The Papillomavirus E7 Oncoprotein Is Ubiquitinated by UbcH7 and Cullin 1- and Skp2-Containing E3 Ligase (original) (raw)
Abstract
Recurrent infections with high-risk human papillomaviruses (HPVs) are associated with human cervical cancers. All HPV-associated cancer tissues express the viral oncoproteins E6 and E7, which stimulate cell growth. The expression of E7 is crucial for both the initiation and the maintenance of HPV-associated cancer. Recent studies showed that the level of E7 in cancer cells is regulated by ubiquitin-dependent proteolysis through the 26S proteasome. In this study, we characterized the enzymes involved in the ubiquitin-dependent proteolysis of E7. We show that UbcH7, an E2 ubiquitin-conjugating enzyme, is specifically involved in the ubiquitination of E7. Furthermore, we show that E7 interacts with the SCF (Skp-Cullin-F box) ubiquitin ligase complex containing Cullin 1 (Cul1) and Skp2 and can be ubiquitinated by the Cul1-containing ubiquitin ligase in vitro. Coimmunoprecipitation analyses revealed that E7 interacts with Skp2 and Cul1 in vivo. Finally, the half-life of E7 was found to be significantly longer in Skp2−/− mouse embryo fibroblasts (MEFs) than in wild-type MEFs. Taken together, these results suggest that the Cul1- and Skp2-containing ubiquitin ligase plays a role in the ubiquitination and proteolysis of E7. In HPV type 16-containing cervical carcinoma cell line Caski, E7 localizes to both the cytoplasm and the nucleus. Brief treatment of Caski cells with MG132 (a proteasome inhibitor) causes the accumulation of E7 in discrete nuclear bodies. These nuclear bodies are detergent insoluble and contain polyubiquitinated E7. We suggest that E7 relocates to specific nuclear bodies for proteolysis in HPV-containing epithelial cells.
Epidemiological studies have established that the high-risk types of human papillomavirus (HPV) are the main etiological factors for cervical cancer (reviewed in references 23, 35, 50, and 58). Significant percentages (20 to 30%) of premalignant and malignant oral and head and neck cancer lesions have also been documented to contain these high-risk HPVs (41). Cervical cancer alone accounts for almost 12% of all cancers in women (58). Therefore, elucidation of viral functions that contribute to malignant conversion is of major importance.
HPVs infect the proliferating epidermal or mucosal epithelial cells. Following persistent infections and after a long latency period, a small percentage of viral lesions progress to carcinoma in situ and squamous cell carcinoma. During this progression to malignancies, the viral genome often integrates into the host chromosome. All HPV-transformed cancer tissues express two HPV-encoded oncoproteins, E6 and E7. Both E6 and E7 possess transformation activity, and they cooperate to transform primary human keratinocytes, fibroblasts, and epithelial cells (reviewed in references 23, 35, 41, 50, and 58). Moreover, continued expression of the E7 protein is necessary for both maintenance of the transformed phenotype and a productive virus life cycle (15, 50, 51). A recent study showed that a reduction in the expression of E7 by RNA interference induces apoptosis in cervical cancer cells (26). Targeted transcriptional repression of the E6 and E7 oncoproteins by HPV E2 protein also induces senescence in HPV-containing cancer cells (17). Taken together, these studies show that a reduction in the level of E7 inhibits the growth of cancer cells.
One of the major biochemical functions of E7 is to induce DNA replication in differentiated epithelial cells (8). In differentiated cells, the retinoblastoma (Rb) family proteins Rb and p130 bind the E2F family transcription factors to repress the expression of the replication enzyme genes (reviewed in references 14 and 54). E7 disrupts the interaction between Rb family proteins and E2F, resulting in a release of the E2F factors in their transcriptionally active forms (7, 54). This E7-mediated conversion of E2F factors to their activated forms stimulates DNA replication and cell division, consistent with the observation that keratinocytes constitutively expressing E7 remain replication competent even after differentiation (8). In addition, it was shown that the E7 protein alone is capable of reactivating cellular DNA replication in differentiated epithelial cells (reviewed in references 15, 23, 35, 50, 51, and 58).
Previous studies showed that E7 induces the proteolytic degradation of Rb (3, 5, 28). E7 induces the degradation of Rb through the ubiquitin-26S proteasome (3, 5, 28). The proteolysis of Rb involves both N- and C-terminal regions of E7 that are also critical for the transforming function of E7, suggesting that the proteolysis of Rb is linked to the transforming function of E7 (3, 16). More recent studies showed that the HPV type 16 (HPV16) E7 protein is also regulated through proteolysis by the ubiquitin-proteasome pathway (16, 45, 53). Wang et al. observed that a significant level of polyubiquitinated E7 accumulates in Caski cervical carcinoma cells following treatment with an inhibitor of the 26S proteasome (53). Also, E7 interacts with and modifies the function of the S4 ATPase, a component of the 19S subunit of the proteasome (2).
In this report, we further analyzed E7 ubiquitination and studied the cellular enzymes involved in that process. Using a cell-free reconstituted ubiquitination assay with recombinant HPV16 E7, we identified UbcH7 as the E2-conjugating enzyme for E7 ubiquitination. To detect the E3 ligase activity of E7 ubiquitination, we looked for an association between the known E3 ligases and E7. SCF (Skp-Cullin-F box) ubiquitin ligases are involved in the ubiquitination of many cell cycle-regulated proteins, including E2F-1, p27, Orc1, and cyclin D1 (reviewed in reference 12). SCF-Skp2 is a multiprotein complex composed of Cullin 1 (Cul1), Skp1, Rbx1, and the F-box protein Skp2 (12). It is believed that the F-box protein Skp2 associates with the substrate. We show that E7 interacts with the Cul1- and Skp2-containing ubiquitin ligase complex both in vivo and in vitro. Furthermore, using combined immunoprecipitation and ubiquitination assays, we demonstrate that the Cul1-containing ubiquitin ligase and UbcH7 can ubiquitinate E7. Finally, we show that E7 proteolysis is impaired in Skp2−/− cells. Immunocytology and cell fractionation assays suggest that E7 proteolysis occurs in the nuclei of Caski cervical carcinoma cells. Treatment with a proteasome inhibitor leads to the accumulation of E7 in detergent-insoluble discrete nuclear bodies. These E7-containing nuclear bodies contain ubiquitinated E7, and we suggest that E7 is proteolysed by the nuclear proteasome in Caski cervical carcinoma cells.
MATERIALS AND METHODS
Plasmids and reagents.
The cytomegalovirus-driven E7 expression plasmid was described previously (3). The V5 epitope-tagged Cul1 expression plasmid was a kind gift from Pradip Raychaudhuri of the University of Illinois at Chicago. The His-tagged HPV-16 E7 plasmid was a generous gift from Denise Galloway of the Fred Hutchinson Cancer Center, Seattle, Wash. The purified recombinant yeast ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and ubiquitin aldehyde were from Boston Biochem, Cambridge, Mass. The protease inhibitor cocktail, leupeptin, and the proteasome inhibitor MG132 were from Calbiochem. Ubiquitin was from Sigma.
Antibodies.
The anti-V5 monoclonal antibody was from Invitrogen. The anti-Cul1 antibody was from Neomarker. Antiubiquitin monoclonal antibody P4D1, anti-Skp1 and anti-Skp2 antibodies, anti-E7 monoclonal antibody ED17, and anti-promyelocytic leukemia antibody PG-M3 were from Santa Cruz Biotechnology. Anti-E7 monoclonal antibody TVG-701Y and the antitubulin antibody were from Oncogene Science. The anti-UbcH7 antibody was from Boston Biochem. T7-tagged antibody-agarose beads were from Novagen.
Cell culture and fractionation of HeLa cell extracts.
The cell lines used in this study were HeLa (Spinner), Caski, and C33A. HeLa cells were grown in suspension in S-MEM (Invitrogen) supplemented with 10% bovine serum. The cell pellet was washed once with phosphate-buffered saline (PBS) and twice with lysis buffer, which contained 10 mM HEPES (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). The cells were resuspended in 1.5 volumes of lysis buffer and allowed to swell on ice for 20 min. The cells then were disrupted by Dounce homogenization (20 strokes), and the cell suspension was centrifuged at 50,000 × g for 30 min. The supernatant was adjusted to 100 mM NaCl and concentrated by 0 to 80% ammonium sulfate precipitation. Aliquots of the concentrated cytosolic extract were stored at −80°C and used for E7 ubiquitination assays. The HeLa cell cytosolic extract was further fractionated with a DEAE-Sepharose column (Pharmacia) into unabsorbed material (fraction I [FrI]) and a 500 mM NaCl eluate (fraction II [FrII]) by a previously described method (4, 9, 29). Fraction II was further fractionated with ammonium sulfate into fraction IIA (FrIIA) (0 to 38%) and fraction IIB (FrIIB) (40 to 80%) as described previously (4, 21).
Cell fractionation analysis.
Caski cell pellets treated with dimethyl sulfoxide (DMSO) or MG132 were washed twice with PBS and lysed in ice-cold IPB buffer, which contained 10 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 150 mM NaCl plus protease inhibitor cocktail (Calbiochem), and 1 mM PMSF for 30 min on ice (27). The cells were vortexed in IPB buffer multiple times during lysis. The soluble material was recovered by centrifugation at 20,000 × g for 15 min, and the pellet was resuspended in 10 mM Tris-HCl (pH 7.5)-1% sodium dodecyl sulfate (SDS) for 10 min at room temperature. After the addition of 5 volumes of IPB buffer, the samples were sonicated for 20 s and centrifuged for 10 min at 20,000 × g. The supernatant was saved as the insoluble nuclear fraction.
Transfection.
U2OS-E7 cells were grown in Dulbecco minimal essential medium (DMEM; GIBCO-BRL) supplemented with 10% fetal bovine serum. The cells then were transfected at 60% confluence with a V5 epitope-tagged Cul1 expression plasmid (5 μg), an Skp2 expression plasmid (5 μg), or both Cul1 (2.5 μg) and Skp2 (2.5 μg) expression plasmids by the calcium phosphate coprecipitation method as described previously (10). DNA precipitates were removed at 18 h after transfection, and the cells were replenished with fresh medium and harvested 30 h later. The cells were treated with tetracycline (1 μg/ml) and MG132 (5 μM) for 6 h prior to being harvested to induce E7 expression and to block E7 proteolysis. Cell lysates were prepared by using extraction buffer, which contained 50 mM Tris (pH 7.4), 0.1% Triton X-100, 0.25 M NaCl, 5 mM EDTA, 50 mM NaF, 0.1 mM Na orthovanadate, aprotinin (4 μg/ml), leupeptin (10 μg/ml), pepstatin (4 μg/ml), and 2 mM PMSF. Cell lysates (1.0 mg) were precleaned with 1 μg of mouse immunoglobulin G and protein G-Sepharose and immunoprecipitated with T7-tagged antibody-agarose beads (20 μl). The immunoprecipitates were washed five times with buffer containing 50 mM Tris (pH 7.4), 0.1 M NaCl, and 0.5% NP-40, resolved by SDS-11% polyacrylamide gel electrophoresis (PAGE), and immunoblotted with V5, Skp2, and E7 antibodies.
In vivo ubiquitination of E7.
The cell lysates were subjected to Western blot assays with various antibodies by following standard procedures (10). For the detection of E7-ubiquitin conjugates in Caski cells, Caski cells were treated with DMSO or MG132 (10 μM) for 6 h, and cell extracts were made in IPB buffer as soluble and insoluble extracts (27). Soluble extracts (1.5 mg) and insoluble extracts (500 μg) were diluted once with 0.5% NP-40-containing buffer and immunoprecipitated with the E7 antibody. The immunoprecipitates were washed three times with 0.5% NP-40-containing buffer, resolved by SDS-10% PAGE, and immunoblotted with the ubiquitin antibody or the E7 antibody.
In vitro ubiquitination assays.
A HeLa cell cytosolic extract and purified His-tagged HPV16 E7 were used for in vitro ubiquitination assays. HeLa cell S-100 contains all of the necessary enzymes for the ubiquitination of E7. Recombinant His-tagged E7 protein (His-E7) (0.1 to 0.6 μg) was incubated with HeLa cell S-100 (50 μg) at 30°C in a reaction mixture (30 μl) containing ubiquitin (5 μg), MgCl2 (5 mM), DTT (1 mM), ATP (2 mM), an ATP-generating system, and ubiquitin aldehyde (20 μM). E7-ubiquitin conjugates were purified with an Ni affinity column, resolved by SDS-12% PAGE, and probed with either the E7 antibody or the ubiquitin antibody in Western blot assays. We also used [35S]methionine-labeled E7 made by in vitro transcription-translation in rabbit reticulocyte lysates for E7 ubiquitination.
Ubiquitin ligase activity assays.
Ubiquitin ligase activity assays with the immunoprecipitated SCF complex were performed by previously described procedures (31, 43). Cul1 immunocomplexes were purified from transfected cell extracts (2 mg) with monoclonal antibodies (2.5 μg) against the V5 epitope and protein G-Sepharose beads (30 μl) and washed once with a buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1% NP-40, and 10% glycerol (wash buffer). The washed immunocomplexes were added to a ubiquitination reaction mixture (30 μl) containing 20 mM Tris (pH 7.5), MgCl2 (5 mM), ATP (2 mM), DTT (1 mM), E1 (30 ng), UbcH7 (300 ng), Roc-1 (20 ng), purified His-E7 (0.15 μg), and bovine ubiquitin (5 μg). The reaction mixture was incubated for 1 h at 30°C. After the reaction, the beads were separated by centrifugation at 3,000 rpm for 5 min and washed twice with the wash buffer described above. The beads and the supernatant were boiled separately for 5 min with SDS sample buffer containing β-mercaptoethanol, resolved by SDS-10% PAGE, and immunoblotted with either E7 or ubiquitin monoclonal antibodies. As a control for immunoprecipitation, we used a control antibody.
Analysis of the half-lives of E7 in wild-type and Skp2−/− MEFs.
Wild-type and Skp2−/− mouse embryo fibroblasts (MEFs) were cultured as described previously (39). Nonsenescent MEFs (at passages 2 and 3) were infected with a recombinant adenovirus expressing E7 (Ad-E7) for 18 h as described previously (2). The infected cells were treated with 50 μg of cycloheximide/ml for various times from 30 min to 2 h. The cells were lysed in radioimmunoprecipitation buffer containing protease inhibitors and phosphatase inhibitors, and cell extracts (400 to 500 μg) were separated by SDS-11% PAGE and probed with E7 and tubulin antibodies.
Immunofluorescence staining.
The cells were grown overnight on glass coverslips and incubated for 5 h in the presence of DMSO or MG132 (10 μM). The cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature, washed once with 0.1 M glycine in PBS, and permeabilized for 5 min with 0.2% Triton X-100 in PBS. After fixation, the cells were washed four times with PBS (5 min each time) and blocked with 5% goat serum for 1 h at room temperature. The cells were incubated with an E7 monoclonal antibody (1:200) for 2 h at room temperature. The cells were washed five times with PBS (5 min each time), incubated with a 1:500 dilution of tetramethyl rhodamine isothiocyanate-conjugated donkey anti-mouse antibody for 1 h at room temperature, and washed five times with PBS (5 min each time). For immunostaining of PML, the cells were incubated with a PML monoclonal antibody (1:200) for 2 h at room temperature. The cell nuclei were labeled with bisbenzamide (2 μg/ml) in PBS for 3 min at room temperature. For actin staining, the cells were counterstained with fluorescein isothiocyanate-conjugated phalloidin after E7 immunostaining. After a final wash with PBS, the cells were mounted on slides with Vectashield (Vector) mounting medium and viewed by confocal microscopy.
RESULTS
In vitro ubiquitination of E7.
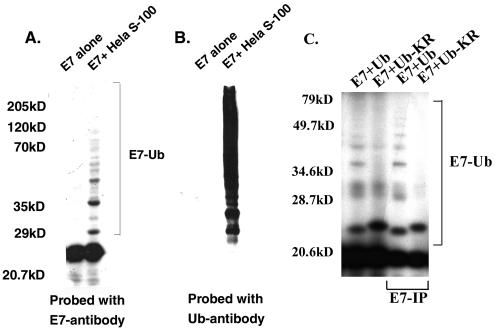
Previous studies showed that the E7 protein is degraded by the 26S proteasome and that ubiquitinated E7 can be detected in HPV-transformed Caski cell lysates (45, 53). Polyubiquitination involves three major enzymes: E1, which activates ubiquitin; E2, which transfers the ubiquitin from E1 to the ubiquitin ligase E3; and the substrate-specific ubiquitin ligase E3, which attaches ubiquitin molecules to the substrate (reviewed in references 9, 12, 21, 22, and 25). To learn more about the enzymes involved in E7 ubiquitination, we sought to identify the ubiquitin-conjugating enzyme E2 and the ubiquitin ligase E3 for E7. We used His-E7 purified from bacterial lysates as a substrate and a HeLa cell cytosolic extract as the source of ubiquitin enzymes. The His-E7 protein (0.1 to 0.5 μg) was incubated with the HeLa cell cytosolic extract (50 μg) at 30°C for 1 h in a reaction mixture containing ubiquitin, MgCl2, DTT, an ATP-generating system, and ubiquitin aldehyde. The reaction mixture was divided into two halves and was resolved by SDS-11% PAGE. Proteins in each half were probed with either E7 or ubiquitin antibody in Western blot assays. A ladder of high-molecular-weight proteins recognized by both E7 and ubiquitin monoclonal antibodies represents ubiquitinated E7 (Fig. 1A and B).
FIG. 1.
Ubiquitination of E7 in cell-free lysates. (A and B) Purified His-E7 protein (0.5 μg) was ubiquitinated by using HeLa cell cytosolic extracts (50 μg) in a reaction mixture containing ubiquitin and ATP as described in Materials and Methods. The E7-ubiquitin conjugates were purified with an Ni affinity column, divided into two parts, and resolved by SDS-10% PAGE. One part (A) was probed with an E7 antibody and the other part (B) was probed with a ubiquitin (Ub) antibody in Western blot assays. (C) 35S-labeled nontagged HPV16 E7 made in rabbit reticulocyte lysates was ubiquitinated with HeLa cell S-100 (50 μg) in the presence of wild-type ubiquitin or ubiquitin-KR. The ubiquitinated proteins were immunoprecipitated (IP) with the E7 antibody, resolved by SDS-10% PAGE, and detected by fluorography.
To further confirm that the high-molecular-weight proteins are indeed ubiquitinated E7 and not other ubiquitinated proteins associated with E7, we used untagged 35S-labeled HPV16 E7 made by in vitro transcription-translation in ubiquitination assays. To demonstrate specificity, modified recombinant ubiquitin, ubiquitin-KR, containing a replacement of lysine 48 by an arginine, was used in the ubiquitination reaction. After ubiquitination, the ubiquitinated E7 was immunoprecipitated from the reaction mixture by using E7 antibody and analyzed by SDS-PAGE and autoradiography. As shown in Fig. 1C, the reaction mixture produced polyubiquitinated E7, and the polyubiquitination was inhibited by ubiquitin-KR. Methyl-ubiquitin added to the ubiquitination reaction also did not allow the formation of polyubiquitinated E7 (data not shown). Taken together, these results showed that E7 is efficiently polyubiquitinated in a cell-free ubiquitination system and that the HeLa cell cytosolic extract contains all of the necessary enzymes for E7 ubiquitination.
Isolation of enzymes catalyzing E7 ubiquitination.
To identify the enzymes required for the formation of E7-ubiquitin conjugates, HeLa cell S-100 was fractionated through a DEAE-Sepharose column by a previously described procedure (4, 9, 21). The flowthrough fraction was collected as FrI, and the bound proteins eluted from the column with 0.5 M KCl were collected as FrII. After extensive dialysis, FrII was further resolved by ammonium sulfate precipitation. Proteins precipitating at 0 to 40% ammonium sulfate were designated FrIIA, and those precipitating at 40 to 80% were designated FrIIB. These fractions were enriched in different components of the ubiquitin pathway (4, 9, 21). To assay their ability to support E7 ubiquitination, each fraction or a combination was analyzed in ubiquitination assays in the presence of recombinant E1 protein, wild-type ubiquitin, and His-E7. Neither fraction alone supported E7 ubiquitination (Fig. 2, lanes 2 to 4). A combination of FrI and FrIIA was found to be efficient for the ubiquitination of E7 (Fig. 2, lane 5).
FIG. 2.
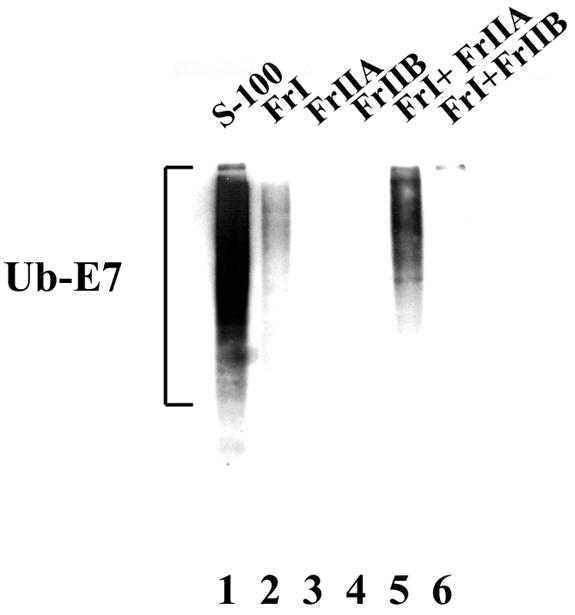
HeLa cell FrI and FrIIA together promote the formation of E7-ubiquitin conjugates. Ubiquitination reactions were carried out as described in Materials and Methods with His-E7 (0.3 μg), ubiquitin (5 μg), recombinant E1 (30 ng) and, when indicated, HeLa cell Fr1 (25 μg), FrIIA (25 μg), FrIIB (25 μg), or S-100 (30 μg). The E7-ubiquitin (Ub-E7) conjugates were purified with an Ni affinity column, resolved by SDS-10% PAGE, and probed with a ubiquitin antibody in Western blot assays.
The E2-conjugating enzyme UbcH7 specifically ubiquitinates E7 in vitro.
A previous study showed that FrI and FrIIA contain multiple E2 proteins, including Ubc5A, Ubc5B, Ubc5C, UbcH6, UbcH7, UbcH2, UbcH3, and UbcH10. FrIIA contains many ubiquitin ligases (4). To identify the specific E2-conjugating enzyme for E7 ubiquitination, we carried out a ubiquitination reaction with FrIIA (25 μg), recombinant E1, and 300 ng of purified recombinant E2 protein. The ubiquitination reaction mixture also contained ubiquitin, MgCl2, DTT, an ATP-generating system, and ubiquitin aldehyde and was incubated at 30°C for 1 h. The ubiquitinated E7 proteins were detected by probing with E7 antibody in Western blot assays. As shown in Fig. 3A, UbcH7 specifically conjugated E7. Similar ubiquitination with FrI or FrIIB as the source of E3 did not produce ubiquitinated E7 (data not shown). To confirm that UbcH7 is the specific ubiquitin carrier protein for E7, we used a 35S-labeled HPV16 E7 protein made in rabbit reticulocyte lysates. FrIIA was used as the source of E3, and recombinant UbcH5A, UbcH5B, UbcH5C, UbcH6, and UbcH7 proteins were compared. Clearly, UbcH7 was found to be the only E2 protein that efficiently ubiquitinated E7 (Fig. 3B). SDS-PAGE of FrI and FrIIA showed that UbcH7 is present exclusively in FrI (Fig. 3C); therefore, we predicted that HeLa cell FrIIA would contain the ubiquitin ligase for E7.
FIG. 3.
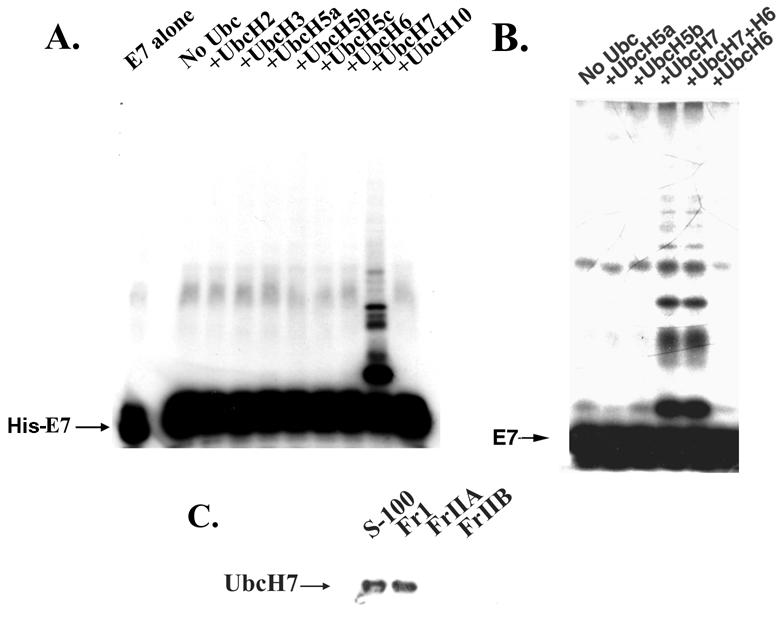
UbcH7 is involved in the conjugation of ubiquitin to E7. (A) His-E7 protein (0.65 μg) was ubiquitinated by using HeLa cell FrIIA (50 μg), recombinant E1 protein (30 ng), and the indicated recombinant E2 protein (300 ng) in ubiquitination assays as described in Materials and Methods. The E7-ubiquitin conjugates were purified with an Ni affinity column, resolved by SDS-11% PAGE, and probed with an E7 antibody. (B) 35S-labeled E7 made in rabbit reticulocyte lysates was ubiquitinated with recombinant E1 protein (30 ng), HeLa cell FrIIA (25 μg), and the indicated recombinant E2 protein (300 ng). The reaction mixtures were resolved by SDS-10% PAGE, and the proteins were developed by fluorography. (C) Proteins (200 μg) from HeLa cell S-100, FrI, FrIIA, or FrIIB were resolved by SDS-12% PAGE and probed with a UbcH7 antibody in Western blot assays.
E7 associates with the Cul1- and Skp2-containing ubiquitin ligase complex.
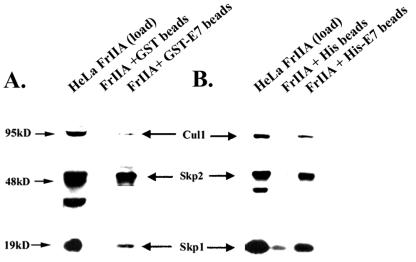
SCF ubiquitin ligases regulate proteolytic events that drive cells through the G1-S phase (reviewed in reference 12). SCF-Skp2 is a multiprotein complex composed of Cul1, Skp1, Rbx1, and the F-box protein Skp2. The F-box protein is responsible for substrate recognition, and Skp2 is responsible for the ubiquitination of many cell cycle-regulated proteins, including E2F-1, p27, Orc1, and cyclin A/Cdk2 (6, 32, 36, 40, 55). SCF-Skp2 was fractionated in HeLa cell FrIIA (Fig. 4). Since FrIIA contained the ubiquitin ligase for E7, we tested whether Skp2 could interact with E7. FrIIA (0.5 mg) was allowed to bind to beads containing glutathione _S_-transferase (GST) (1 μg) or GST-E7 (1 μg) for 1 h at 4°C. After extensive washing, the bound proteins were released from the beads by boiling with SDS sample buffer, separated by SDS-10% PAGE, and analyzed by Western blot assays with antibodies against Cul1, Skp1, and Skp2. As shown in Fig. 4A, GST-E7 bound to the Cul1-Skp1-Skp2 complex. We performed the same experiment with His-E7 bound to Ni-agarose beads, and as shown in Fig. 4B, His-E7 bound to the Cul1-Skp2 complex.
FIG. 4.
E7 binds to the SCF-Cul1-Skp2-Skp1 complex. (A) GST-E7 (1 μg) immobilized on glutathione-Sepharose beads was incubated with 500 μg of HeLa cell FrIIA. The bound proteins were separated by SDS-10% PAGE and probed with Cul1, Skp1, and Skp2 antibodies. (B) His-E7 (1 μg) immobilized on Ni-agarose beads was incubated with 500 μg of HeLa cell FrIIA. After extensive washing, the bound proteins were separated by SDS-10% PAGE and probed with Cul1, Skp1, and Skp2 antibodies.
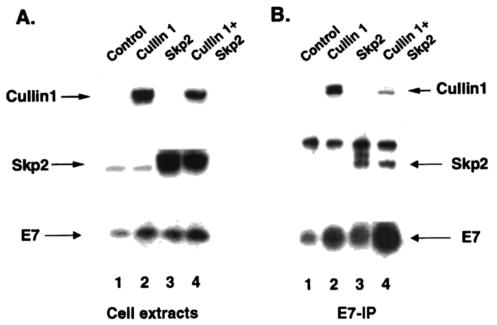
For analysis of the in vivo association between E7 and the Cul1-Skp2 complex, we cotransfected U2OS-E7 cells, which allow the expression of T7-tagged HPV16 E7 protein following the addition of tetracycline (44). The E7 expressed in these cells is regulated by proteasomal degradation (44; data not shown). U2OS-E7 cells were transfected with V5 epitope-tagged Cul1 and Skp2 expression plasmids. The transfected cells were treated with both tetracycline (1 μg/ml) and MG132 (5 μM) for 6 h to induce E7 and block E7 proteolysis. Cell lysates (1 mg) from the transfected cells were immunoprecipitated with monoclonal antibody against the T7 tag, and the immunoprecipitates were probed with V5 antibody to detect Cul1 and Skp2 antibody to detect Skp2 in Western blot assays. The immunoprecipitates were also probed with E7 monoclonal antibody to detect the level of immunoprecipitated E7. The T7-tagged antibody coprecipitated Cul1 and Skp2 from the transfected cell lysates (Fig. 5B). We performed similar transfections with an Skp1 expression plasmid but could not detect any specific signal due to the interfering signals from the immunoglobulin G light chain (data not shown). Based on both in vivo and in vitro binding of Cul1 and Skp2 to E7, we suggest that E7 associates with the SCF-Skp2 complex.
FIG. 5.
E7 associates with Skp2 and Cul1 in vivo. U2OS-E7 cells were transfected with plasmids expressing V5 epitope-tagged Cul1 (5 μg) or Skp2 (5 μg) or a combination of V5 epitope-tagged Cul1 (2 μg) and Skp2 (2.5 μg) expression plasmids. Prior to being harvested, the cells were treated for 6 h with tetracycline (1 μg/ml) to induce the expression of E7 and with MG132 (5 μM) to prevent E7 proteolysis. (A) Total cell lysates (200 μg) were resolved by SDS-11% PAGE and analyzed for E7, Cul1, and Skp2 in Western blot assays. (B) Cell extracts (1 mg) were immunoprecipitated (IP) with T7-tagged antibody-agarose beads as described in Materials and Methods. The immunoprecipitated proteins were separated by SDS-11% PAGE and probed with V5, Skp2, and E7 antibodies in Western blot assays.
Cul1-containing E3 ligase and UbcH7 ubiquitinate E7 in vitro.
Next, we used coupled immunoprecipitation and in vitro ubiquitination assays to analyze whether the Cul1-containing ubiquitin ligase could ubiquitinate E7. For this experiment, C33A cells were transfected with a V5 epitope-tagged Cul1 expression plasmid, lysates from the transfected cells were immunoprecipitated with an antibody against the V5 epitope (V5-Ab) or a control antibody, and the immune complexes were collected with protein G-Sepharose beads. The beads (20 μl) containing the immune complexes were added to a ubiquitination reaction containing recombinant His-E7 (50 ng), recombinant E1 (30 ng), recombinant UbcH7 (300 ng), and Rbx (20 ng). The reaction mixture also contained ubiquitin, MgCl2, DTT, and an ATP-generating system and was incubated at 30°C for 1 h. The beads were separated from the reaction mixture by quick spinning and were washed briefly with low-salt buffer, and the eluted proteins were resolved by SDS-11% PAGE followed by Western blot analysis with E7 antibody. An aliquot of the supernatant (after spinning of the beads) was also analyzed. High-molecular-weight proteins representing ubiquitinated E7 were recognized by the E7 monoclonal antibody (Fig. 6). Interestingly, the ubiquitinated E7 proteins were detected only in the bead fraction and not in the supernatant fraction. This result suggests that only the fraction of E7 in association with the Cul1 complex is ubiquitinated. Similar immunoprecipitation experiments with the control antibody did not show ubiquitination of E7.
FIG. 6.
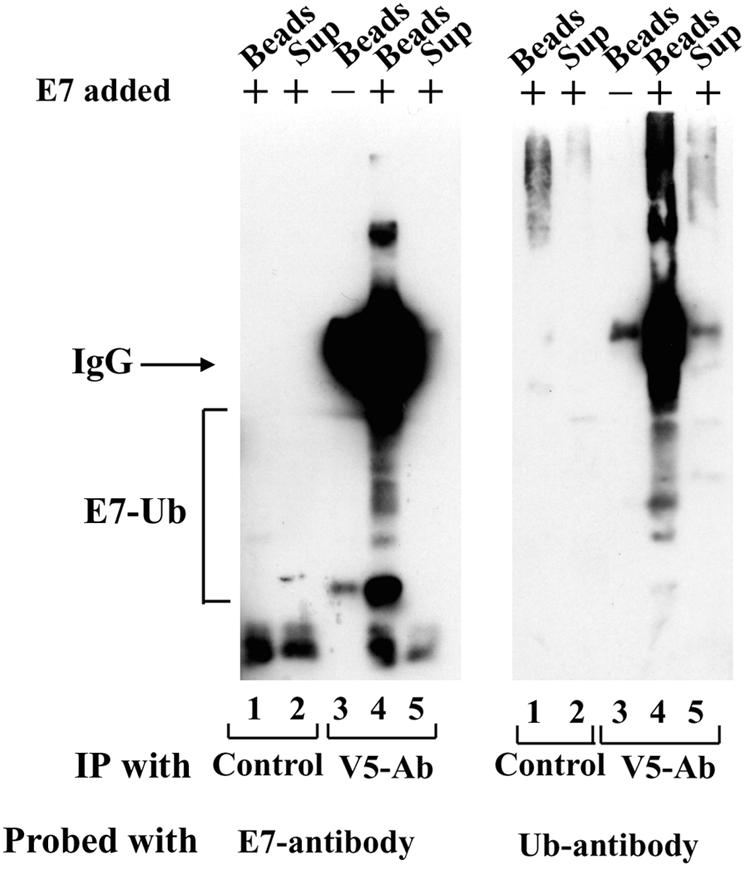
E7 is ubiquitinated by the Cul1 complex and UbcH7 in vitro. C33A cells were transfected with a V5 epitope-tagged Cul1 expression plasmid, and the transfected cell extracts (2 mg) were immunoprecipitated (IP) with a control antibody or a V5 antibody (V5-Ab) and protein G-Sepharose beads. After brief washing, the beads with the immune complexes were used to ubiquitinate His-E7 (0.15 μg) as described in Materials and Methods. After the reaction, the beads were separated from the supernatants (Sup) by centrifugation. Proteins associated with the beads and in the supernatants were resolved separately by SDS-10% PAGE and probed with E7 and ubiquitin (Ub) monoclonal antibodies. IgG, immunoglobulin G; E7-Ub, E7-ubiquitin.
Impaired proteolysis of HPV16 E7 in Skp2−/− MEFs.
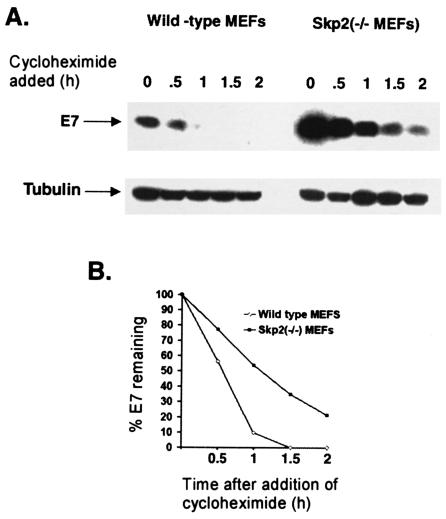
To investigate the role of Skp2 in E7 proteolysis, we compared the half-lives of HPV16 E7 in MEFs prepared from wild-type and Skp2−/− mice (39). Both wild-type and Skp2−/− MEFs at passage 3 were infected with recombinant Ad-E7 as described previously (2). After 18 h of infection, the cells were treated with cycloheximide (50 μg/ml) to block protein synthesis. The cells were harvested at different times after cycloheximide treatment, and the extracts were analyzed for the E7 protein by Western blotting. As shown in Fig. 7, E7 exhibited a very short half-life, approximately 30 min, in wild-type MEFs and was completely degraded within 1 h in wild-type MEFs. The abundance of E7 was found to be consistently higher (more than twofold) in Skp2−/− MEFs than in wild-type MEFs at the same level of infection (Fig. 7A). Moreover, as shown in Fig. 7, the half-life of E7 was significantly longer in Skp2−/− MEFs. Densitometric scanning revealed that the half-life of E7 was more than 1 h in Skp2−/− cells (Fig. 7B). Taken together, these results show that Skp2 is involved in the proteasomal degradation of E7. Interestingly, E7 proteolysis was not completely blocked in Skp2−/− MEFs, suggesting that E7 can also be degraded by an Skp2-independent pathway.
FIG. 7.
E7 expressed in Skp2−/− MEFs showed a longer half-life than E7 expressed in wild-type MEFs. Wild-type and Skp2−/− MEFs (at passages 2 and 3) were infected with recombinant Ad-E7 as described in Materials and Methods. After 18 h, cycloheximide (50 μg/ml) was added to the medium, and the cells were harvested at various times. (A) Cell lysates from wild-type MEFs (500 μg) and Skp2−/− MEFs (250 μg) were separated by SDS-11% PAGE and probed with E7 and tubulin antibodies. (B) The band intensities in the Western blot were determined by densitometric scanning. For each plot, the band intensity corresponding to the 0-h time point was taken as 100%, and the relative intensities of the bands at various time points were represented as percent E7 remaining.
Ubiquinated E7 accumulates in the nuclei of Caski cells.
Exposure to the proteasome inhibitor MG132 (or lactacystin) led to a huge increase in the level of E7 protein in Caski cells, consistent with the fact that E7 is proteolysed by the 26S proteasome (16, 45, 53). To determine the site of E7 proteolysis, we compared the subcellular distributions of E7 in DMSO-treated and MG132-treated Caski cells. Extracts of Caski cells were fractionated as described in Materials and Methods. In DMSO-treated Caski cells, more than 70% of steady-state E7 was recovered in the detergent-soluble cytosolic extract, suggesting that E7 is predominantly cytoplasmic in these cells (Fig. 8A). In contrast, when Caski cells were treated with MG132 for 6 h, more than 50% of E7 was found in the detergent-insoluble fraction (Fig. 8A). To test the localization of polyubiquitinated E7, the detergent-soluble and -insoluble Caski cell proteins were immunoprecipitated with an E7 antibody, and the immunoprecipitated proteins were subjected to Western blot analysis with E7 antibody and ubiquitin antibody (Fig. 8B and C). Interestingly, the majority of the polyubiquitinated E7 in MG132-treated cells was recovered in the detergent-insoluble fraction (Fig. 8C, lane 12).
FIG. 8.
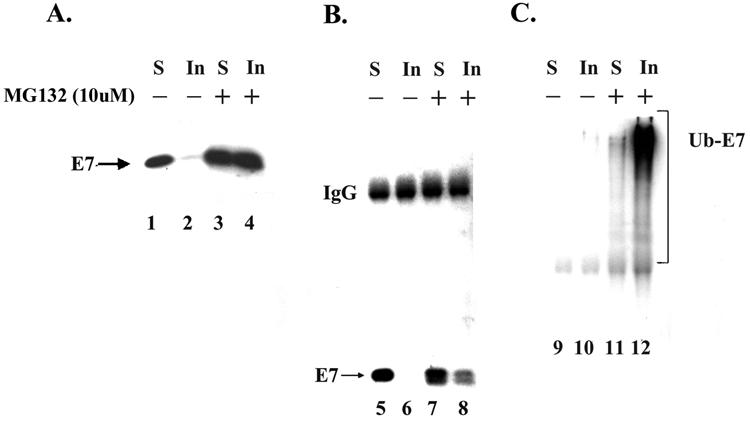
Ubiquitinated E7 is accumulated in the detergent-insoluble nuclear fraction of MG132-treated Caski cells. (A) Caski cells were treated with DMSO or MG132 (10 μM) for 5 h before being harvested. The cells were lysed and separated into detergent-soluble (S) and detergent-insoluble (In) extracts as described in Materials and Methods. Detergent-soluble (300 μg) and detergent-insoluble (300 μg) cell extracts were separated by SDS-10% PAGE and probed by immunoblotting with an E7 antibody. (B and C) Detergent-soluble (1.5 mg) and detergent-insoluble (0.41 mg) cell extracts were immunoprecipitated with an E7 polyclonal antibody. The immunoprecipitated proteins were collected with protein A-Sepharose beads, separated by SDS-10% PAGE, and probed with an E7 monoclonal antibody (B) or a ubiquitin antibody (C). IgG, immunoglobulin G; Ub-E7, ubiquitin-E7.
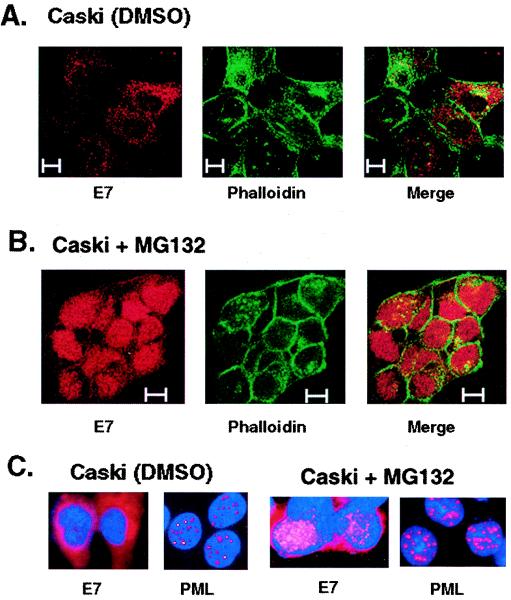
A previous study characterized the presence of pericentriolar membrane-free distinct cytoplasmic inclusions containing deposits of aggregated, ubiquitinated proteins known as aggresomes (27). To test whether multiubiquitinated E7 localizes to aggresomes in proteasome inhibitor-treated cells, we used fluorescence microscopy to monitor the localization of E7 in untreated and MG132-treated Caski cells. In agreement with the biochemical cell fractionation analysis, fluorescence for E7 was detected mainly in the cytosolic and perinuclear regions in untreated Caski cells (Fig. 9A). Some E7 immunostaining was also detected in nuclei. Strikingly, after exposure to 10 μM MG132 for 6 h, a significant increase in E7 immunofluorescence was detected, and the majority of the E7 fluorescence was localized to discrete nuclear foci (Fig. 9B and C). Nuclear immunostaining for E7 was associated with discrete dot-like structures. Probing with the PML antibody showed that E7 immunostaining and PML immunostaining partially overlapped in MG132-treated Caski cells (Fig. 9C). Taken together, our results suggest that ubiquitinated E7 accumulates in distinct nuclear bodies for degradation in Caski cells.
FIG. 9.
Accumulation of E7 protein in discrete nuclear foci in MG132-treated Caski cells. (A and B) Caski cells were treated with DMSO (A) or MG132 (10 μM) (B) for 6 h. The cells were fixed and processed for immunostaining with an E7 monoclonal antibody and tetramethyl rhodamine isothiocyanate-labeled (red) anti-mouse antibodies to detect E7. The cells were counterstained with fluorescein isothiocyanate-conjugated phalloidin (green) to detect actin. Immunofluorescence was detected by confocal microscopy. (C) DMSO-treated and MG132-treated Caski cells were separately immunostained with an E7 monoclonal antibody and a PML antibody and counterstained with bisbenzamide (2 μg/ml) to label cell nuclei. The images were visualized with a Zeiss microscope.
DISCUSSION
The HPV E7 oncoprotein is unstable and is a target of the 26S proteasome. E7 functions as a transcription regulator and induces S-phase synthesis in HPV-transformed epithelial cells (reviewed in references 23, 35, 50, and 58). The function of E7 is critical for both induction and maintenance of HPV-associated cancer. Thus, it is important to understand the molecular mechanisms that govern the cellular stability of this oncoprotein. In this study, we described the E2 and E3 ubiquitin ligases involved in the ubiquitination of E7. Using in vitro ubiquitination assays, we showed that UbcH7 is the specific ubiquitin-conjugating enzyme involved in E7 ubiquitination. Furthermore, we provided evidence that E7 interacted with and was ubiquitinated by the Cul1- and Skp2-containing ubiquitin ligase complex. Immunofluorescence and cell fractionation studies demonstrated that multiubiquitinated E7 accumulates predominantly in discrete nuclear foci upon inhibition of proteasome-dependent degradation.
Dissection of the enzymatic components involved in the ubiquitination of E7 revealed a role for the E2-conjugating enzyme UbcH7. The E2-conjugating enzymes are a closely related family of proteins (25, 48). Several E2 enzymes have been characterized (25). Often, a common E2 enzyme carries out the ubiquitination of multiple substrates. Our results showed that the other E2 enzymes, including the closest homologues UbcH6 and UbcH5a, are significantly less active in E7 ubiquitination (42). All of the recombinant E2 enzymes used in this study are functionally active, as they can form active thiol esters with ubiquitin (data not shown). Furthermore, UbcH7 is fractionated in FrI of HeLa cell S-100, which was essential for the ubiquitination of E7 in vitro. UbcH7 can function in conjugation with different types of E3 ubiquitin ligases, including a hect-E3, such as E6AP, or a ring-E3, such as c-Cbl (24, 57). The results presented in this study show that for the ubiquitination of E7, UbcH7 cooperates with the Cul1-Skp2 (SCF) complex, which belongs to the ring-E3 family of ligases (29, 57).
The SCF complexes are well-studied ubiquitin ligases that mediate the ubiquitination of diverse cell cycle regulatory and signaling proteins (reviewed in reference 12). Our results showed that E7 could interact with the Cul1-Skp2 complex both in vitro and in vivo. Moreover, E7 could be ubiquitinated by purified UbcH7 and the Cul1 complex. Finally, we showed that the half-life of E7 was increased from less than 30 min in wild-type MEFs to more than 1 h in Skp2−/− MEFs. Taken together, these results suggest that the Cul1- and Skp2-containing ubiquitin ligase plays a major role in the ubiquitination and proteolysis of E7. Interestingly, we observed delayed but not complete blockage of E7 proteolysis in Skp2−/− cells, suggesting that E7 can also be degraded by an Skp2-independent pathway. Similar Skp2-independent proteolysis has been reported for other Skp2 targets, including p27 and E2F-1 (20, 43). Furthermore, many cellular proteins, including p53, p21, and p27, are targeted by multiple ubiquitin ligases (12). Among them, p21 is degraded by the proteasome through both ubiquitination-dependent and ubiquitination-independent pathways (49). The ubiquitination-independent proteolysis of p21 involves its association with the C8 subunit of the proteasome (52). Previous results showed that E7 could bind to the S4 subunit of the 26S proteasome (2). The role of the E7-S4 interaction in E7 proteolysis is not clear. Future investigations will provide insight into other mechanisms of E7 proteolysis.
SCF-dependent ubiquitination often requires specific phosphorylation of the substrate (13, 37, 43, 56). Both p27Kip1and E2F-1 are phosphorylated by cyclin-dependent kinases prior to ubiquitination by SCF-Skp2 (37, 43). E7 is phosphorylated by casein kinase II at serine 30 or 31 and by an unknown kinase at serine 71 (33). E7 also associates with cyclin E/Cdk2 kinase and cyclin A/Cdk2 kinase, but it is not known whether E7 is phosphorylated by these cyclin-dependent kinases (1, 11, 34, 38). Although our result show that recombinant His-E7 can be efficiently ubiquitinated in vitro, they does not rule out the possibility that the ubiquitination of E7 is regulated by specific phosphorylation. HeLa cell FrIIA used for E7 ubiquitination contained a kinase(s) for E7 phosphorylation, and it is possible that E7 is phosphorylated during in vitro ubiquitination. We observed a doublet corresponding to E7 in extracts of Caski cells after treatment with the proteasome inhibitor MG132 (Fig. 8). Further studies will be required to determine whether phosphorylation regulates the ubiquitination of E7 in vivo.
The results presented in this study suggest that E7 proteolysis occurs in the nucleus. There are conflicting reports about the subcellular localization of the HPV E7 oncoprotein (19, 46, 47, 60). E7 has been reported to be predominantly nuclear in transiently transfected cells (18, 19). Others have reported the cytoplasmic localization of E7 in CV1 cells (60) and in normal oral keratinocyte cells (46). Using biochemical fractionation experiments, we found that a major portion of the E7 protein in Caski cells is cytoplasmic. This is a surprising observation, because multiple nuclear functions of E7, including the transactivation of E2F-regulated genes, binding with the Rb family of transcription regulators, and interactions with the transcription factors Fos, E2F-1, and others, predict the nuclear localization of E7. It has been convincingly demonstrated that these nuclear interactions of E7 are critical for its transformation function. However, interactions of E7 with cytoplasmic proteins, such as actin, M2 pyruvate kinase, and acid α-glucosidase, have also been reported (46, 59, 60). Interestingly, we recovered a significant level of E7 in the nuclear fraction of Caski cells treated with the proteasome inhibitor MG132. This result suggests that E7 in the nuclear compartment is more labile than cytoplasmic E7. It is also possible that MG132 triggers some modification of E7 that induces its nuclear localization. Immunofluorescence with the E7 monoclonal antibody further confirmed the cell fractionation results. E7 immunofluorescence was readily detected in the cytosolic and perinuclear regions of asynchronously growing Caski cells (Fig. 9A and C). These cells also showed some E7 staining in discrete foci in the nuclei.
A previous study showed that the inhibition of proteasome activity causes the deposition of protein aggregates as cytoplasmic inclusion bodies known as aggresomes (27). Aggresomes generally contain ubiquitin-rich cytoplasmic proteins and are linked to the pathogenesis of many diseases (30). Following proteasomal inhibition, E7 immunostaining was observed specifically in discrete nuclear foci, not in cytosolic aggresome-like structures, suggesting that E7 proteolysis occurs in the nucleus. In support of this hypothesis, we recovered most of the high-molecular-weight polyubiquitinated E7 in the detergent-insoluble nuclear pellet of MG132-treated Caski cells. The E7 immunostaining was found to partially overlap the PML immunostaining in MG132-treated Caski cells. This result suggests that E7 proteolysis occurs in defined nuclear bodies (Fig. 9C).
Many questions remain unanswered. (i) What triggers the localization of E7 to the nucleus? (ii) Is E7 ubiquitination regulated by specific phosphorylation? (iii) Are nuclear E7 and cytoplasmic E7 differentially modified? E7 strongly activates the transcription of E2F- and AP1-dependent genes. The major known biochemical function of E7 is the activation of E2F to stimulate DNA replication and cell division. It is intriguing to speculate that in cervical cancer cells, E7 is sequestered in the cytoplasm in a transcriptionally inactive form. Ubiquitin-dependent proteolysis actively regulates the level of E7 in the nuclei of HPV-containing tumor cells and may be linked to the survival mechanism of the tumor cells.
Acknowledgments
We thank Pradip Raychaudhuri for generously providing the V5 epitope-tagged Cul1 expression plasmid and critically reading the manuscript. We thank Denise Galloway (Fred Hutchinson Cancer Center) for providing the plasmid expressing His-E7. We thank Tania Bonder for providing purified recombinant Roc-1 used in ubiquitin ligase activity assays.
Support for this research was provided by grant DE12506 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Arroyo, M., S. Bagchi, and P. Raychaudhuri. 1993. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex. Mol. Cell. Biol. 13**:**6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berezutskaya, E., and S. Bagchi. 1997. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J. Biol. Chem. 272**:**30135-30140. [DOI] [PubMed] [Google Scholar]
- 3.Berezutskaya, E., B. Yu, A. Morozov, P. Raychaudhuri, and S. Bagchi. 1997. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 8**:**1277-1286. [PubMed] [Google Scholar]
- 4.Blumenfeld, N., H. Gonen, A. Mayer, C. E. Smith, N. R. Siegel, A. L. Schwartz, and A. Ciechanover. 1994. Purification and characterization of a novel species of ubiquitin-carrier protein, E2, that is involved in degradation of non-“N-end rule” protein substrates. J. Biol. Chem. 269**:**9574-9581. [PubMed] [Google Scholar]
- 5.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56**:**4620-4624. [PubMed] [Google Scholar]
- 6.Carrano, A. C., E. Eytan, A., Hershko, and M. Pagano. 1999. Skp2 is required for the ubiquitin-mediated degradation of the Cdk-inhibitor p27. Nat. Cell Biol. 1**:**193-199. [DOI] [PubMed] [Google Scholar]
- 7.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89**:**4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9**:**2335-2349. [DOI] [PubMed] [Google Scholar]
- 9.Coux, O., and A. L. Goldberg. 1998. Enzymes catalyzing ubiquitination and proteolytic processing of the p105 precursor of nuclear factor kappa B1. J. Biol. Chem. 273**:**8820-8828. [DOI] [PubMed] [Google Scholar]
- 10.Datta, P. K., P. Raychaudhuri, and S. Bagchi. 1995. Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription. Mol. Cell. Biol. 15**:**5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, R., R. Hicks, T. Crook, J. Morris, and K. Vousden. 1993. Human papillomavirus type 16 E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. J. Virol. 67**:**2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15**:**435-467. [DOI] [PubMed] [Google Scholar]
- 13.Diehl, J. A., F. Zindy, and C. J. Sherr. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11**:**957-972. [DOI] [PubMed] [Google Scholar]
- 14.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12**:**2245-2262. [DOI] [PubMed] [Google Scholar]
- 15.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74**:**6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75**:**7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin, E. C., and D. DiMaio. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 97**:**12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenfield, I., J. Nickerson, S. Penman, and M. Stanley. 1991. Human papillomavirus 16 E7 protein is associated with the nuclear matrix. Proc. Natl. Acad. Sci. USA 88**:**11217-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guccione, E., P. Massimi, A. Bernat, and L. Banks. 2002. Comparative analysis of the intracellular location of the high- and low-risk human papillomavirus oncoproteins. Virology 293**:**20-25. [DOI] [PubMed] [Google Scholar]
- 20.Hara, T., T. Kamura, K. Nakayama, K. Oshikawa, S. Hatakeyama, and K.-I. Nakayama. 2001. Degradation of p27Kip1 at the G0-G1 transition mediated by a Skp2-independent ubiquitination pathway. J. Biol. Chem. 276**:**48937-48943. [DOI] [PubMed] [Google Scholar]
- 21.Hershko, A., H. Heller, S. Elias, and A. Ciechanover. 1983. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258**:**8206-8214. [PubMed] [Google Scholar]
- 22.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30**:**405-439. [DOI] [PubMed] [Google Scholar]
- 23.Howley, P. M. 1996. The viruses and their replication, p. 947-978. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 24.Huang, L., E. Kinnucan, G. Wang, S. Beaudenon, P. M. Howley, J. M. Huibregtse, and N. P. Pavletich. 1999. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286**:**1321-1326. [DOI] [PubMed] [Google Scholar]
- 25.Jentsch, S. 1992. The ubiquitin-conjugation system. Annu. Rev. Genet. 26**:**179-207. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, M., and J. Milner. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21**:**6041-6048. [DOI] [PubMed] [Google Scholar]
- 27.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143**:**1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, D. L., D. A. Thompson, and K. Munger. 1997. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology 239**:**97-107. [DOI] [PubMed] [Google Scholar]
- 29.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284**:**657-661. [DOI] [PubMed] [Google Scholar]
- 30.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10**:**524-530. [DOI] [PubMed] [Google Scholar]
- 31.Maeda, I., T. Ohta, H. Koizumi, and M. Fukuda. 2001. In vitro ubiquitination of cyclin D1 by ROC1-CUL1 and ROC1-CUL3. FEBS Lett. 494**:**181-185. [DOI] [PubMed] [Google Scholar]
- 32.Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. 1999. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1**:**14-19. [DOI] [PubMed] [Google Scholar]
- 33.Massimi, P., and L. Banks. 2000. Differential phosphorylation of the HPV-16 E7 oncoprotein during cell cycle. Virology 276**:**388-394. [DOI] [PubMed] [Google Scholar]
- 34.McIntyre, M. C., M. N. Ruesch, and L. A. Laimins. 1996. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology 215**:**73-82. [DOI] [PubMed] [Google Scholar]
- 35.McMurray, H. R., D. Nguyen, T. F. Westbrook, and D. J. McCance. 2001. Biology of human papillomaviruses. Int. J. Exp. Pathol. 82**:**15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendez, J., H. Zou-Yang, K. So-Young, M. Hidaka, W. P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9**:**481-491. [DOI] [PubMed] [Google Scholar]
- 37.Montagnoli, A., F. Fiore, E. Eytan, A. C. Carrano, G. F. Draetta, A. Hershko, and M. Pagano. 1999. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 13**:**1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morozov, A., P. Shiyanov, E. Barr, J. M. Leiden, and P. Raychaudhuri. 1997. Human papillomavirus type 16 E7 bypasses G1 arrest induced by serum starvation and by the expression of p21. J. Virol. 71**:**3451-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama, K., H. Nagahama, Y. A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu T. Tsukiyama, N. Ishida, M. Kitagawa, K. Nakayama, and S. Hatakeyama. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27 Kip1, polypoidy and centrosome overduplication. EMBO J. 19**:**2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, H., D. M. Gitig, and A. Koff. 1999. Cell-free degradation of p27kip1, a G1 cyclin-dependent kinase inhibitor, is dependent on CDK2 activity and the proteasome. Mol. Cell. Biol. 19**:**1190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niv, A., N. Sion-Vardi, A. Gatot, M. Nash, and D. M. Fliss. 2000. Identification and typing of human papillomavirus (HPV) in squamous cell carcinoma of the oral cavity and oropharynx. J. Laryngol. Otol. 114**:**41-46. [DOI] [PubMed] [Google Scholar]
- 42.Nuber, U., S. Schwarz, P. Kaiser, R. Schneider, and M. Scheffner. 1996. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J. Biol. Chem. 271**:**2795-2800. [DOI] [PubMed] [Google Scholar]
- 43.Ohta, T., and Y. Xiong. 2001. Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-Cullin ligases. Cancer Res. 61**:**1347-1353. [PubMed] [Google Scholar]
- 44.Pan, W., A. Datta, G. R. Adami, P. Raychaudhuri, and S. Bagchi. 2003. p19 ARF inhibits the functions of the HPV16 E7 oncoprotein. Oncogene 22**:**5496-5503. [DOI] [PubMed] [Google Scholar]
- 45.Reinstein, E., M. Scheffner, M. Oren, A. Ciechanover, and A. Schwartz. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19**:**5944-5950. [DOI] [PubMed] [Google Scholar]
- 46.Rey, O., S. Lee, M. A. Baluda, J. Swee, B. Ackerson, R. Chiu, and N. H. Park. 2000. The E7 oncoprotein of human papillomavirus type 16 interacts with F-actin in vitro and in vivo. Virology 268**:**372-381. [DOI] [PubMed] [Google Scholar]
- 47.Sato, H., S. Watanabe, A. Furuno, and K. Yoshiike. 1989. Human papillomavirus type 16 E7 protein expressed in Escherichia coli and monkey COS-1 cells: immunofluorescence detection of the nuclear E7 protein. Virology 170**:**311-315. [DOI] [PubMed] [Google Scholar]
- 48.Scheffner, M., S. Smith, and S. Jentsch. 1998. The ubiquitin conjugating system, p. 65-98. In J.-M. Peters, J. R. Harris, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum Press, Inc., New York, N.Y.
- 49.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5**:**403-410. [DOI] [PubMed] [Google Scholar]
- 50.Stubenrauch, F., and L. A. Laimins. 1999. Human papillomavirus life cycle: active and latent phases. Semin. Cancer Biol. 9**:**379-386. [DOI] [PubMed] [Google Scholar]
- 51.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimonis. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal keratinocytes. Proc. Natl. Acad. Sci. USA 96**:**8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20**:**2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, J., A. Sampath, P. Raychaudhuri, and S. Bagchi. 2001. Both Rb and E7 are regulated by the ubiquitin proteasome pathway in HPV-containing cervical tumor cells. Oncogene 20**:**4740-4749. [DOI] [PubMed] [Google Scholar]
- 54.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81**:**323-330. [DOI] [PubMed] [Google Scholar]
- 55.Yam, C. H., R. W. Ng, W. Y. Siu, A. W. Lau, and R. Y. Poon. 1999. Regulation of cyclin A-cdk2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol. 19**:**635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, Z. K., J. L. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. USA 95**:**11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102**:**533-539. [DOI] [PubMed] [Google Scholar]
- 58.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2**:**342-350. [DOI] [PubMed] [Google Scholar]
- 59.Zwerschke, W., B. Mannhardt, P. Massimi, S. Nauenburg, D. Pim, W. Nickel, L. Banks, A. J. Reuser, and P. Jansen-Durr. 2000. Allosteric activation of acid alpha-glucosidase by the human papillomavirus E7 protein. J. Biol. Chem. 275**:**9534-9541. [DOI] [PubMed] [Google Scholar]
- 60.Zwerschke, W., S. Mazurek, P. Massimi, L. Banks, E. Eigenbrodt, and P. Jansen-Durr. 1999. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16E7 oncoprotein. Proc. Natl. Acad. Sci. USA 96**:**1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]