developed or are currently developing into experimentally tractable model organisms, from which informative data can be collected to complement the inferences derived from comparative genomics. These two methodologies can reinforce and iteratively promote one another as we explore the novel and conserved cell biology of the endocytic system.
Broadening the scope of analysis to models outside the standard animal/fungal set and taking an evolutionary perspective has been valuable for the understanding of endocytosis. This same principle, of finding homologies and making comparisons, can also be applied to other organelles, with interesting work now showing functional insight between endolysosomes and the secretory granules of ecologically important organisms like ciliates (Elde et al. 2005) and dinoflagellates (Gubbels and Duraisingh 2012) and medically important organisms like the malaria parasite Plasmodium spp. or the related parasite T. gondii (Gubbels and Duraisingh 2012; Klinger et al. 2013). Finally, the clear structural relationships that have emerged for the paralogous Rab and SNARE families, and which provided some of the impetus for the OPH, have clearly been of great utility. However, even more fundamental connections can be drawn in this manner and were encapsulated by the protocoatomer hypothesis, which was originally inspired by the recognition of similar folds between clathrin and nuclear pore proteins (Devos et al. 2004). This view has been extended, and confirmed by many subsequent studies, and provided a mechanism that unites membrane deformation activities in vesicle coats, the flagellum, and the nuclear envelope. Furthermore, the proto-coatomer insight suggests that although paralogy was easy to recognize for Rabs and SNAREs, deep evolutionary relationships may require more structure-based methodology; the implication is that additional connections between systems, as now being shown for the tethers and vesicle coats (Dokudovskaya et al. 2009), probably await to be uncovered.
The evolutionary cell biology approach is not just applicable to endocytosis or membrane trafficking but a powerful strategy right across cell biology. Although this article has given some examples of the types of discoveries made in endocytic machinery, as the conceptual shift of adding comparative context to elegant in-depth work in tractable model organisms is taken on in diverse areas, the benefits to understanding both evolutionary and functional cell biology are very exciting to anticipate.
ACKNOWLEDGMENTS
We thank A. Turkewitz for useful discussions and for sharing data before publication. Thanks also to L. Barlow for assistance with Figure 2. This work is supported by a Natural Sciences and Engineering Research Council discovery grant to J.B.D. and by grants 090007/Z/09/Z and MR/K008749/1 from the Wellcome Trust and the Medical Research Council, respectively, to M.C.F. J.B.D. is the Canada Research Chair in Evolutionary Cell Biology.
Footnotes
Editors: Sandra L. Schmid, Alexander Sorkin, and Marino Zerial
REFERENCES
*Reference is also in this collection.
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol 59: 429–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adung’a VO, Gadelha C, Field MC 2013. Proteomic analysis of clathrin interactions in trypanosomes reveals dynamic evolution of endocytosis. Traffic 14: 440–457 [DOI] [PubMed] [Google Scholar]
- Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F 2009. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic 10: 161–171 [DOI] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS 2001. Adaptins: The final recount. Mol Biol Cell 12: 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan JS 2012. Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem 81: 507–532 [DOI] [PubMed] [Google Scholar]
- Briguglio JS, Kumar S, Turkewitz AP 2013. Lysosomal sorting receptors are essential for secretory granule biogenesis in Tetrahymena. J Cell Biol 203: 537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM, Thattai M, Mayor S 2012. Evolutionary cell biology: Lessons from diversity. Nat Cell Biol 14: 651. [DOI] [PubMed] [Google Scholar]
- Burki F, Okamoto N, Pombert JF, Keeling PJ 2012. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc Biol Sci 279: 2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1987. Eukaryotes with no mitochondria. Nature 326: 332–333 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52: 297–354 [DOI] [PubMed] [Google Scholar]
- Conibear E 2002. An ESCRT into the endosome. Mol Cell 10: 215–216 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2002. Novel syntaxin gene sequences from Giardia, Trypanosoma and algae: Implications for the ancient evolution of the eukaryotic endomembrane system. J Cell Sci 115: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Doolittle WF 2004. Molecular and phylogenetic characterization of syntaxin genes from parasitic protozoa. Mol Biochem Parasitol 136: 123–136 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Field MC 2007. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J Cell Sci 120: 2977–2985 [DOI] [PubMed] [Google Scholar]
- Dacks JB, Poon PP, Field MC 2008. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci 105: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann Y, Seixas E, Gouw M, Tavares-Cadete F, Seabra MC, Pereira-Leal JB 2011. Thousands of rab GTPases for the cell biologist. PLoS Comput Biol 7: e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S, Waharte F, Schlessinger A, Pieper U, Devos DP, Cristea IM, Williams R, Salamero J, Chait BT, Sali A, et al. 2009. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics 10: M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP 2005. Elucidation of clathrin-mediated endocytosis in Tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet 1: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M, Brighouse A, Gabernet-Castello C, Field MC, Dacks JB 2012. Sculpting the endomembrane system in deep time: High resolution phylogenetics of Rab GTPases. J Cell Sci 125: 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Carrington M 2009. The trypanosome flagellar pocket. Nat Rev Microbiol 7: 775–786 [DOI] [PubMed] [Google Scholar]
- Field H, Farjah M, Pal A, Gull K, Field MC 1998. Complexity of trypanosomatid endocytosis pathways revealed by Rab4 and Rab5 isoforms in Trypanosoma brucei. J Biol Chem 273: 32102–32110 [DOI] [PubMed] [Google Scholar]
- Field MC, Gabernet-Castello C, Dacks JB 2007. Reconstructing the evolution of the endocytic system: Insights from genomics and molecular cell biology. Adv Exp Med Biol 607: 84–96 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140: 631–642 [DOI] [PubMed] [Google Scholar]
- Gabernet-Castello C, O’Reilly AJ, Dacks JB, Field MC 2013. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol Biol Cell 24: 1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Duraisingh MT 2012. Evolution of apicomplexan secretory organelles. Int J Parasitol 42: 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martinez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS 2007. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19: 1295–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JF, Lang BF, Simpson AG, Roger AJ 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic supergroups. Proc Natl Acad Sci 106: 3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD 2011. The ESCRT pathway. Dev Cell 21: 77–91 [DOI] [PubMed] [Google Scholar]
- *.Henne WM, Stenmark H, Emr SD 2013. Molecular mechanisms of the membrane scultpting ESCRT pathway. Cold Spring Harb Perspect Biol 5: a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EK, Walker G, van der Giezen M, Dacks JB 2011. Multivesicular bodies in the enigmatic flagellate Breviata anathema and the evolution of ESCRT-0. J Cell Sci 124: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MN, Dacks JB, Robinson MS 2011. The fifth adaptor protein complex. PLoS Biol 9: e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Irving C, Borner GH 2012. Adaptor protein complexes AP4 and AP5: New players in endosomal trafficking and progressive spastic paraplegia. Traffic 14: 153–164 [DOI] [PubMed] [Google Scholar]
- Hirst J, Borner GH, Edgar J, Hein MY, Mann M, Buchholz F, Antrobus R, Robinson MS 2013. Interaction between AP5 and the hereditary spastic paraplegia proteins SPG11 and SPG15. Mol Biol Cell 24: 2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH 2008. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 20: 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger CM, Nisbet RE, Ouologuem DT, Roos DS, Dacks JB 2013. Cryptic organelle homology in apicomplexan parasites: Insights from evolutionary cell biology. Curr Opin Microbiol 16: 424–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Klute MJ, Herman EK, Nunez-Miguel R, Dacks JB, Field MC 2011. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J Cell Sci 124: 1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol 48: 373–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Leedale GF, Bradbury P, eds. 2000. Illustrated guide to the protozoa, 2nd ed Society of Protozoologists (International Society of Protistologists), Allen Press, Lawrence, KS [Google Scholar]
- Leung KF, Dacks JB, Field MC 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9: 1698–1716 [DOI] [PubMed] [Google Scholar]
- Manna PT, Kelly S, Field MC 2013. Adaptin evolution in kinetoplastids and emergence of the variant surface glycoprotein coat in African trypanosomatids. Mol Phylogenet Evol 67: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GW, Goulding D, Field MC 2004. The single dynamin-like protein of Trypanosoma brucei regulates mitochondrial division and is not required for endocytosis. J Biol Chem 279: 10692–10701 [DOI] [PubMed] [Google Scholar]
- Mowbrey K, Dacks JB 2009. Evolution and diversity of the Golgi body. FEBS Lett 583: 3738–3745 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC 2001. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313: 889–901 [DOI] [PubMed] [Google Scholar]
- Puertollano R 2005. Interactions of TOM1L1 with the multivesicular body sorting machinery. J Biol Chem 280: 9258–9264 [DOI] [PubMed] [Google Scholar]
- Ramadas R, Thattai M 2013. New organelles by gene duplication in a biophysical model of eukaryote endomembrane evolution. Biophys J 104: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol 14: 167–174 [DOI] [PubMed] [Google Scholar]
- Roger AJ 1999. Reconstructing early events in eukaryotic evolution. Am Nat 154: S146–S163 [DOI] [PubMed] [Google Scholar]
- Scheuring D, Künzl F, Viotti C, Yan MS, Jiang L, Schellmann S, Robinson DG, Pimpl P 2012. Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol 12: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacht A, Mowbrey K, Elias M, Kahn RA, Dacks JB 2013. Ancient complexity, opisthokont plasticity, and discovery of the 11th subfamily of Arf GAP proteins. Traffic 14: 636–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledzewski K, Brinkmann H, Mendel RR 1999. Phylogenetic analysis of components of the eukaryotic vesicle transport system reveals a common origin of adaptor protein complexes 1, 2, and 3 and the F subcomplex of the coatomer COPI. J Mol Evol 48: 770–778 [DOI] [PubMed] [Google Scholar]
- Silverman JS, Muratore KA, Bangs JD 2013. Characterization of the late endosomal ESCRT machinery in Trypanosoma brucei. Traffic 14: 1078–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słabicki M, Theis M, Krastev DB, Samsonov S, Mundwiller E, Junqueira M, Paszkowski-Rogacz M, Teyra J, Heninger AK, Poser I, et al. 2010. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol 8: e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Dacks JB, Martin Embley T 2006. Ultrastructural description of Breviata anathema, n. gen., n. sp., the organism previously studied as Mastigamoeba invertens. J Eukaryot Microbiol 53: 65–78 [DOI] [PubMed] [Google Scholar]
- Walker G, Dorrell RG, Schlacht A, Dacks JB 2011. Eukaryotic systematics: A user’s guide for cell biologists and parasitologists. Parasitology 138: 1–26 [DOI] [PubMed] [Google Scholar]
- Yang M, Coppens I, Wormsley S, Baevova P, Hoppe HC, Joiner KA 2004. The Plasmodium falciparum Vps4 homolog mediates multivesicular body formation. J Cell Sci 117: 3831–3838 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattcharya D 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21: 809–818 [DOI] [PubMed] [Google Scholar]
- Zhao S, Burki F, Bráte J, Keeling PJ, Klaveness D, Shalchian-Tabrizi K 2012. Collodictyon—An ancient lineage in the tree of eukaryotes. Mol Biol Evol 29: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]