Bacterial flagellin is a dominant antigen in Crohn disease (original) (raw)
Abstract
Chronic intestinal inflammation, as seen in inflammatory bowel disease (IBD), results from an aberrant and poorly understood mucosal immune response to the microbiota of the gastrointestinal tract in genetically susceptible individuals. Here we used serological expression cloning to identify commensal bacterial proteins that could contribute to the pathogenesis of IBD. The dominant antigens identified were flagellins, molecules known to activate innate immunity via Toll-like receptor 5 (TLR5), and critical targets of the acquired immune system in host defense. Multiple strains of colitic mice had elevated serum anti-flagellin IgG2a responses and Th1 T cell responses to flagellin. In addition, flagellin-specific CD4+ T cells induced severe colitis when adoptively transferred into naive SCID mice. Serum IgG to these flagellins, but not to the dissimilar Salmonella muenchen flagellin, was elevated in patients with Crohn disease, but not in patients with ulcerative colitis or in controls. These results identify flagellins as a class of immunodominant antigens that stimulate pathogenic intestinal immune reactions in genetically diverse hosts and suggest new avenues for the diagnosis and antigen-directed therapy of patients with IBD.
Introduction
Crohn disease (CD) and ulcerative colitis (UC), collectively referred to as inflammatory bowel disease (IBD), are relatively common inflammatory diseases of the gastrointestinal (GI) tract. Histopathologically and anatomically, these two conditions are distinct, with CD characterized by transmural inflammation that can occur throughout the GI tract, and UC characterized by more superficial inflammation confined to the colon and rectum. Interestingly, both diseases are dependent upon factors present within the complex intestinal microbiota. Indeed, a unifying hypothesis has emerged that proposes that IBD results from a dysregulated mucosal immune response to the intestinal microbiota in genetically susceptible individuals (reviewed in refs. 1, 2).
While the dependence of IBD on intestinal microbes is increasingly clear, the molecular mechanisms underlying this dependence are not. The intestinal mucosa is exposed to the largest concentration of foreign bacterial antigens of any tissue in the body, estimated to be up to 1012 organisms per gram of stool in the normal colon. An emerging concept is that there is an active “dialogue” between the microbiota, intestinal epithelial cells, and mucosal immune cells, with each partner communicating with the others (3). In this context, “innate” immune responses, which recognize conserved microbial products such as lipopolysaccharide (LPS) and peptidoglycan (PG), are likely to be important in these microbial-host interactions and intestinal homeostasis. Critical to the host’s “sensing” of microbes are members of the Toll-like receptor (TLR) family that, alone or in combination, recognize a wide array of microbe-associated molecular patterns on either pathogens or commensals (reviewed in refs. 4–6). Various TLRs are expressed on intestinal epithelial cells (7–10) and more broadly on macrophages and dendritic cells in the lamina propria. In addition, the identification of Nod2, an intracellular protein that recognizes muramyl dipeptide, as a susceptibility gene for CD highlights the potential role of pattern-recognition receptors and their ligands in diseases such as IBD (11).
Given the involvement of innate immune mechanisms in the modulation of T cell responses, the bacterial dependence of IBD is likely to involve both bacterial products such as LPS, PG, and other TLR ligands, and specific bacterial antigens capable of stimulating CD4+ T cell responses. CD4+ T lymphocytes have been identified as the crucial effector cells in experimental models of IBD (12–14), and these pathogenic CD4+ T cell responses are directed against the enteric microbiota. Enteric bacterial antigen–reactive CD4+ T cells are able to induce colitis when adoptively transferred into immunodeficient recipients (14). The in vitro data to date suggest that there is a relatively small number of immunodominant antigens that stimulate the pathogenic T cell responses (15), but the complexity of the intestinal microflora has posed a significant challenge to their identification. One notable successful example is the identification of “I2” by Braun and colleagues (16). This antigen, derived from a Pseudomonas species present within the intestinal microflora, was discovered using a molecular technique, representational difference analysis (RDA), to identify DNA sequences present in intestinal tissue from IBD patients but not in normal control (NC) tissue (17). These data highlight the utility of using unbiased molecular approaches to address this challenging problem.
Accordingly, we used a molecular technique known as serological expression cloning (SEC) to identify specific bacterial antigens driving experimental IBD. SEC involves the screening of DNA expression libraries in lambda phage with defined antisera. In clinical or experimental systems of infectious diseases, in which entire microbial genomes can be screened with defined reactive sera, SEC has proven to be extremely useful in the identification of antigens and vaccine targets (18, 19).
In this manuscript, we describe the molecular cloning of antigens by SEC using sera from colitic C3H/HeJBir mice. Interestingly, the dominant antigens identified were a family of related, novel flagellins. Strong reactivity against specific flagellins was seen in multiple models of experimental colitis across several distinct MHC haplotypes. There was a clear IgG2a predominance to the anti-flagellin response, suggesting a concurrent Th1-biased T cell response against flagellin. Indeed, marked reactivity against flagellin was seen in mesenteric and splenic T cell cultures from colitic animals, and flagellin-specific T cells were able to induce colitis when adoptively transferred into immunodeficient animals. Surprisingly, the reactivity against these flagellins (but not against the dissimilar Salmonella flagellin) was also seen in human IBD sera, with significant reactivity in patients with CD but not UC or control patients. Given the recognition that flagellin is the ligand for TLR5 (20), these data not only provide insight into antigens that may be involved in the pathogenesis of IBD but also highlight a link between IBD and innate immunity.
Results
Seroreactivity in mice is directed mainly against a specific group of flagellins.
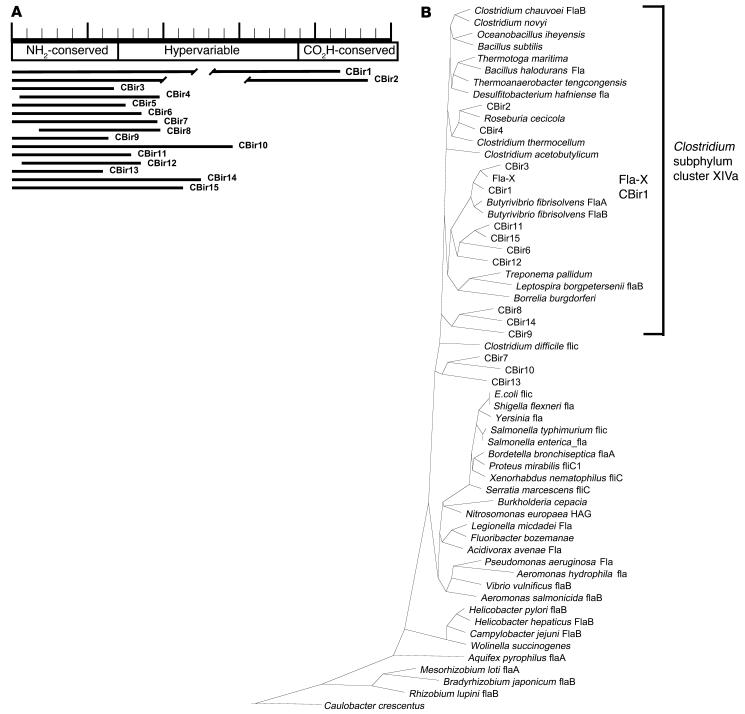
Serologic expression cloning resulted in 55 clones that were sequenced and identified. Using the basic local alignment search tool to search the GenBank databases demonstrated that 15 (26.8%) of these clones were flagellin-like sequences. None of the sequences directly matched any flagellin in the GenBank database, and all flagellin sequences identified represented unique clones. Given the average insert size of 0.8 kb in the library, no full-length flagellin clones were identified. However, all of the flagellin clones contained sequences derived from the conserved NH2 terminus, with varying amounts of the hypervariable central domain, and only two clones contained partial sequence from the conserved carboxy domain. Sequences from the 15 flagellin clones identified (CBir1–CBir15) were aligned at the protein level to flagellin sequences available in the public domain using the Clustal program in DNAStar. As shown in Figure 1B, these flagellins are most closely related to flagellins from Butyrivibrio, Roseburia, Thermotoga, and Clostridium species and appear to align, by similarity, in the Clostridium subphylum cluster XIVa of Gram-positive bacteria. Sequences from the remaining 40 clones (see Table 1) were also unique and were either related to known proteins (33 clones) or without significant homology to known proteins (7 clones).
Figure 1.
Flagellin clone identity and similarity to known flagellin sequences. (A) Schematic of CBir flagellin clones from serological expression screening. The predicted amino acid sequences from the flagellin expression clones (CBir1_CBir15) are mapped in relation to the representation of the B. fibrisolvens sequence at the top. Ruler length equals 500 amino acids. Similarity in the NH2 conserved sequence between these flagellin clones and B. fibrisolvens sequences ranged from 45 to 84% (mean, 60.3%). Breaks in the lines representing clones CBir1 and CBir2 indicate differences in sequence length in the hypervariable region. NH2-conserved, conserved NH2 sequence; CO2H-conserved, conserved carboxy sequence. (B) Phylogenetic tree showing relatedness at the conserved NH2 termini of CBir1_CBir15 clones to flagellin sequences in the GenBank database. The dendrogram was constructed using the Clustal program in DNAStar and reflects similarity at the amino acid level. The approximate location of the Clostridium subphylum cluster XIVa is indicated with a bracket.
Table 1.
Identity of serological expression clones
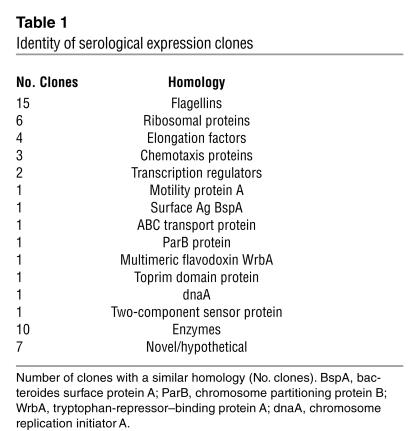
Because of strong serum antibody reactivity to one particular flagellin clone, called CBir1, we cloned and expressed its full-length gene. During this effort, we also cloned a second, highly homologous and reactive flagellin (83.5% similarity to CBir1 at the NH2 conserved domain) and refer to it here as Fla-X (Figure 2B). Recombinant proteins representing full-length sequence and NH2 and carboxy fragments of both CBir1 and Fla-X were subsequently expressed in E. coli with a six-histidine tag to aid in protein purification (Figure 2, C and D, respectively).
Figure 2.
Schematic of recombinant flagellin constructs with percent similarity to related flagellin B from the anaerobe B. fibrisolvens (GenBank accession number AAB82613). (A) Structure of B. fibrisolvens flagellin B showing conserved NH2 and carboxy (CO2H-conserved) regions and the hypervariable central domain. (B) Diagram of the full-length amino acid sequence of mouse cecal bacteria flagellins CBir1 and Fla-X, indicating the similarity of the three domains with the respective B. fibrisolvens domains. (C and D) Schematics of recombinant flagellin proteins and fragments for CBir1 (C) and Fla-X (D) expressed in E. coli and purified by six-histidine tag affinity to nickel-nitrilotriacetic acid columns.
Antibody reactivity to flagellin directed against the NH2 terminus is of the IgG2a subclass and correlates with disease.
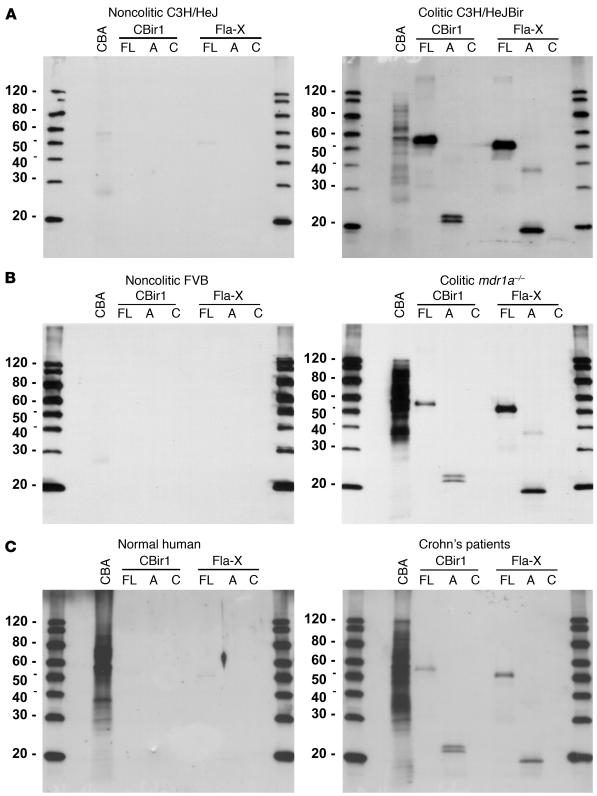
Western blot analyses using these purified recombinant flagellins and fragments with sera from the diseased C3H/HeJBir mice demonstrated the strong reactivity to flagellin, predominantly to the NH2-terminal fragments (Figure 3A). Little or no antibody reactivity was seen to the carboxy-terminal CBir1 or Fla-X recombinant fragments in the sera tested (Figure 3). This selective reactivity to the NH2 domain is consistent with the presence of an NH2 domain in all flagellin clones identified in the initial serological screen (Figure 1). In addition, strong reactivity to both flagellins was seen using sera from two additional experimental models of colitis: mdr1a–/– mice (Figure 3B) and B6.IL-10–/– mice (data not shown). These last two models are on strains with different haplotypes from each other (H-2s and H-2b, respectively) and from the C3H/HeJBir strain (H-2k), and the sera were obtained from mice from geographically different mouse facilities. In addition, these additional colitic strains have very different mechanisms underlying the genetic predisposition to develop IBD; that is, epithelial barrier dysfunction in the _mdr1a_–/– mice and a defect in regulatory T cells in the B6.IL-10–/– mice. Little or no reactivity was seen to CBir1 or Fla-X in noncolitic mouse serum from control MHC haplotype–matched noncolitic mice (Figure 3, A and B). Interestingly, we also saw a similar pattern of reactivity to the NH2 termini of both CBir1 and Fla-X with a serum pool from patients with CD (Figure 3C).
Figure 3.
Western blot analysis of the serum antibody response to recombinant flagellins CBir1 and Fla-X and their fragments. (A) Noncolitic C3H/HeJ (pool of two) versus colitic C3H/HeJBir (pool of five) mice. (B) Noncolitic FVB (pool of five) versus colitic mdr1a–/– (pool of five) mice. (C) Random human blood donor (Normal human) versus a pool of CD patients with severe disease. Protein samples include mouse CBA, full-length recombinant proteins (FL), the NH2 conserved region (A) and the conserved carboxy region (C) of flagellin (see Figure 2, C and D).
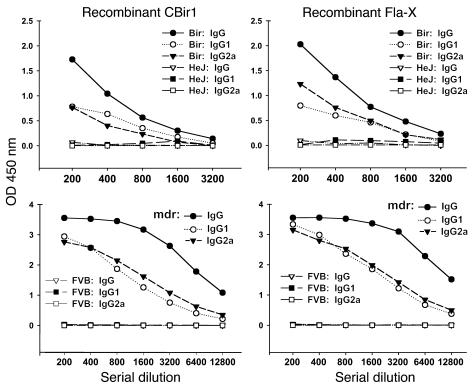
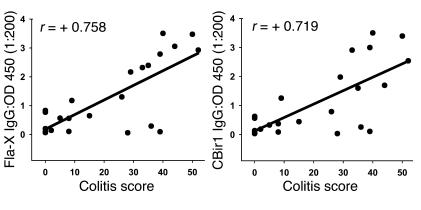
In order to generate more quantitative data with multiple IBD models at various time points in the course of disease, we developed an antibody subclass ELISA against full-length or fragments of CBir1 or Fla-X (Figure 2, C and D). With this assay, we confirmed Western blot data showing that the antibody reactivity observed was predominantly to the NH2 terminus (data not shown) and of the IgG2a subclass. High titers of anti-flagellin antibody were seen in the four genetically distinct models of IBD tested (colitic mice: C3H/HeJBir [Figure 4], mdr1a–/– [Figure 4], BALB/c.IL-10–/– [not shown] and B6.IL-10–/– [not shown]), while minimal to no reactivity was seen in serum from the H-2–matched, control, noncolitic mouse strains. Given the nonuniform incidence of colitis in the mdr1a–/– colony at varying time points, we randomly chose 23 animals in the colony and assigned quantitative histopathological scores using a scale (from 0 to 60) that incorporates both the degree and extent of inflammation in the large intestine (21). We measured antibodies against Fla-X and CBir1 by ELISA in a “blinded” manner using sera from these animals and found that an increased titer of anti-flagellin IgG correlated positively with worsening IBD histopathology (r = +0.758 and +0.719, respectively; Figure 5). Weak correlations were found between antibody and mouse age (r = +0.325; data not shown) and between colitis score and mouse age (r = +0.372; data not shown).
Figure 4.
ELISA titration of mouse serum anti-flagellin against recombinant flagellins CBir1 and Fla-X with secondary antibodies specific for mouse IgG, IgG1, and IgG2a antibodies. Colitic C3H/HeJBir serum (pool of five) versus noncolitic C3H/HeJ serum (pool of two) was used in the upper panel and colitic mdr1a–/– serum (pool of five) versus noncolitic FVB serum (pool of five) was used in the lower panel.
Figure 5.
Correlation of colitis histopathology score (0_60) with serum anti_Fla-X and anti-CBir1. Twenty-three mdr1a–/– mice, ranging in age from 6 to 13 weeks, were randomly chosen for assignment of quantitative histopathology scores. Serum anti-flagellin from these mice was quantified by ELISA. Colitis scores of 0_2 represent no disease; 3_15, mild disease, 16_35, moderate disease, and more than 35, severe disease (22). Similar results were obtained for both recombinant flagellins: Fla-X (left panel) and CBir1 (right panel).
Anti-CBir1 reactivity in CD patient sera but not normal or UC patient sera.
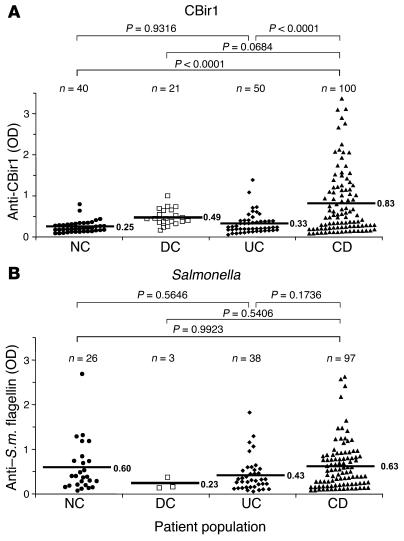
Subsequently, we tested a large panel of sera from controls and patients with IBD for reactivity against CBir1 and Fla-X using antigen-specific ELISAs. We found a significantly higher level of serum anti–CBir1/Fla-X flagellin in CD patients than in NCs, disease controls (DCs), and UC patients (Figure 6A). It should be noted that more than 50% of the UC sera were from patients with a modified Truelove and Witts severity index greater than 7, indicating moderate to active disease. The observation of serum responses to flagellins CBir1 and Fla-X in a group of CD patients, but not UC patients (Figure 6A), highlights the possibility that anti-flagellin responses may be valuable in the diagnosis of IBD, in particular with regards to the more precise discrimination between UC and CD, and the definition of CD patient subsets.
Figure 6.
Association of anti-flagellin antibodies with human IBDs. Human sera, well characterized for CD and UC, were tested by ELISA for reactivity to flagellin CBir1 (A) and Salmonella muenchen (S.m.) flagellin (B). Statistical analysis was performed with the Tukey-Kramer test; the resulting statistics (P values) as well as population size (n) are shown above the graphs. Mean OD450 values are indicated by horizontal bars.
Reactivity to the Salmonella muenchen flagellin (which is highly similar to the flagellin of Eschericia coli [84–91% at the conserved NH2 end]), however, showed no significant correlation to disease (Figure 6B). As shown in Figure 6B, the mean values for the anti-Salmonella response were nearly identical in the control, CD, and UC populations. In all populations, there appeared to be a minority of samples without significant reactivity and a majority of samples that were “positive.” While these data may reflect the random exposure to Salmonella in humans due to prior infection (possibly subclinical) or a cross-reactivity to an undefined but closely related flagellin (probably from the Enterobacteriaceae family), it is clear that the serological response to the Salmonella flagellin molecule does not correlate with IBD. Similarly, there was no correlation between reactivity to Salmonella flagellin and colitis in the C3H/HeJBir or _mdr1a_–/– strains compared with the MHC haplotype–matched controls (data not shown).
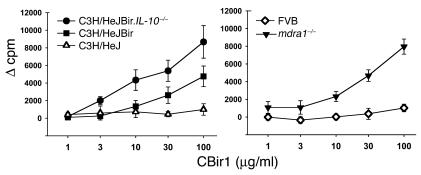
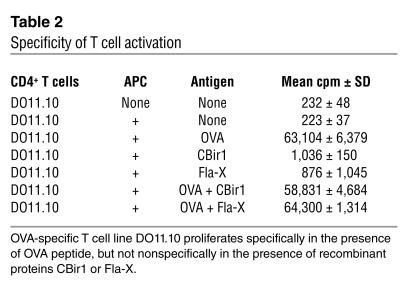
Marked reactivity against flagellin is seen at the T cell level, and flagellin-specific T cells are able to induce colitis when adoptively transferred. Given the strong IgG2a bias seen in the antibody response in the mouse IBD strains, we hypothesized that flagellin-specific Th1 T cells would be present in mice with IBD. To address this possibility, we purified CD4+ T cells from pooled spleens and mesenteric lymph nodes from colitic mdr1a–/–, C3H/HeJBir, and C3H/HeJ.IL-10–/– mice (and haplotype-matched, noncolitic FVB and C3H/HeJ mice) and tested the cells for reactivity against purified CBir1 and Fla-X in vitro in the presence of antigen-presenting cells (APCs). CD4+ T cells from the colitic mdr1a–/–, C3H/HeJBir, and C3H/HeJ.IL-10–/– mice, but not from age-matched control FVB or C3H/HeJ mice raised in the same mouse facility, responded to CBir1, as assessed by proliferation (Figure 7). It was possible that the responses seen were due to the fact that the flagellin molecule was nonspecifically activating the cultured T cells via TLR5 or TLR4 activation (through endotoxin contamination of the recombinant protein). This possibility was excluded by the lack of stimulation in both the noncolitic T cell cultures (Figure 7) and in an independent T cell culture system that showed no influence of Fla-X or CBir1 on the ovalbumin-specific proliferation of CD4+ T cells from DO11.10 ovalbumin-specific T cell receptor–transgenic animals (Table 2).
Figure 7.
Dose response of CD4+ T cell proliferation to CBir1 and Fla-X in multiple strains of mice. Left panel: C3H/HeJ (open triangles), C3H/HeJBir (squares), and C3H/HeJBir.IL-10–/– (circles). Right panel: FVB (diamonds) and mdr1a–/– (filled triangles). The y axes indicate sample counts per minute (cpm) minus control T cell plus APC cpm (Ø cpm) for each experimental group. The x axes indicate the dose (∝g/ml) of recombinant flagellin used in each assay. Vertical bars indicate plus or minus one standard deviation of the mean value.
Table 2.
Specificity of T cell activation
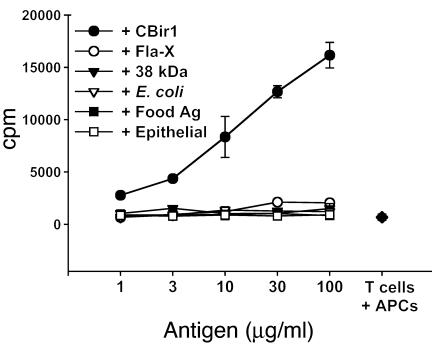
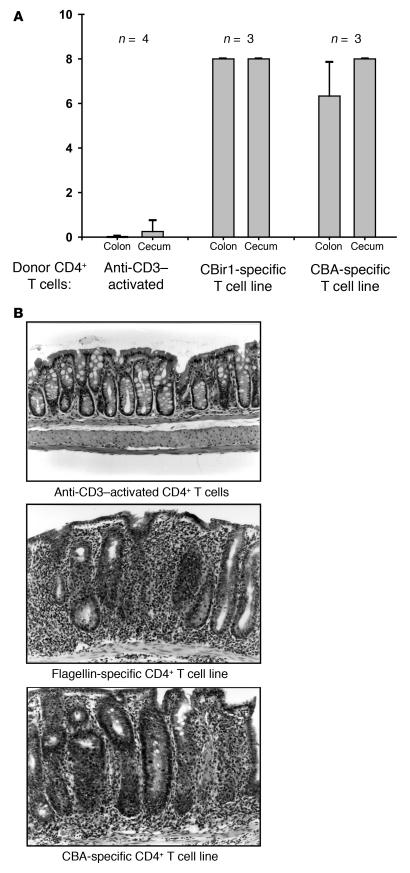
We have previously shown that a T cell line specific for cecal bacterial protein/antigen (CBA), but not CD4+ T cells polyclonally activated by anti-CD3, could induce mucosal inflammation when adoptively transferred into H-2–matched immunodeficient scid/scid mice (14). To address the potentially pathogenic role of flagellin-specific T cells in the initiation of mucosal inflammation, we generated a CD4+ T cell line reactive with CBir1 flagellin from C3H/HeJBir mice by repeated stimulation with antigen and APCs. This CD4+ T cell line strongly responded to CBir1 but not to Fla-X or a variety of other microbial, food, and epithelial antigens (Figure 8). These CBir1-specific CD4+ T cells were adoptively transferred into C3H/HeJ-scid/scid recipients. Control SCID mice received anti-CD3–activated CD4+ T cells as a negative control or a CD4+ T cell line reactive to CBA as a positive control. Quantitative histopathological scores were assigned at 8 weeks after transfer (Figure 9A). The CBir1-specific CD4+ T cell line induced colitis in all recipients of an intensity that was similar to or greater than that induced by the CBA-specific CD4+ T cell line, whereas none of the recipients given anti-CD3–activated C3H/HeJBir CD4+ T cells developed disease (representative histology is shown in Figure 9B).
Figure 8.
Dose response and specificity of C3H/HeJBir CD4+ CBir1-specific T cell line. T cell line CBir-1B1 proliferated specifically in response to recombinant flagellin protein CBir1. Antigens used in the assay include recombinant flagellins CBir1 (filled circles) and Fla-X (open circles); the 38-kDa antigen of M. tuberculosis (p38 antigen; 38 kDa: filled triangles); lysate of E. coli antigens (E. coli; open triangles); protein antigens extracted from mouse food pellets (Food Ag; filled squares); and a lysate of the ModeK epithelial cell line, of C3H origin (epithelial: open squares). Several randomly expressed recombinant commensal bacterial antigens were also tested and were negative (including randomly cloned C3H/HeJ mouse cecal bacterial antigens 99 [rIB99] and 32 [rIB32]). T cells plus APCs only are indicated by a filled diamond.
Figure 9.
Adoptive transfer of C3H/HeJBir CD4+ CBir1-specific T cell line into C3H/HeJ scid/scid recipients. (A) Two months after transfer, cecal and colon histopathology was assigned scores with a quantitative system (14). CD4+ T cells activated polyclonally with mAb against CD3 prior to transfer were used as a negative control (Anti-CD3_activated). A CBA-specific CD4+ T cell line reactive with unselected cecal bacterial antigens was used as a positive control (CBA-specific T cell line); the CBir1-specific CD4+ T cell line corresponds to the flagellin-specific T cell line in Figure 8. Sample size (n) is indicated at the top. (B) Representative histopathology of the groups shown in A: Anti-CD3_activated CD4+ T cells (top panel), CBir1 flagellin_specific CD4+ T cells (middle panel), and CBA-specific CD4+ T cells (bottom panel). Magnification, ∞200.
Discussion
Data from both humans and experimental animals underscore the critical role of intestinal bacteria in the establishment and maintenance of IBD (2). The molecular mechanisms underlying this clear bacterial dependence on mucosal inflammation are likely to relate to the intrinsic ability of these organisms to trigger both acquired and innate immune responses in the gut. Using an unbiased molecular screen to search for bacterial antigens relevant to IBD, the dominant antigens identified were a family of related, novel flagellins. A strong, IgG2a-biased serological response to these specific flagellins was seen in multiple models of experimental colitis across several distinct MHC haplotypes. In addition, marked reactivity against these flagellins was seen at the T cell level, and flagellin-specific T cells were able to induce colitis when adoptively transferred into immunodeficient animals. Interestingly, while these flagellins were identified from mouse cecal bacteria, there was clear, specific reactivity against these molecules in patients with CD (but not in patients with UC or in NCs). Given the activity of flagellin as a specific ligand for TLR5, these data provide a potentially important link between adaptive and innate immune responses in mucosa in the pathogenesis of IBD.
Among bacterial antigens, flagellin is an interesting candidate to play a role in mucosal immune responses. Specifically, flagellin is a common bacterial antigen present on most motile bacteria in the gut (22). Moreover, flagellin is highly antigenic; indeed, responses against flagellin are protective in Salmonella infections in mice (23, 24). Furthermore, flagellin has been identified as a pathogen-associated molecular pattern and has been identified as the specific ligand for TLR5. Monomeric, translocated flagellin is capable of stimulating cytokine and chemokine production by TLR5-positive intestinal epithelial cells (8), and flagellin has been demonstrated to induce the maturation of dendritic cells via TLR5 (25, 26). Indeed, we have observed that full-length flagellin Fla-X (endotoxin free) is capable of stimulating TNF-α production by human macrophages in vitro (M.J. Lodes and R.M. Hershberg, data not shown). It is tempting to speculate that the intrinsic “adjuvanticity” of flagellin is likely to contribute to its antigenicity. While flagellin molecules clearly have the capacity to stimulate the production of proinflammatory cytokines via TLR5, we favor the hypothesis that the B and T cell responses to flagellin contribute more directly to the chronic intestinal inflammation seen in IBD.
Our clinical data (Figure 6) are consistent with the fact that the aberrant response in patients with CD is specific to the subgroup of flagellins identified in our molecular screen. Specifically, there was no correlation between IBD and a response to flagellin from Salmonella muenchen, which is very similar (84–91%) in the NH2 conserved region to the flagellin from the commensal organism Escherichia coli. It must be emphasized that the flagellins identified were from a source of material devoid of known bacterial pathogens. The bacteria with genes that “encode” the flagellins CBir1 and Fla-X (the two dominant flagellins tested) are unknown; however, preliminary phylogenetic data suggest that these flagellins are most closely related to the flagellins of bacteria in the genera Butyrivibrio, Rosburia, Thermotoga, and Clostridium and fall within the Clostridium subphylum XIVa cluster of Gram-positive bacteria (Figure 1B). This group of bacteria contains many of the butyrate-producing bacteria that are found in the human gut (27, 28). We speculate that the aberrant response to the flagellin molecule(s) from these organisms is related to a combination of the intrinsic property of the molecules themselves (as immunogens and adjuvants) and an underlying genetic susceptibility. The question of the basis for selective responses to an antigen present in the normal commensal flora of most (if not all) individuals is a challenging one. Using monoclonal antibodies directed against CBir1, we have demonstrated that this antigen is present in the stool of wild-type strains (FVB, C57BL/6, BALB/c, and C3H/HeJ) and colitic strains (_mdr1a_–/–, B6.IL-10–/–, and C3H/HeJBir) (data not shown). These data indicate that the presence of the antigen itself does not strictly correlate with colitis. Still, the widespread presence of these antigens does not preclude the possibility of enhanced colonization of organisms expressing these flagellins in CD lesions. This idea is certainly consistent with the recent observations by Swidsinski and colleagues (29). Alternatively, alterations in barrier function (30, 31) or intrinsic innate immune responses (32, 33) may enhance the likelihood of the response to these flagellin molecules in patients prone to develop IBD.
In general, the data are consistent with the hypothesis that IBD is associated with a defect in tolerance to commensal organisms (34). The IgG2a-biased antibody against Fla-X and CBir1 highlights the Th1 bias of the T cell responses seen. The broad recognition of these flagellins in several different mouse models and in humans with CD indicates that these flagellins are among the immunodominant antigens of the microbiota. However, the exact role of these flagellins in the pathogenesis of IBD (e.g., whether they are predominant or obligatory for disease) compared with that of other microbial antigens remains to be defined. We favor a model in which a T regulatory response to specific flagellins (and/or other bacterial antigens) may be selectively impaired in IBD. In this context, specific flagellin molecules may represent novel targets for antigen-directed therapy in IBD. Experiments are currently underway to address this possibility.
Clearly, flagellin is not the only bacterial antigen involved in the pathogenesis of IBD. It is likely that there will be specific subsets of patients with IBD (both CD and UC) that display patterns of seroreactivity against the molecular determinants that we have identified, similar to the patterns currently seen with responses to oligomannans from Saccharomyces cerevisiae, I2, and ompC for E. coli (35). Indeed, the identification of flagellin complements the efforts Braun and co-workers in discovering bacterial antigens relevant to the pathogenesis of IBD. One interesting example is I2, an antigen derived from Pseudomonas fluorescens that appears to have “superantigen-like” activity (16, 17). As we observed with the specific flagellins identified here, only a subset of patients with CD show specific seroreactivity against I2. It should be noted that an unbiased molecular approach, RDA, was also used to identify I2. In contrast to the technique of SEC, which depends upon the selective antibody responses to a given antigen, RDA depends upon the selective presence of bacteria in anatomic sections of IBD. Given the heterogenous nature of IBD and the complexity of the intestinal microflora, we are continuing our efforts to identify additional antigens in patients that do not respond to flagellin. It will be of considerable interest to determine how the reactivity against these antigens correlates with genetic determinants, clinical phenotypes, and responses to treatment.
Methods
Isolation of genomic DNA of mouse cecal bacterium.
Pelleted bacteria from C3H/HeJBir mouse ceca were inactivated at 80°C for 20 minutes and then were treated with 2 ml lysozyme (20 mg/ml in Tris-EDTA [TE] buffer) for 1 hour at 37°C. This solution was rocked at room temperature for 10 minutes with 40 ∝l proteinase K (10 mg/ml) and 140 ∝l 20% SDS (Sigma-Aldrich, St. Louis, Missouri, USA) and then incubated for 15 minutes at 65°C, then 0.4 ml of 5 M NaCl and 0.32 ml of a 10% cetyltrimethylammonium bromide (CTAB) solution (1 g CTAB [Sigma-Aldrich], 1.4 ml 5M NaCl, and 8.6 ml distilled H2O) was added, followed by incubation at 65°C for 10 minutes. DNA was then extracted twice with phenol, followed by extraction with phenol/chloroform/isoamyl alcohol (24:24:2), and then with chloroform. Finally the DNA was precipitated with 0.6 volumes of isopropanol and resuspended in TE buffer.
Genomic expression library construction.
A detailed description of library construction can be found elsewhere (19). Briefly, 20 ∝g of genomic DNA of mouse cecal bacterium was resuspended in 400 ∝l of TE buffer and was sonicated for five seconds at 30% continuous power with a Sonic Dismembrator (Fisher Scientific, Pittsburgh, Pennsylvania, USA) to generate fragments of approximately 0.5–5.0 kb. DNA fragments were blunted with T4 DNA polymerase (Invitrogen, Carlsbad, California, USA) and were ligated to _Eco_RI adaptors (Stratagene, La Jolla, California, USA) with T4 DNA ligase (Stratagene). Adapted inserts were then phosphorylated with T4 polynucleotide kinase (Stratagene) and were selected by size with a Sephacryl 400-HR column (Sigma-Aldrich). Approximately 0.25 ∝g of insert was ligated to 1.0 ∝g Lambda ZAP Express Vector treated with _Eco_RI and calf intestinal alkaline phosphatase (Stratagene), and the ligation mix was packaged with Gigapack III Gold packaging extract (Stratagene) following the manufacturer’s instructions.
Expression screening.
Immunoreactive proteins were screened from approximately 6 ∞ 105 plaque-forming units (PFU) of the unamplified cecal bacterium expression lambda library. Briefly, twenty 150-mm petri dishes were plated with E. coli XL1-Blue MRF′ host cells (Stratagene) and approximately 3 ∞ 104 PFU of the unamplified library and were incubated at 42°C until plaques formed. Dry nitrocellulose filters (Schleicher and Schuell, Keene, New Hampshire, USA), pre-wet with 10 mM isopropyl β-thiogalactopyranoside (IPTG), were placed on the plates, which were then incubated overnight at 37°C. Filters were removed and washed three times with PBS containing 0.1% Tween 20 (PBST) (Sigma-Aldrich), blocked with 1.0% BSA (Sigma-Aldrich) in PBST, and washed three times with PBST. Filters were next incubated overnight with E. coli lysate–adsorbed C3H/HeJ Bir mouse serum (1:200 dilution in PBST), washed three times with PBST, and incubated with a goat anti-mouse IgG + IgA + IgM (heavy and light chain) alkaline phosphatase–conjugated secondary antibody (diluted 1:10,000 with PBST; Jackson Laboratories, West Grove, Pennsylvania, USA) for 1 hour. Filters were finally washed three times with PBST and two times with alkaline phosphatase buffer (pH 9.5) and were developed with nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolylphosphate _p_-toluidine salt (Invitrogen). Reactive plaques were then isolated and a second or third plaque purification was performed. Excision of phagemid followed the Stratagene Lambda ZAP Express protocol, and the resulting plasmid DNA was sequenced with an automated sequencer (ABI, Foster City, California, USA) using M13 forward, reverse, and sequence-specific internal DNA sequencing primers. Nucleic acid and predicted protein sequences were used to search the GenBank nucleotide and translated databases. Protein analysis was performed with the PSORT program (National Institute for Basic Biology, Okazaki, Japan) and with the IDENTIFY program of EMOTIF (Department of Biochemistry, Stanford University). Sequence alignments were produced with the MegAlign program (Clustal) of DNAStar (Madison, Wisconsin, USA). Note that 20 random clones from the lambda library were picked and sequenced prior to serolological expression cloning. None of the 20 were found to be derived from mouse DNA and no flagellins were identified.
Cloning of full-length flagellins representing clones CBir1 and Fla-X.
Clone CBir1 contains the conserved NH2 and variable regions of an unknown immunoreactive flagellin. The full-length sequence was obtained by first amplifying the unknown CBir1 carboxy terminus from total genomic cecal bacterium DNA with Expand polymerase (Roche, Indianapolis, Indiana, USA) and the primers CBir1var1 (designed from the variable region of CBir1; CACAATCACAACATCTACCCAG) and CBir1 Carb Z (designed from the carboxy terminus of the related flagellin B of Butyrivibrio fibrisolvens, GenBank accession number AF026812; 5′-TTACTGTAAGAGCTGAAGTACACCCTG-3′). This PCR product was cloned with a Zero Blunt TOPO PCR Cloning Kit (Invitrogen), digested with _Eco_RI, and gel-isolated (carboxy end of CBir1). Clone CBir1 plasmid DNA, which represents the NH2 terminus plus flagellin central variable region and overlaps with the cloned carboxy region, was digested with _Sca_I and then gel-isolated. Both overlapping (181-bp) DNA fragments (approximately 20 ng each) were added to a PCR reaction with the primers CBir1 HIS and CBir1 TERMX (see below), and the amplification product was cloned and expressed as described below.
Fla-X is an immunoreactive full-length flagellin sequence with no known identity in the public databases. Full-length flagellin Fla-X was cloned from total cecal bacterium genomic DNA by PCR amplification with the primers CBir Fla-X HIS (5′-CAATTACATATGCATCACCATCACCATCACGTAGTACAGCACAATC-3′) and CBir1 TERMX (5′-ATAGACTAAGCTTACTGTAAGAGCTGAAGTACACCCTG-3′), and was expressed as described below. The amplification product was cloned with a Zero Blunt TOPO PCR Cloning Kit (Invitrogen), and several clones were sequenced.
Recombinant protein.
Recombinant Salmonella muenchen flagellin (≥95% pure by SDS-PAGE) was obtained from Calbiochem (La Jolla, California, USA). Expression of other recombinant flagellin proteins and deletion constructs was accomplished by amplification from the cloned plasmid or genomic DNA (full length Fla-X) with Pfu polymerase (Stratagene) and the following primer pairs: for full-length CBir1, CBir1 HIS (5′-CAATTACATATG_CATCACCATCACCATCAC_GTAGTACAGCACAATTTACAGGC-3′) and CBir1 TERMX (5′-ATAGACTAAGCTTACTGTAAGAGCTGAAGTACACCCTG-3′); for the CBir1 NH2 plus variable regions, CBir1 HIS and CBir1 AV TERM (5′-ATAGACTAAGCTTAAGAAACCTTCTTGATAGCGCCAG-3′); for the CBir1 NH2 terminus, CBir1 HIS and CBir1 A TERM (5′-TAGACTGAATTCTAGTCCATAGCGTCAACGTTCTTTGTGTC-3′); for the CBir1 carboxy terminus, CBir1 C HIS (5′-CAATTACATATG_CATCACCATCACCATCAC_AAGATGAACTTCCATGTAGGTGC-3′) and CBir1 TERMX; for full-length Fla-X, CBir Fla-X HIS (5′-CAATTACATATG_CATCACCATCACCATCAC_GTAGTACAGCACAATC-3′) and CBir1 TERMX (ATAGACTAAGCTTACTGTAAGAGCTGAAGTACACCCTG-3′); for the Fla-X NH2 plus variable regions, Fla-X HIS (5′-CAATTACATATG_CATCACCATCACCATCAC_GTAGTACAGCACAATCTTAGAGC-3′) and Fla-X AV TERM (5′-ATAGACTAAGCTTAGAGGCTGAAATCAATGTCCTCG-3′); for the Fla-X NH2 terminus, Fla-X HIS and Fla-X A TERM (5′-ATAGACTAAGCTTAATGTGCTGAAAGATATCTTGTCAC-3′); and for the Fla-X carboxy terminus, Fla-X C HIS (5′-CAATTACATATG_CATCACCATCACCATCAC_TTCAGCCTCCATGTAGGTGCAGATGC-3′) and CBir1 TERMX. Primers contained restriction sites for cloning (in bold) and a six-histidine tag (in italics) for protein purification (NH2 terminus). The amplification products were digested with the restriction enzymes _Nde_I and _Hin_dIII or _Eco_RI, depending on the primer set used, gel-isolated, and ligated to a pET 17b plasmid vector (Novagen, Madison, Wisconsin, USA) previously cut with _Nde_I and with _Hin_dIII or _Eco_RI and dephosphorylated with alkaline phosphatase (MB grade; Roche). The ligation mix was transformed into XL1 Blue competent cells (Stratagene) and plasmid DNA was prepared for sequencing (Qiagen, Valencia, California, USA). Recombinant protein was expressed by transformation of plasmid DNA into BL21 pLysS competent cells (Novagen) and induction of a single-colony cell culture with 2 mM IPTG (Sigma-Aldrich). Recombinant protein was recovered from cell lysate with nickel-nitrilotriacetic acid agarose beads (Qiagen), following the manufacturer’s instructions, and was dialyzed in 10 mM Tris, pH 4–11 depending on predicted recombinant pI characteristics. Recombinant proteins were “quality-checked” for purity by SDS-PAGE followed by staining with Coomassie blue and by NH2-terminal protein sequencing, and were quantified with a Micro BCA assay (Pierce, Rockford, Illinois, USA). Recombinants were assayed for endotoxin contamination with the Limulus assay (Bio Whittaker, Walkersville, Maryland, USA). Production of the Mycobacterium tuberculosis 38-kDa protein has been described previously (36).
ELISA.
Ninety-six–well EIA/RIA microtiter plates (3369; Corning Costar, Cambridge, Massachusetts, USA) were coated overnight at 4°C with 100 ng/well of the recombinant proteins. Solutions were then aspirated from the plates, which were then blocked for 2 hours at room temperature with PBS containing 1% (weight/volume) BSA. This was followed by washing in PBST. Serum diluted in PBS containing 0.1% BSA was added to wells and incubated for 30 minutes at room temperature, followed by washing six times with PBST and then incubation with secondary antibody–HRP conjugate (1:10,000 dilution) for 30 minutes. Plates were then washed six times in PBST and then were incubated with tetramethylbenzidine (TMB) substrate (Kirkegaard and Perry, Gaithersburg, Maryland, USA) for 15 minutes. The reaction was stopped by the addition of 1 N sulfuric acid, and plates were “read” at 450 nm using an ELISA plate reader (Biotek instrument EL311, Hyland Park VA). Background values were determined by reading of reactions that lacked the primary antibody step.
Western blot analysis.
Recombinant antigens (50–200 ng/lane) were subjected to SDS-PAGE analysis using 15% polyacrylamide “minigels.” The antigens were transferred to nitrocellulose BA-85 (Schleicher & Schuell, Keene, New Hampshire, USA) and were blocked for 1 hour at room temperature with PBS containing 1% Tween 20. Blots were then washed three times, 10 minutes each wash, in PBST. Next, blots were probed for 1 hour at room temperature with serum diluted 1:500 in PBST followed by washing three times, 10 minutes each wash, in PBST. Blots were then incubated for 30 minutes at room temperature with secondary antibody–HRP diluted 1:10,000 in wash buffer and were again washed three times for 10 minutes each wash in PBST containing 0.5 M sodium chloride. Finally, blots were incubated in chemiluminescent substrate for ECL (Amersham Plc, Little Charlton, UK) for about 1 minute and then were exposed to X-ray film (XAR5) for 10–60 seconds, as required.
CD4+ T cell isolation and culture, and generation of a Cbir1-specific T cell line.
CD4+ T cells were isolated from mesenteric lymph nodes (MLNs) of mice with BD IMAG anti-mouse CD4 beads according to the manufacturer’s instructions (BD Biosciences Pharmingen, San Diego, California, USA). Briefly, MLN cells were labeled with anti-CD4 beads and then were placed within the magnetic field of the BD Imagnet. The unlabeled cells in suspension were removed and the cells binding to beads were washed and used in the CD4+ T cell culture. More than 99% of cells were CD4+, as shown by flow cytometry (data not shown). For the generation of a T cell line reactive to CBir1, CD4+ T cells were isolated from MLNs of C3H/HeJBir mice as described above and were cultured with splenic APCs that were pulsed with CBir1 (100 mg/ml) overnight. The cells were restimulated every 10–14 days.
Antigen-specific proliferation of T cells.
Spleen and MLN CD4+ T cells, isolated as described above, or a CBir1 flagellin–specific T cell line (4 ∞ 105 cells/well) were incubated in triplicate in the presence of antigen-pulsed, irradiated APCs (4 ∞ 105 cells per well; treated with 1–100 ∝g/ml antigen for 18 hours at 37°C) in 96-well flat-bottomed tissue culture plates (Falcon, Lincoln Park, New Jersey, USA) at 37°C in 5% CO2 humidified air. [3H]thymidine (0.5 ∝Ci) (New England Nuclear, Boston, Massachusetts, USA) was added at day 3 of culture and the cells were harvested at 16 hours after the pulse. The cells were harvested on glass fiber filters on a PHD cell harvester (Cambridge Technology Inc., Watertown, Massachusetts, USA), washed with distilled water, and dried. Proliferation was assessed as the amount of incorporation of [3H]thymidine into cell DNA, as measured by beta scintillation counting (Beckman Instruments, Palo Alto, California, USA) of the harvested samples, and was expressed as cpm ± SD. The preparation of epithelial cell proteins and food antigens has been described previously (14). Ethical approval for animal studies was obtained from the Institutional Animal Care and Use Committee at the University of Alabama (Birmingham, Alabama) and from Corixa Corporation.
Specificity of CD4+ T cell stimulation.
APCs were BALB/c spleen cells that were pulsed for 24 hours with nothing, OVA peptide at 2 ∝g/ml, CBir1 at 100 ∝g/ml, or Fla-X at 100 ∝g/ml, alone or in combinations as shown in Table 2. These APCs were washed and irradiated with 3,000 rads prior to culture. CD4+ T cells were isolated from DO11.10 mice and were cultured at a density of 1 ∞ 105 with 4 ∞ 105 prepulsed APCs. [3H]TdR was added at day 3 of culture and the cells were harvested after 16 hours.
Adoptive transfer.
CD4+ T cells were cultured with cecal bacterial antigen–pulsed and irradiated C3H/HeJ splenic cells in complete medium at 37°C for 4 days in 5% CO2 air before being transferred intravenously into C3H/HeSnJ scid/scid recipients. Three months later, the recipients were killed and then the cecum and the proximal, medial, and distal portions of the colon were fixed in formalin. Fixed tissues were embedded in paraffin, and sections were stained with hematoxylin and eosin for histological examination. All slides were “read” by an experienced pathologist (A. Lazenby, Department of Pathology, University of Alabama at Birmingham) without knowledge of their origin.
Human subjects.
Serum samples from 212 subjects (50 UC patients, 100 CD patients, 22 DCs, and 40 NCs) were obtained from the serum archive of the Cedars-Sinai IBD Research Center. Sera were produced from standard phlebotomy blood specimens and were given an “anonymous” number code, divided into aliquots, and stored at –80°C until use. The UC and CD patient specimens were obtained from a genetic case-control study (37, 38). Each patient’s diagnosis was confirmed by clinical history, endoscopic and radiologic examination, and histopathology findings. The NC group is a collection of environmental controls that contain sera from individuals with no symptoms/signs of disease (i.e., spouses). DC samples include sera from patients with presumed infectious enteritis (stool culture negative for specific pathogens), blastocystis, celiac disease, collagenous colitis, irritable bowel syndrome, radiation proctitis, and acute schistosomiasis. The UC group includes both pANCA-positive and -negative specimens, while the CD group contains samples that are marker-negative, ASCA+; I2+; OmpC+ (I2-positive); OmpC+, I2+, and ASCA+; and pANCA+. Ethical approval for human studies was obtained from the institution review board at Cedars-Sinai Medical Center.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the flagellins CBir1 and Fla-X have been assigned GenBank accession numbers AY551005 and AY551006, respectively.
Acknowledgments
This work was supported by Corixa Corp., the Broad Medical Foundation, NIH PO1 DK44240 to C.O. Elson, and NIH PO1 DK46763 to S.R. Targan. We thank Audrey Lazenby for review and grading of the histopathology.
Footnotes
Michael J. Lodes and Yingzi Cong contributed equally to this work.
Michael J. Lodes’ present address is: CombiMatrix Corp., Mukilteo, Washington, USA.
Nonstandard abbreviations used: cecal bacterial protein/antigen (CBA); cetyltrimethylammonium bromide (CTAB); Crohn disease (CD); disease control (DC); gastrointestinal (GI); inflammatory bowel disease (IBD); isopropyl β-D-thiogalacto-pyranoside (IPTG); mesenteric lymph node (MLN); normal control (NC); PBS containing 0.1% Tween 20 (PBST); peptidoglycan (PG); representational difference analysis (RDA); serological expression cloning (SEC); tetramethylbenzidine (TMB); Toll-like receptor (TLR); Tris-EDTA (TE); ulcerative colitis (UC).
Conflict of interest: M.J. Lodes, R. Mohamath, M. Fort, and R.M. Hershberg, were previously employed by Corixa Corp., a publicly traded biotechnology company, which has sponsored a portion of this work. The majority of this work was funded by a grant from the Eli Broad Foundation for Medical Research to R. Hershberg, C. Elson, and S. Targan. C. Elson, who funded a portion of this work, was previously a consultant for Corixa Corp. S. Targan is a founding scientist, director, and consultant for Prometheus Laboratories.
References
- 1.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 2.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 3.McCracken VJ, Lorenz RG. The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell. Microbiol. 2001;3:1–11. doi: 10.1046/j.1462-5822.2001.00090.x. [DOI] [PubMed] [Google Scholar]
- 4.Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Curr. Opin. Immunol. 2003;15:396–401. doi: 10.1016/s0952-7915(03)00080-3. [DOI] [PubMed] [Google Scholar]
- 5.Akira S. Mammalian Toll-like receptors. Curr. Opin. Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 6.Sieling PA, Modlin RL. Toll-like receptors: mammalian ‘taste receptors’ for a smorgasbord of microbial invaders. Current Opin. Microbiol. 2002;5:70–75. doi: 10.1016/s1369-5274(02)00288-6. [DOI] [PubMed] [Google Scholar]
- 7.Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immunol. 2000;68:7010–7017. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting Edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 9.Abreu MT, et al. TLR4 and MD-2 expression is regulated by immune-mediated signals in human intestinal epithelial cells. J. Biol. Chem. 2002;277:20431–20437. doi: 10.1074/jbc.M110333200. [DOI] [PubMed] [Google Scholar]
- 10.Hershberg RM. The epithelial cell cytoskeleton and intracellular trafficking V. Polarized compartmentalization of antigen processing and Toll-like receptor signaling in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G833–G839. doi: 10.1152/ajpgi.00208.2002. [DOI] [PubMed] [Google Scholar]
- 11.Inohara N, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: implications for Crohn’s disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 12.Berg DJ, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4+ TH1-like responses. J. Clin. Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powrie F, et al. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–562. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 14.Cong Y, et al. CD4+ T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice: increased T helper cell Type 1 response and ability to transfer disease. J. Exp. Med. 1998;187:855–864. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandwein SL, et al. Spontaneously colitic C3H/HeJBir mice demonstrate selective antibody reactivity to antigens of the enteric bacterial flora. J. Immunol. 1997;159:44–52. [PubMed] [Google Scholar]
- 16.Dalwadi H, Wei B, Kronenberg M, Sutton CL, Braun J. The Crohn’s disease-associated bacterial protein I2 is a novel enteric T cell superantigen. Immunity. 2001;15:149–158. doi: 10.1016/s1074-7613(01)00164-9. [DOI] [PubMed] [Google Scholar]
- 17.Sutton CL, et al. Identification of a novel bacterial sequence associated with Crohn’s disease. Gastroenterology. 2000;119:23–31. doi: 10.1053/gast.2000.8519. [DOI] [PubMed] [Google Scholar]
- 18.Skeiky YAW, et al. Cloning, expression, and immunological evaluation of two putative secreted serine protease antigens of Mycobacterium tuberculosis. Infect. Immun. 1999;67:3998–4007. doi: 10.1128/iai.67.8.3998-4007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodes, M.J., Dillon, D.C., Houghton, R.L., and Skeiky, Y.A.W. 2004. Expression cloning. In Molecular diagnosis of infectious diseases. 2nd edition. J. Walker, series editor; J. Decker and U. Reischl, volume editors. Humana Press. Totowa, New Jersey, USA. 91–106.
- 20.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2000;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 21.Burich A, et al. Helicobacter-induced inflammatory bowel disease in IL-10- and T cell-deficient mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;281:G764–G778. doi: 10.1152/ajpgi.2001.281.3.G764. [DOI] [PubMed] [Google Scholar]
- 22.Winstanley C, Morgan JAW. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology. 1997;143:3071–3084. doi: 10.1099/00221287-143-10-3071. [DOI] [PubMed] [Google Scholar]
- 23.McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect. Immun. 2000;68:3344–3348. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 2000;164:986–993. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- 25.McSorley SJ, Ehst BD, Yu Y, Gewirtz AT. Bacterial flagellin is an effective adjuvant for CD4+ T cells in vivo. J. Immunol. 2002;169:3914–3919. doi: 10.4049/jimmunol.169.7.3914. [DOI] [PubMed] [Google Scholar]
- 26.McDermott PF, Ciacci-Woolwine F, Snipes JA, Mizel SB. High-affinity interaction between gram-negative flagellin and a cell surface polypeptide results in human monocyte activation. Infect. Immun. 2000;68:5525–5529. doi: 10.1128/iai.68.10.5525-5529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barcenilla A, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 29.Swidsinski A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2003;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 30.Hollander D. Intestinal permeability, leaky gut, and intestinal disorders. Curr. Gastroenterol. Rep. 1999;1:410–416. doi: 10.1007/s11894-999-0023-5. [DOI] [PubMed] [Google Scholar]
- 31.Breslin NP, et al. Intestinal permeability is increased in a proportion of spouses of patients with Crohn’s disease. Am. J. Gastroenterol. 2001;96:2934–2938. doi: 10.1111/j.1572-0241.2001.04684.x. [DOI] [PubMed] [Google Scholar]
- 32.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 33.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 34.Duchmann R, et al. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease. Clin. Exp. Immunol. 1995;102:448–455. doi: 10.1111/j.1365-2249.1995.tb03836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landers CJ, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 36.Lodes MJ, et al. Serodiagnosis of human granulocytic ehrlichiosis by using novel combinations of immunoreactive recombinant proteins. J. Clin. Microbiol. 2001;39:2466–2476. doi: 10.1128/JCM.39.7.2466-2476.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyoda H, et al. Distinct associations of HLA class II genes with inflammatory bowel disease. Gastroenterology. 1993;104:741–748. doi: 10.1016/0016-5085(93)91009-7. [DOI] [PubMed] [Google Scholar]
- 38.Yang H-Y, et al. Ulcerative colitis: a genetically heterogeneous disorder defined by genetic (HLA class II) and subclinical (antineutrophil cytoplasmic antibodies) markers. J. Clin. Invest. 1993;92:1080–1084. doi: 10.1172/JCI116613. [DOI] [PMC free article] [PubMed] [Google Scholar]