Unspliced Rous Sarcoma Virus Genomic RNAs Are Translated and Subjected to Nonsense-Mediated mRNA Decay before Packaging (original) (raw)
Abstract
Retroviruses package full-length, unspliced RNAs into progeny virions as dimerized RNA genomes. They also use unspliced RNAs as mRNAs to produce the gag and pol gene products. We asked whether a single Rous sarcoma virus (RSV) RNA can be translated and subsequently packaged or whether genomic packaging requires a nontranslated population of RNAs. We addressed this issue by utilizing the translation-dependent nonsense-mediated mRNA decay (NMD) pathway. NMD is the selective destruction of mRNAs bearing premature termination codons (PTCs). The pathway has been shown to be associated with splicing in higher eukaryotes. Here, we demonstrate that both translation and the cellular factor Upf1 are required for the decay of unspliced, PTC-bearing RSV RNA by the NMD pathway. To address the relationship between RNA translation and packaging, we examined virus produced in cells cotransfected with PTC-bearing retroviral clones and wild-type viral clones. We observed that PTC-bearing transcripts are packaged into viral particles at levels three- to fivefold less than those of control RNAs. Since PTC-mediated degradation requires translation, we conclude that RSV can package progeny virion particles using previously translated RNAs.
After reverse transcription and integration of their genomes, retroviruses use cellular machinery to produce new viral RNA and protein products (39, 46). Some of these viral RNAs are exported from the nucleus without first being spliced. Complex retroviruses export their unspliced RNA with the help of virus-encoded accessory proteins, such as Rev for human immunodeficiency virus type 1 (HIV-1) (28, 29). Simple retroviruses use _cis_-acting RNA elements, such as the constitutive transport element (CTE) for Mason-Pfizer monkey virus (9) or the direct repeats for Rous sarcoma virus (RSV) (37, 38), which recruit cellular export factors to the transcripts (8). After export, either these unspliced RNAs can be translated to produce Gag and Gag-Pol polyproteins (46) or they can be dimerized and packaged as genomes in progeny virions (6, 17).
Currently, there is no unified view regarding the source of retroviral RNAs that are packaged in virions. Levin and colleagues (21, 22) studied the effects of actinomycin D (actD) on Rauscher murine leukemia virus (MLV) full-length RNA metabolism. After 6 h of treatment with actD to inhibit RNA synthesis, MLV-infected cells still make viral proteins and bud viral particles. However, packaging of genomic RNA into these particles drops 10-fold, suggesting that the population of unspliced viral RNAs used for packaging has been depleted. Translation persists because the viral mRNA is stable in the cytoplasm. This suggests that MLV employs two functionally distinct pools of unspliced RNA.
In contrast to MLV, HIV-1 and HIV-2 continue to package genomic RNAs in infected cells after treatment with actD, apparently due to a single population of RNAs that can function interchangeably as mRNA and genomic virion RNA (10, 16). Further, neither active translation nor prior translation of the genomic RNA is a prerequisite for HIV-1 RNA packaging (10, 16). However, HIV-2 encapsidates genomic RNAs cotranslationally, favoring the cis packaging of RNAs that have been translated (16).
Through actD treatment of cells containing radiolabeled RNAs, Stoltzfus et al. (44) determined the half-life of unspliced RSV RNAs to be 7.5 h both in the cytoplasm and in virions. Thus, they propose that RSV also employs a single population of RNAs to serve as genomic RNAs as well as mRNAs. In contrast, Sonstegard and Hackett (43) propose that ribosomes and the Gag precursor protein compete for binding to the third small open reading frame (uORF3) upstream of the gag gene. Translation of gag results in an increase in Gag protein concentration within the cell, eventually leading to its binding at uORF3, which contains part of the encapsidation signal, Ψ (1, 3, 24); this results in increased RNA packaging. In this model, polyribosome-bound RNAs are not substrates for packaging, and RSV's unspliced RNA is segregated into two functionally distinct pools (43).
To clarify the issue of whether translated RSV RNAs are packaged, we used a conserved cellular pathway known as nonsense-mediated mRNA decay (NMD). NMD is the selective destruction of mRNA transcripts bearing premature termination codons (PTCs) (25, 30, 32, 36). This pathway serves as a cellular quality control mechanism to ensure that mRNAs are processed properly. It has been estimated that 30% of all genetic disorders are caused by PTCs (13). The NMD pathway is directly coupled to the process of translation, and inhibition of translation blocks NMD (7, 12).
The NMD pathway is also associated with splicing (48, 49). Concurrent with the splicing event, a protein complex is deposited on the mRNA 20 to 24 nucleotides upstream of every exon junction (19). This complex, known as the exon junction complex (EJC), indicates the splicing history of each mRNA (20). During translation, ribosomes displace these complexes as they traverse the mRNA. When a ribosome encounters a termination codon located upstream of an EJC, the mRNA is degraded (25, 30). In support of the EJC-mediated model, naturally intronless genes, such as those for histone H4 and Hsp70, are immune to decay despite the presence of PTCs (31).
Earlier reports from our lab (2, 4, 5), which predate the proposed EJC-mediated model for NMD, demonstrate that nonsense codons at various positions in the gag gene of RSV lead to instability of the unspliced RNA in primary chicken embryo fibroblasts (CEFs). In light of recent findings linking NMD to splicing, this result is somewhat surprising, since unspliced RSV RNAs would have been predicted to be immune to decay. This investigation sought to determine if the PTC-induced instability was actually due to NMD. First, using a dominant-negative mutant of hUpf1 that abrogates NMD (45), we demonstrated the stabilization of RSV RNAs bearing PTCs. Secondly, inhibition of translation also led to stabilization of these RNAs. These results suggest that unspliced viral RNAs with PTCs are substrates for the NMD pathway.
We next examined whether translated RSV RNAs could subsequently be packaged. Analysis of packaged viral RNAs, produced in cells cotransfected with wild-type and mutant viral constructs, showed that PTC-bearing RNAs were packaged in progeny virions at a level three- to fivefold lower than that of RNAs bearing control codon insertions. This reduction in viral RNA levels is similar to that seen in cells as a result of NMD. If the packaged RNAs were not translated, they would not be subject to NMD, and no reduction in packaged levels would be expected. Since these PTC-bearing RNAs were subjected to NMD, and therefore at least one round of translation (7, 12), we conclude that RSV is able to package genomic RNAs that have previously been translated.
MATERIALS AND METHODS
Cell culture and transfection.
Secondary cultures of CEFs were maintained in medium 199 (Gibco) supplemented with 2% tryptose phosphate broth (Sigma), 1% chick serum, 1% calf serum, and 1% penicillin-streptomycin (Gibco). Cells were grown at 39°C in the presence of 5% carbon dioxide. Transfections were performed using 200 μg of DEAE-dextran/ml in serum-free medium 199, as described previously (38), with cells that were approximately 80**%** confluent. Transfections done in 6-cm plates (see Fig. 1, 2, and 3) used 4 to 6 μg of DNA in 1 ml of medium 199 containing DEAE-dextran. Transfections done in 10-cm plates used 10 μg of DNA in 3 ml of medium 199 containing DEAE-dextran. Cells were incubated for 4 h at 39°C and then shocked with serum-free medium 199 containing 10% dimethyl sulfoxide for 2 min at room temperature. Cells were then washed twice with serum-free medium 199. Anisomycin (Calbiochem) was used to inhibit cellular translation at a final concentration of 100 μg/ml in serum-containing medium 199.
FIG. 1.
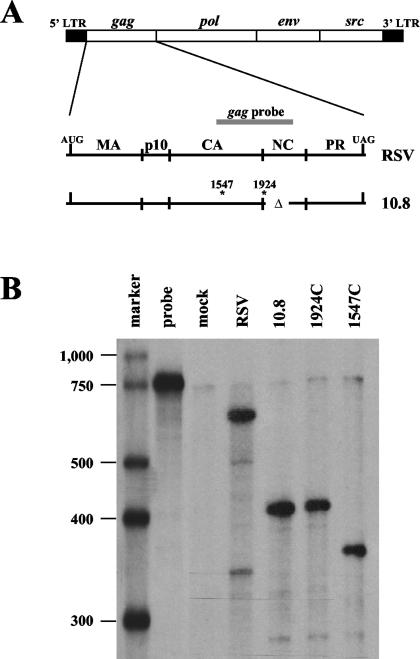
A single antisense riboprobe complementary to RSV gag can differentiate between various mutant RNA species. (A) A detailed schematic of the integrated RSV genome with the gag region enlarged. The gag riboprobe (gray bar) spans nt 1494 to 2121. The deletion in nucleocapsid (nt 1924 to 2047) is indicated by Δ. Sites of oligonucleotide insertions are indicated by asterisks along with their nucleotide number. LTR, long terminal repeat; MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease. (B) RNase protection assay of RNAs isolated from transiently transfected CEFs using the gag probe. 1924C and 1547C lanes represent CEFs transfected with retroviral constructs bearing linker insertions with control codons.
FIG. 2.
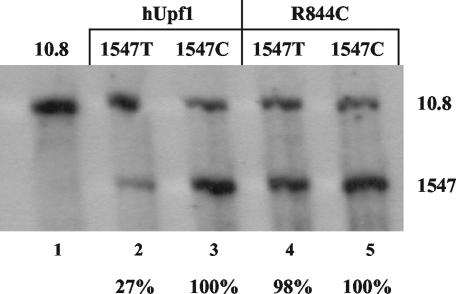
The decay of unspliced RNAs bearing premature termination codons is dependent upon Upf1. CEFs were transiently transfected with equal amounts of three DNA constructs: 10.8 (as a loading control), the retroviral clones with a PTC (1547T) or control codon (1547C) insertion, and either wild-type hUpf1 or a dominant-negative mutant (R844C). CEFs were maintained for 48 h and then harvested for RNA. RNA analysis was done by RPA using the gag probe (shown in Fig. 1A). Percentages indicated represent RNA levels of 1547T RNAs normalized to the respective 1547C RNA (with either hUpf1 or R844C). Results are from five independently performed transfections.
FIG. 3.
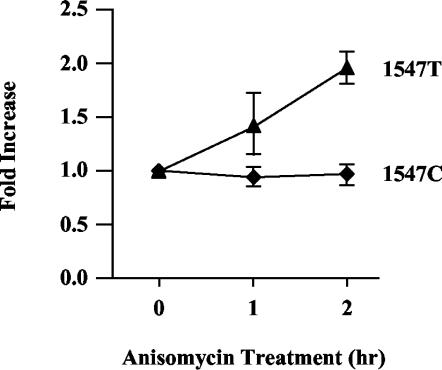
The decay of unspliced RNAs bearing premature termination codons requires translation. CEFs were transiently cotransfected with 1.5 μg of wild-type RSV (as a loading control) and 3 μg of either 1547T or 1547C. Two days later, cells were treated with 100 μg of anisomycin/ml and then harvested for RNA at the indicated times. RNA analysis was carried out by RPA using the gag probe (see Fig. 1A). The ratio of 1547 RNA to RSV RNA was determined, and the ratios were compared to those of samples without anisomycin treatment (0 h). 1547T RNA levels (▴) and 1547C RNA levels (♦) are indicated. Results are from three independently performed transfections.
RNA isolation and RNase protection assays.
Total cellular RNA was harvested 40 to 48 h posttransfection using RNA-Bee (Tel-Test) according to the manufacturer's instructions. The gag probe was generated in vitro using viral sequences under the control of a T7 promoter and incorporating [α-32P]CTP (Perkin-Elmer). RNAs were precipitated and resuspended in 30 μl of 80% formamide hybridization solution (80% [vol/vol] deionized formamide, 40 mM piperazine-N,_N_′-bis(2-ethanesulfonic acid) [pH 6.7], 0.4 mM NaCl, 1 mM EDTA). The gag riboprobe (250,000 cpm) (see Fig. 1A) was added to each sample. RNAs were denatured at 95°C for 5 min and incubated for 16 to 20 h at 42°C. Three hundred microliters of RNase digestion buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA) supplemented with 10 U of RNase T1/ml and 5 μg of RNase A/ml (both from Calbiochem) was added, followed by incubation at 37°C for 1 h. Digestion was halted by the addition of sodium dodecyl sulfate (SDS) and proteinase K (Roche) at final concentrations of 0.6% (vol/vol) and 0.14 mg/ml, respectively. After incubation at 37°C for 15 min, samples were extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with ethanol. RNAs were resuspended in 95% formamide loading dye (95% [vol/vol] deionized formamide, 0.02% bromophenol blue, 0.02% xylene cyanol), denatured for 5 min at 95°C, and incubated on ice for at least 2 min. Samples were loaded on a 6% acrylamide-8 M urea gel and electrophoresed. RNA levels were quantified using an InstantImager (Packard).
Virus purification.
Viral particles were recovered from the culture media approximately 48 h after transfection, after first centrifuging at 2,000 rpm for 10 min in a Damon/IEC clinical centrifuge to pellet cellular debris. The supernatant was then divided into two equal aliquots and subjected to ultracentrifugation for 2 h at 4°C in a Beckman Ultracentrifuge, using an SW-41 rotor at 34,000 rpm. One of each pair of viral pellets was resuspended in RNA-Bee (Tel-Test) for RNA isolation. The other viral pellet was resuspended in Laemmli protein sample buffer (0.05 M Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 10% β-mercaptoethanol) and heated to 95°C for 2 min.
Western blot analysis.
Cells were removed from the plates by incubation with trypsin (Gibco) and pelleted. Both total cellular proteins and virion proteins were subjected to SDS-polyacrylamide gel electrophoresis, using a 10% acrylamide resolving gel. After electrophoresis, proteins were transblotted to Hybond nitrocellulose (Amersham). The membrane was then blocked in phosphate-buffered saline-Tween (0.05%) with 5% dry milk for 1 h at room temperature. RSV Gag proteins were detected using polyclonal rabbit anti-avian myeloblastosis virus p19_gag_ serum, obtained from D. P. Bolognesi (Duke University), and ImmunoPure goat anti-rabbit immunoglobulin G (heavy plus light chains, horseradish peroxidase conjugated; Pierce). Blots were then visualized by autoradiography using the Enhanced Chemiluminescence reagent kit (Amersham).
RESULTS
Generation of a riboprobe that can discriminate between multiple species of mutated RSV RNAs.
Earlier reports from our lab showed that unspliced RSV RNAs are destabilized when PTCs are present in the gag gene (4, 5). This work preceded reports that linked NMD to splicing (48, 49). Since naturally intronless cellular mRNAs are immune to NMD (31), we asked if the instability of the retroviral RNA was due to NMD or to another mechanism.
For this study, we used the Prague C strain of RSV. Mutants were created by inserting oligonucleotide linkers at nucleotide (nt) 1547 or 1924, both located within the gag gene, which has a natural termination codon at nt 2483. These oligonucleotide inserts contained either premature termination codons (1547T and 1924T) or control codons (1547C and 1924C), as described by Barker and Beemon (5). The inserts were placed into a viral vector, termed 10.8 (34), that contains a partial deletion of nucleocapsid (NC) (Fig. 1A). This deletion removes the zinc finger domains of NC and blocks the ability of the 10.8 Gag product to bind and package the retroviral RNA. This allows analysis of steady-state viral RNAs in the cell without the loss of RNAs due to packaging into progeny virions.
We generated a 770-nt 32P-labeled antisense riboprobe that is complementary to nt 1494 to 2121 of wild-type RSV RNA (gag probe) (Fig. 1A). This probe spans both sites of oligonucleotide insertion and also the NC deletion site. To test this probe's ability to distinguish each of these RNAs from one another, CEFs were transiently transfected with retrovirus clones bearing oligonucleotide insertions that lacked PTCs. After 48 h, total cellular RNA was harvested and analyzed by RNase protection assay (RPA).
We found that the probe was able to discriminate the viral RNAs from one another due to probe cleavage at the site of oligonucleotide insertion as well as at the nucleocapsid deletion (Fig. 1B). The wild-type construct (RSV) yielded a protected product of 627 nt. The 10.8 and 1924C constructs yielded a major product of 430 nt (protecting from nt 1494 to 1924). The 1547C construct allowed probe cleavage at the NC deletion as well as cleavage at its unique oligonucleotide insertion site, yielding a major product of 387 nt (1547 to 1924). Since the constructs bearing PTCs (1547T and 1924T) have insertions at the same sites as their control codon counterparts, they generated the same-size products using this probe (data not shown), which was used in RPAs throughout this study.
Unspliced RSV RNAs bearing PTCs are substrates for the NMD pathway.
We first wished to determine if Upf1, a vital cellular protein component of the NMD pathway (18, 26, 27), is involved in the instability of PTC-bearing RSV RNAs. To this end, we obtained a dominant-negative clone of human Upf1 (hUpf1) called R844C, which contains an arginine-to-cysteine mutation in its RNA helicase domain that abrogates NMD (45). CEFs were transiently cotransfected with wild-type hUpf1 or the R844C mutant and either the 1547T or 1547C viral construct. The 10.8 construct was also cotransfected for the purpose of normalization. After 48 h, total cellular RNA was harvested and analyzed by RPA (Fig. 2). When coexpressed with wild-type hUpf1, 1547T RNA levels were reduced to 27% of 1547C expression (Fig. 2, lanes 2 and 3), similar to the reduction seen previously without wild-type hUpf1 overexpression (4, 5). In contrast, the expression of 1547T was 98% of that of 1547C when both were expressed with R844C (Fig. 2, lanes 4 and 5). There was not a significant difference in the expression pattern of 1547C when it was expressed with either hUpf1 or R844C, demonstrating that the Upf1 mutant does not affect non-PTC-bearing RSV transcripts. These results implicate Upf1 involvement in the decay of unspliced retroviral RNAs bearing PTCs. Since NMD requires Upf1, we conclude that the instability is likely due to NMD.
To confirm and extend this result, we also examined the effect of translation inhibition on RSV RNA stability. Since translation is required for the NMD pathway (7, 12), its inhibition should lead to the stabilization of transcripts bearing PTCs. By measuring [35S]methionine incorporation, we observed that 98% of all cellular translation was inhibited in CEFs treated with 100 μg of anisomycin/ml for 1 h (data not shown). To test the effect of anisomycin treatment on expression of PTC-bearing viral RNAs, CEFs were transiently transfected with 1.5 μg of wild-type RSV DNA (as a loading control) and 3 μg of either the 1547T or 1547C viral DNA construct. Cells were maintained for 48 h and then treated with 100 μg of anisomycin/ml. RNA was harvested 1 and 2 h post-drug treatment and assayed by RPA. Expression of the 1547T and 1547C RNAs relative to the RSV loading control was quantified. The untreated samples (0 h) were given a value of 1.0, and the expression of samples treated with anisomycin for 1 or 2 h was determined relative to the respective untreated samples (Fig. 3). Data were averaged from three experiments. While the anisomycin treatment did not affect the stability of nonsense-free transcripts (1547C), the stability of PTC-bearing RNAs (1547T) was reproducibly increased twofold over the course of 2 h of drug exposure (Fig. 3). This result suggests that translation is required for the decay of unspliced retroviral RNAs bearing nonsense codons. In combination with the dominant-negative hUpf1 experiment, this demonstrates that unspliced viral RNAs bearing PTCs are substrates for the NMD pathway.
Use of NMD to determine if virion RNAs have been previously translated.
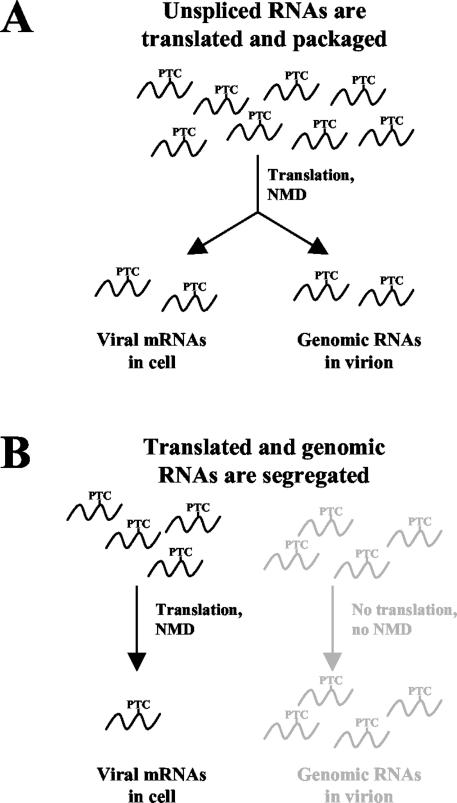
If viral RNAs used for Gag translation and packaging are the same, meaning they are capable of being both translated and packaged, they will all be subjected to the NMD pathway (Fig. 4A). Packaged RNAs should be sensitive to NMD, and this effect should be seen in viral particles. In other words, viral RNAs with PTCs would be found packaged at a lower level than non-PTC-bearing transcripts because they undergo NMD.
FIG. 4.
Using nonsense-mediated mRNA decay to determine if RSV packages previously translated RNA genomes. (A) If RSV packages RNAs that are translated, the NMD effect (degradation of PTC-containing RNA) should be seen both in the cell and in progeny virions. (B) Segregation into two populations means that certain unspliced RNAs are specific for translating viral proteins while others are specific for encapsidation. Since the RNAs are restricted to either the translation or the packaging activities, the NMD effect should be seen only in the cellular RNAs. Virion RNAs would be unaffected by PTCs since translation would not take place.
The alternative hypothesis is presented in Fig. 4B. According to this translation-independent model, RNAs are specifically segregated into two populations: one for the translation of viral proteins and one for the genomic packaging of viral particles. Since the RNAs never cross from one population to the other, the genomic RNAs would not be translated, NMD could not occur, and PTC-bearing viral RNAs would be immune to decay. As such, the amount of PTC-bearing RNA packaged into new viral particles should be equal to the amount of non-PTC-bearing RNAs.
Unspliced RSV RNAs bearing PTCs remain unstable in the presence of wild-type Gag.
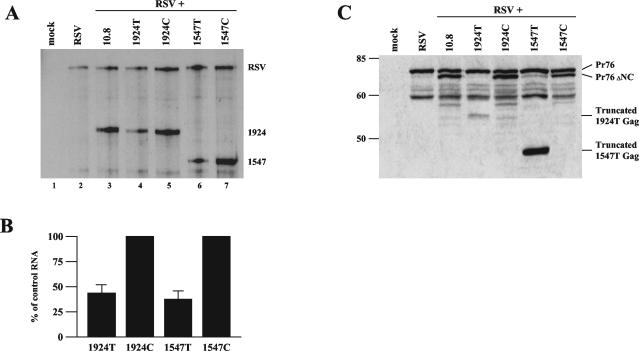
It was previously shown that expression of PTC-bearing RSV RNAs could be partially suppressed when cotransfected with a sixfold molar excess of a wild-type gag minigene (4). To allow for viral packaging of the NC deletion constructs, we transiently transfected CEFs with equal amounts of wild-type RSV and the NC deletion (10.8) clones with or without PTCs. Cotransfection of wild-type RSV allows packaging of the mutant RNAs by supplying wild-type Gag protein in trans. In addition, the wild-type RNA serves as a transfection and loading control. After 2 days, the medium was harvested and cells were treated with trypsin to remove budding viral particles. Total cellular RNA was then isolated and analyzed by RPA (Fig. 5A). Using RSV as a loading control reference, 1547T and 1924T viral RNAs were specifically down-regulated to 36 and 42%, respectively, of their non-PTC-containing counterparts, 1547C and 1924C (compare lanes 6 and 7 and lanes 4 and 5).
FIG. 5.
Decay of unspliced retroviral transcripts with nonsense codons occurs in the presence of wild-type Gag. CEFs in 10-cm plates were transiently transfected with equivalent amounts (5 μg) of the wild-type and mutant RSV clones indicated. (A) Total cellular RNA was harvested and analyzed by RPA using the gag probe. (B) Bar graph quantification of 1924T and 1547T RNA levels normalized to their control codon counterpart, set at 100%. Results are from six independently performed transfections. (C) Western blot analysis of intracellular viral proteins. Cells were trypsinized, resuspended in 1× Laemmli buffer, and used for SDS-polyacrylamide gel electrophoresis. Antibodies were targeted against the viral MA protein.
Additionally, we examined the intracellular viral proteins that were being generated by the wild-type and mutant viral clones. Western blot analysis was carried out, using an antibody against the matrix (MA) protein, which is at the N terminus of the Gag polyprotein (Fig. 5C). We found that Pr76gag, the Gag polyprotein precursor, is produced in all samples, while a slightly smaller Gag product (Pr76ΔNC) is made in the 10.8 samples, including 1547C and 1924C, with only trace amounts in 1547T and 1924T. A minor amount of the 1924T truncated protein product was detected, consistent with the decay of its corresponding mRNA. Interestingly, the truncated product from 1547T was consistently expressed at a much higher level than the other Gag proteins. This is puzzling, since the amount of protein produced does not correlate with the reduced 1547T mRNA expression level seen (Fig. 5A). This smaller truncated product may not be suitable for use in the assembly of progeny virions since it does not contain a full capsid (CA) domain, and thus, the 1547T Gag precursor may simply accumulate in the cell. In contrast, the 1924T products, which terminate after the CA domain, may be assembled into hybrid particles with wild-type Gag proteins.
These results confirm that the NMD pathway, even in the presence of wild-type Gag protein, selectively degrades unspliced viral transcripts bearing PTCs. Having established that the NMD effect was not suppressed with wild-type Gag expression, we can now examine the effects of PTCs on viral packaging.
RSV genomic RNAs are sensitive to NMD, indicating prior translation has occurred.
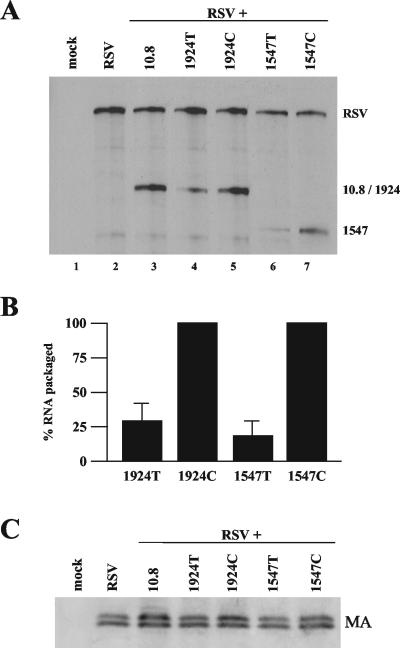
The virion particles present in the supernatant from the previous experiment (Fig. 5) were purified for analysis of both viral RNA and protein levels. The supernatant was split into two equal portions. Viral particles in each were pelleted by ultracentrifugation. One half was harvested for RNA, which was then analyzed by RPA using the gag riboprobe (Fig. 6A). NC deletion 10.8 RNAs were packaged efficiently into viral particles, indicating no preference for cis packaging (Fig. 6B, lane 3). To test for potential differences in virus production, the other half was used for Western blot analysis, employing antibodies directed against MA (Fig. 6C). No appreciable difference in viral MA levels produced from the different constructs was observed by use of Western blotting (Fig. 6C).
FIG. 6.
RSV packages genomic RNAs that have been translated. (A) A representative RPA, using the gag probe, of RNA extracted from pelleted viral particles from the medium of the CEFs transfected as described for Fig. 5. (B) Bar graph quantification of RPA data from six independent experiments. 1924T and 1547T RNA levels were normalized to their control codon counterpart, set at 100%. Results are from six independently performed transfections. (C) Western blot analysis of viral particles, using antibodies targeted against the viral MA protein.
Further, 1547T and 1924T RNAs were packaged in virions 19 and 30% as often as their non-PTC-bearing counterparts 1547C and 1924C (Fig. 6A, compare lanes 6 and 7 and lanes 4 and 5). These results are similar to the relative amounts of PTC-containing and -lacking RNAs observed within the cell (Fig. 5). The reduced amount of packaging of PTC-bearing RNAs relative to that of their control codon counterparts suggests that most of the genomic RNAs that RSV packages are first translated and then packaged as depicted in the model in Fig. 4A.
Some (19 to 30%) of the PTC-bearing viral RNAs were not degraded by the NMD pathway and were packaged into progeny virion particles. It is possible that this RNA was never translated, and that is why it was not degraded. If so, these RNAs represent the amount of genomic RNA that RSV packages without translation. Therefore, the remaining 70 to 80% of the non-PTC-bearing RNAs that are packaged represent RNAs that have been translated and subsequently packaged as genomes. We therefore conclude that most of the genomic RNAs that RSV packages have been translated previously, at least once. However, a small fraction of the genomic RNAs could be transcribed and packaged directly without being first translated.
DISCUSSION
This study focuses on two distinct events that occur within the cell regarding the metabolism of retroviral RNA. Our first finding is that unspliced RSV transcripts are subject to NMD, a process that was previously reported to require a splicing event in higher eukaryotes (48, 49). The second is that RSV uses the same unspliced RNAs for both translation and packaging. Since PTC-bearing RNAs are not packaged as frequently as non-PTC-bearing RNAs, we conclude that most packaged RNAs are first translated.
RSV unspliced RNA undergoes nonsense-mediated mRNA decay.
The first half of the study examines the susceptibility of unspliced retroviral RNAs to NMD. We demonstrated that the decay requires Upf1, a protein known to be essential for NMD (18, 26, 27). Additionally, we showed that this decay was dependent upon translation, another required event in the NMD pathway (7, 12). This evidence is consistent with the NMD pathway being responsible for the decay of RSV RNA. However, it remains to be understood how this retroviral RNA is vulnerable to this decay, since NMD is thought to require a splicing event (31, 48, 49).
In mammalian systems, naturally intronless genes are immune to NMD, and NMD is restored when an actively spliced intron is inserted into the gene (31). Further, splicing results in the deposition of an EJC upstream of every splice junction (19, 20). Translating ribosomes subsequently displace EJCs. The failure of the ribosome to remove EJCs bound to the transcript because of premature termination activates NMD (32). Analogously, it was previously reported that a PTC 165 nt upstream of the natural termination codon for the RSV gag gene caused decay while one only 30 nt upstream did not (5). If EJC components (or some other unknown NMD-promoting complex) are recruited to the unspliced RNA messages, at least one complex is likely located within this 135-nt region upstream of the natural termination codon.
Alternatively, the virus may utilize a novel pathway that does not require splicing but does require the activity of Upf1. Gatfield et al. (15) recently demonstrated that unspliced messages in Drosophila undergo NMD; further, this decay requires all three Upf proteins but does not require EJC components. Intronless HEXA minigenes transfected into Chinese hamster ovary cells are also susceptible to NMD (40). It was also found that PTCs within the src gene of RSV, introduced by frameshift mutations, caused down-regulation of src mRNA (42). This is particularly interesting since the src coding region, including its initiating AUG, lies completely downstream of the splice junction. These examples, as well as our data regarding unspliced RSV RNA, indicate that NMD may function by utilizing more than one specific recognition mechanism. The idea that Upf1, but not the EJC proteins, is involved is possible given that Upf1 has separable roles in the cell (33). Since RSV uses an unknown nuclear export pathway for unspliced RNAs (38), NMD-promoting factors that are recognized by Upf1 may be added during export rather than during splicing. It will be of interest to determine the composition and location of this NMD-promoting complex, as well as how Upf1 interacts with it.
In this study, and in most others, the effect of NMD is not absolute; PTC-bearing transcripts are not fully turned over (5, 26, 31, 40). There are several possible reasons for this. One possibility is that a fraction of the RNAs may not be translated, allowing them to escape decay. Second, the cellular site of NMD seems to vary from transcript to transcript. Some reports indicate that NMD occurs in association with the nucleus (23), while others indicate cytoplasmic NMD (5, 40). If NMD occurs in the cytoplasm, the examination of total cellular RNA levels will include nuclear RNA fractions that have yet to be subjected to NMD. Interestingly, it was reported that some RNAs bearing PTCs accumulate at the site of transcription in the nucleus (35). Third, the NMD pathway may be inefficient, allowing some PTC-bearing transcripts it detects to escape decay.
Genomic RSV RNAs are translated prior to packaging.
The second half of this report employed the translational dependency of NMD to investigate whether retroviral genomic RNAs are translated prior to packaging. PTC-bearing RNAs were packaged into progeny virions at levels three- to fivefold less than those of non-PTC-bearing RNAs, indicating that the genomic RNAs were subjected to the NMD pathway and were therefore translated prior to packaging. Consistent with a previous report (44), we conclude that the RNAs used to package RSV virions come from the same pool of RNAs that are translated to synthesize the structural and enzymatic viral proteins. Observations supporting this same idea have been described for HIV-1 (10, 11, 14). It should be noted that this conclusion is not universal and does not apply to all retroviruses, since MLV packages from two distinctly segregated pools (14, 21, 22).
In contrast to our findings, Sonstegard and Hackett (43) propose that RSV RNA is segregated into two populations, based on competition between Pr76gag and ribosome binding to uORF3. However, a more recent report consistent with our findings dispels the relationship between packaging and translation. Banks and Linial (3) report that the translation of uORF3 plays a minimal role in packaging. They propose that the uORF3 initiation codon is critical for packaging but not for translation.
Parent and colleagues (41) provide evidence depicting a transient localization of the Gag precursor protein to the nucleus, suggesting that nuclear Gag-RNA interactions may exist in transformed quail cells. They report that blocking Gag entry into the nucleus interferes with proper packaging of genomic dimers. Since RSV RNAs are subject to NMD (this study) in the cytoplasmic fraction of CEFs (5), we suggest that they are translated before packaging. Primary CEFs may support viral packaging and localize Gag differently than transformed quail cells. Based on our results, we believe that unspliced RNAs are first exported to the cytoplasm, translated, and then made available for packaging. However, it is possible that Gag localization in the nucleus could result in direct packaging, thereby shielding the RNA from translational machineries.
Our study also provides some insight into cis versus trans packaging. Our study used packaging-deficient viral clones bearing NC deletions. These RNAs encode Gag proteins incapable of binding and packaging viral RNAs. However, coexpression of wild-type clones in CEFs yielded viral progeny with RNAs containing the NC deletion, demonstrating that trans packaging for RSV is very efficient (Fig. 6B). Kaye and Lever (16) present a contrary model for HIV-2, suggesting that RNAs are packaged cotranslationally, with cis packaging being favored.
Our evidence provides viral strategies that may serve to promote the production of error-free infectious particles. Since the retroviral reverse transcriptase is error prone (47), the introduction of mutations, including PTCs, into the unspliced transcripts is probable. Through the NMD pathway, the virus confers a selective advantage to RNAs without PTCs. As a result, this will increase the concentration of Gag proteins that become structural proteins in progeny virions. Second, requiring genomic viral RNAs to be translated prior to packaging provides a quality control mechanism for the virus to help promote successful infection. By packaging RNAs that have been translated and therefore subjected to NMD, the virus promotes the selection of genomic RNAs lacking gag PTCs that will be used to infect new host cells.
Acknowledgments
This work was supported by NIH grant R01 CA48746. J.J.L. was supported in part by NIH predoctoral training grant T32GM07231.
We thank D. P. Bolognesi for the p19 antibody and Hal Dietz for the hUpf1 (Rent1) and R844C plasmids and for helpful discussions. We also thank Jason Weil and Tatjana Polony for reviews of the manuscript.
REFERENCES
- 1.Aronoff, R., and M. Linial. 1991. Specificity of retroviral RNA packaging. J. Virol. 65**:**71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 8**:**4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, J. D., and M. L. Linial. 2000. Secondary structure analysis of a minimal avian-leukosis-sarcoma virus packaging signal. J. Virol. 74**:**456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, G. F., and K. Beemon. 1991. Nonsense codons within the Rous sarcoma virus gag gene decrease stability of unspliced viral RNA. Mol. Cell. Biol. 11**:**2760-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, G. F., and K. Beemon. 1994. Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol. Cell. Biol. 14**:**1986-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beemon, K., P. Duesberg, and P. Vogt. 1974. Evidence for crossing-over between avian tumor viruses based on analysis of viral RNAs. Proc. Natl. Acad. Sci. USA 71**:**4254-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belgrader, P., J. Cheng, and L. E. Maquat. 1993. Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosphosphate isomerase mRNA. Proc. Natl. Acad. Sci. USA 90**:**482-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, I. C., E. Rohrbach, C. Schmitt, and E. Izaurralde. 1999. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 18**:**1953-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M. L. Hammarskjold. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 91**:**1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butsch, M., and K. Boris-Lawrie. 2000. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type-1-infected T cells. J. Virol. 74**:**11531-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butsch, M., and K. Boris-Lawrie. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 76**:**3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter, M. S., J. Doskow, P. Morris, S. Li, R. P. Nhim, S. Sandstedt, and M. F. Wilkinson. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270**:**28995-29003. [DOI] [PubMed] [Google Scholar]
- 13.Dietz, H. C. 1997. Nonsense mutations and altered splice-site selection. Am. J. Hum. Genet. 60**:**729-730. [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman, N., and A. Lever. 2000. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 74**:**11413-11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork, and E. Izaurralde. 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22**:**3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaye, J. F., and A. M. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73**:**3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kung, H. J., J. M. Bailey, N. Davidson, P. K. Vogt, M. O. Nicolson, and R. M. McAllister. 1975. Electron microscope studies of tumor virus RNA. Cold Spring Harb. Symp. Quant. Biol. 39**:**827-834. [DOI] [PubMed] [Google Scholar]
- 18.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5**:**2303-2314. [DOI] [PubMed] [Google Scholar]
- 19.Le Hir, H., M. J. Moore, and L. E. Maquat. 2000. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 14**:**1098-1108. [PMC free article] [PubMed] [Google Scholar]
- 20.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins upstream of mRNA exon-exon junctions. EMBO J. 19**:**6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin, J. G., P. M. Grimley, J. M. Ramseur, and I. K. Berezesky. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14**:**152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin, J. G., and M. J. Rosenak. 1976. Synthesis of murine leukemia virus proteins association with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 73**:**1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, S., D. Leonard, and M. F. Wilkinson. 1997. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med. 185**:**985-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linial, M., E. Medeiros, and S. W. Hayward. 1978. An avian oncovirus mutant (_SE_21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell 15**:**1371-1381. [DOI] [PubMed] [Google Scholar]
- 25.Lykke-Andersen, J. 2001. mRNA quality control: marking the message for life or death. Curr. Biol. 11**:**R88-R91. [DOI] [PubMed] [Google Scholar]
- 26.Lykke-Andersen, J., M.-D. Shu, and J. A. Steitz. 2000. Human Upf proteins target an mRNA for nonsense-mediated mRNA decay when bound downstream of a termination codon. Cell 103**:**1121-1131. [DOI] [PubMed] [Google Scholar]
- 27.Maderazo, A. B., F. He, D. A. Mangus, and A. Jacobson. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20**:**4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maize, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338**:**254-257. [DOI] [PubMed] [Google Scholar]
- 29.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60**:**675-683. [DOI] [PubMed] [Google Scholar]
- 30.Maquat, L. E., and G. C. Carmichael. 2001. Quality control of mRNA function. Cell 104**:**173-176. [DOI] [PubMed] [Google Scholar]
- 31.Maquat, L. E., and X. Li. 2001. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA 7**:**445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendell, J. T., and H. C. Dietz. 2001. When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell 107**:**411-414. [DOI] [PubMed] [Google Scholar]
- 33.Mendell, J. T., C. M. J. ap Rhys, and H. C. Dietz. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298**:**419-422. [DOI] [PubMed] [Google Scholar]
- 34.Meric, C., and P. F. Spahr. 1986. Rous sarcoma virus nucleic acid binding protein p12 is necessary for viral 70S dimer formation and packaging. J. Virol. 60**:**450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muhlemann, O., C. S. Mock-Casagrande, J. Wang, S. Li, N. Custodio, M. Carmo-Fonseca, M. F. Wilkinson, and M. J. Moore. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8**:**33-43. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, E., and L. E. Maquat. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23**:**198-199. [DOI] [PubMed] [Google Scholar]
- 37.Ogert, R. A., L. H. Lee, and K. L. Beemon. 1996. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 70**:**3834-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paca, R. E., R. E. Ogert, C. S. Hibbert, E. Izaurralde, and K. L. Beemon. 2000. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 74**:**9507-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabson, A. B., and B. J. Graves. 1997. Synthesis and processing of viral RNA, p. 205-262. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Woodbury, N.Y. [PubMed]
- 40.Rajavel, K. S., and E. F. Neufeld. 2001. Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 21**:**5512-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheifele, L. Z., R. A. Garbitt, J. D. Rhodes, and L. J. Parent. 2002. Nuclear entry and CRM-1 dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99**:**3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson, S. B., and C. M. Stoltzfus. 1994. Frameshift mutations in the v-src gene of avian sarcoma virus act in cis to specifically reduce v-src mRNA levels. J. Virol. 14**:**1835-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonstegard, T. S., and P. B. Hackett. 1996. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 70**:**6642-6652. [PMC free article] [PubMed] [Google Scholar]
- 44.Stoltzfus, C. M., K. Dimock, S. Horikami, and T. A. Ficht. 1983. Stabilities of avian sarcoma virus RNAs: comparison of subgenomic and genomic species with cellular mRNAs. J. Gen. Virol. 64**:**2191-2202. [DOI] [PubMed] [Google Scholar]
- 45.Sun, X., H. A. Perlick, H. C. Dietz, and L. E. Maquat. 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 95**:**10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral proteins. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Woodbury, N.Y. [PubMed]
- 47.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Woodbury, N.Y. [PubMed]
- 48.Zhang, J., and L. E. Maquat. 1996. Evidence that the decay of nucleus-associated nonsense mRNA for human triosephosphate isomerase involves nonsense codon recognition after splicing. RNA 2**:**235-243. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, J., X. Sun, Y. Qian, J. P. LaDuca, and L. E. Maquat. 1998. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 18**:**5272-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]