Alveolar progenitor and stem cells in lung development, renewal and cancer (original) (raw)
. Author manuscript; available in PMC: 2014 Sep 13.
Published in final edited form as: Nature. 2014 Feb 5;507(7491):190–194. doi: 10.1038/nature12930
Abstract
Alveoli are gas-exchange sacs lined by squamous alveolar type (AT) 1 cells and cuboidal, surfactant-secreting AT2 cells. Classical studies suggested AT1 arise from AT2 cells, but recent studies propose other sources. Here we use molecular markers, lineage tracing, and clonal analysis to map alveolar progenitors throughout the mouse lifespan. We show that during development AT1 and AT2 cells arise directly from a bipotent progenitor, whereas after birth new AT1 derive from rare, self-renewing, long-lived, mature AT2 cells that produce slowly expanding clonal foci of alveolar renewal. This stem cell function is broadly activated by AT1 injury, and AT2 self-renewal is selectively induced by EGF ligands in vitro and oncogenic KrasG12D in vivo, efficiently generating multifocal, clonal adenomas. Thus, there is a switch after birth, when AT2 cells function as stem cells that contribute to alveolar renewal, repair, and cancer. We propose that local signals regulate AT2 stem cell activity: a signal transduced by EGFR-KRAS controls self-renewal and is hijacked during oncogenesis, while another signal controls reprogramming to AT1 fate.
Introduction
Pulmonary gas exchange occurs in delicate alveolar sacs lined by two epithelial cell types1 (Extended Data Figure 1). Squamous alveolar type (AT) 1 cells mediate gas exchange, whereas cuboidal AT2 cells secrete surfactant that prevents alveolar collapse; AT2 cells are one of the medically most important cells in the neonate and, as described below, one of the most dangerous in adults. Serious diseases including Respiratory Distress Syndrome and Idiopathic Pulmonary Fibrosis involve a failure to establish or maintain AT1 and AT2 cells, and alveoli are a major site of lung cancer, the leading cause of cancer death2.
Despite their importance, the identity of alveolar progenitor and stem cells is controversial and their activity throughout life uncharted3,4. Classical morphologic and radioautographic studies in rodents suggested that during development progenitors mature into AT2 cells, some of which differentiate into AT1 cells5. Maintenance is difficult to study because of slow turnover6,7, but lineage tag expression in isolated AT1 cells is observed following bulk labeling of AT2 cell populations8,9. To circumvent slow turnover, lung injury models have been employed, and provide evidence that AT2 cells can contribute to alveolar repair8,10,11. Currently, six different cell populations have been proposed as alveolar stem cells based on their capacity for clonal propagation and multilineage differentiation in culture9,12–14. Transplantation assays have also been employed, but because many cells are implanted they cannot assess whether individual cells self-renew and undergo multilineage differentiation4,15. When one of these putative stem cell populations was fate-mapped in vivo, it failed to demonstrate the multilineage differentiation achieved in culture16; similar disparity between ex vivo and in vivo behavior of putative stem cells has been found for other organs17. Here we use a battery of alveolar markers, lineage tracing, and clonal analysis in mice to identify alveolar progenitor and stem cells in vivo and map their locations and activity during lung development, maintenance, and cancer.
AT1 and AT2 cells arise independently during development from a bipotent progenitor
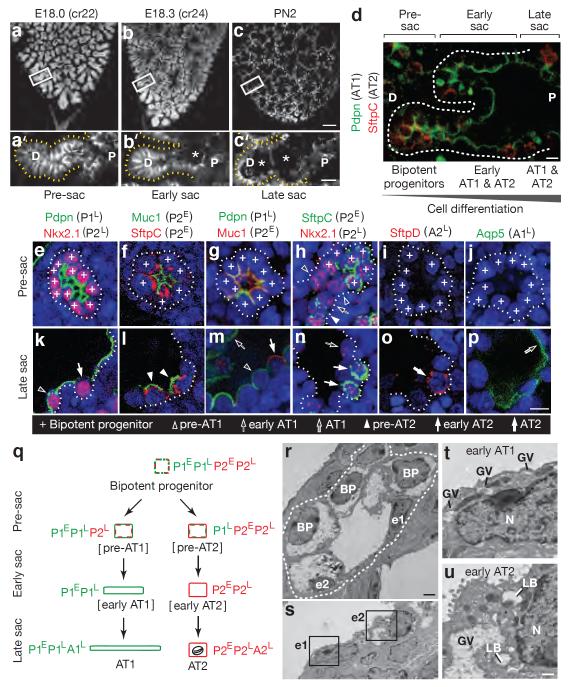
Mature AT1 and AT2 cells appear about one day before birth, when distal tubules begin to dilate (“sacculation”, Figure 1b–d)18,19. We mapped progression of sacculation in three dimensions by analyzing finely staged whole mount lungs immunostained for E-Cadherin (Cdh1) to visualize individual cells (Figure 1a–c and Extended Data Figure 1e–f). Dilation begins at the bronchoalveolar junction then progresses distally towards the airway tip (Figure 1a–c).
Figure 1. Development of alveolar type 1 (AT1) and AT2 cells from bipotent progenitors.
(a–c) Mouse lung lobe tips stained for E-cadherin (E, embryonic day; cr, crown-rump length in mm; PN, postnatal day). (a'–c') Sacculation (sac; asterisks) proceeds proximally (P) to distally (D) along the airway. Bars, 100um (a–c), 20um (a'–c'). (d) Staining for AT1 (Pdpn) and AT2 (SftpC) markers at E18.3 shows co-expression in pre-sacculation zone and restriction in late sacculation zone. Bar, 10um. (e–p) Tips imaged in pre-sacculation (e–j) and late sacculation (k–p) zones identifies six classes of marker expression profiles (P1E,P1L,A1L,P2E,P2L,A2L; Extended Data Table 1). Bar, 20um. (q) Inferred differentiation pathways showing changes in AT1 (green) and AT2 (red) marker classes. Oval, lamellar body. (r,s) Electron micrographs at E18.3 showing early (u) and late (v) sacculation zones. BP, bipotent progenitors; e1, early AT1 cells; e2, early AT2 cells. Bar, 2um. (t,u) Boxed regions in S showing (e1) an early AT1 (squamous, glycogen vacuoles (GV) without lamellar bodies (LB)) and (e2) an early AT2 (cuboidal, GV, LB). N, nucleus; bar, 0.5um.
The classical model proposing that progenitors in development are pre-AT2 cells is difficult to reconcile with the finding that some AT1 cell markers are expressed up to five days before sacculation20. To molecularly classify progenitors, we validated 15 extant AT1 and AT2 markers (Supplementary Table 1) then analyzed the transition in labeling between distal (progenitors) and proximal (nascent AT1 and AT2 cells) positions in a sacculating airway (Figure 1d) to infer dynamic expression changes during differentiation (Figure 1e–p). Markers fell into six expression classes (Extended Data Table 1) distinguishing seven stages in alveolar development (Figure 1e–p). However, instead of a progenitor-AT2-AT1 progression, our data support a model in which bipotent progenitors (P) expressing a subset of AT1 (1) and AT2 (2) markers (P1E, P1L, P2E, P2L) give rise to either AT1 or AT2 cells by shutting off inappropriate cell type markers early (E) or late (L) in differentiation, then turning on cell type-specific late (L) markers (A1L, A2L) as they complete maturation (Figure 1q). Co-expression of AT1 and AT2 markers by progenitors suggests that these specialized cell types may have evolved from a primordial pneumocyte with features of both, like those in lower vertebrates21.
Three additional lines of evidence support the bipotent progenitor model. First, clonal analysis of individual distal airway epithelial tip cells22 labeled on embryonic day (E) 15 using an inducible Cre recombinase (Shh-Cre-ER) demonstrated localized alveolar lineage clusters with marked AT1 and AT2 cells (Extended Data Figure 2a,b), confirming that individual cells are bipotent. Second, ultrastructural analysis of early sacculation revealed three classes of distal epithelial cells (Figure 1r–u): cuboidal cells with glycogen vacuoles but no lamellar bodies (bipotent progenitors); cuboidal cells with vacuoles and lamellar bodies (early AT2 cells); and partially flattened cells with vacuoles (early AT1 cells). We never observed partially flattened cells with lamellar bodies, the presumed AT2-to-AT1 intermediate predicted by the classical model (Extended Data Figure 1g). Third, lineage tracing of newly-differentiated AT2 cells using a Cre recombinase knock-in (LysM-Cre) at the LysozymeM (Lyz2) locus, a late AT2 marker gene (A2L), labeled many AT2 cells in the embryo but no marked AT1 cells were seen by two weeks postnatally (Extended Data Figure 2c,e).
Little epithelial proliferation was detected during sacculation and for several weeks afterward (Extended Data Figure 3), suggesting that maturation of bipotent progenitors generates most or perhaps all AT1 and AT2 cells in development. Flow cytometry at early sacculation (E18.1) showed bipotent progenitors or pre-AT2 cells (Muc1+/Pdpn+) comprised 8% of distal cells, while early or mature AT2 cells (Muc1+/Pdpn−) made up 5%. Bipotent progenitors appear to be fully exhausted by postnatal day four, when sacculation has completed throughout the lung. In adult lungs few if any bipotent progenitors were found by flow cytometry (0.3%+/−0.2% Muc1+/Pdpn+ distal epithelial cells) or labeling with Shh-Cre-ER, suggesting a transition to other sources.
Alveolar maintenance by infrequent activation of AT2 cells in discrete foci
Classical radioautographic studies suggested alveolar epithelium is a slowly renewing population maintained by diffuse proliferation, presumably of AT2 cells23. We used in vivo lineage tracing to investigate the role of AT2 cells in maintenance, marking them in two complementary ways, with similar results. In one, a Cre-ERT2-rtTA knock-in at Surfactant Protein C (SftpC) locus (SftpC-Cre-ER) was used with a membrane-localized fluorescent reporter (mTmG) to pulse-label AT2 cells by tamoxifen induction at postnatal (PN) 18 days; mice were analyzed 13 or 210 days later. Although SftpC is often used as a mature AT2 marker, the results above (Extended Data Table 1) show that as a class P2E marker it is also expressed by the bipotent progenitor. Hence, we also labeled AT2 cells using LysM-Cre described above, a class A2L marker that initiates expression only in mature AT2 cells (Extended Data Figure 4a,b,d,m). Both lines robustly labeled AT2 cells throughout the lung, and at 2 months after marking (SftpC-Cre-ER) or postnatally (LysM-Cre) showed scattered AT1 cells labeled with the AT2 lineage tag (Extended Data Figure 2d,f and Extended Data Figure 5). AT2 cells did not significantly contribute to renewal of bronchiolar lineages including Clara, ciliated, and neuroendocrine cells (Extended Data Figure 4c,e–m), and although rare cells near bronchoalveolar junctions that co-expressed SftpC and Clara cell marker CCSP/Scgb1a1 were also tagged (“bronchioalveolar stem cells” or BASCs12), they proliferated little if at all (Extended Data Figure 4f–h,j–m).
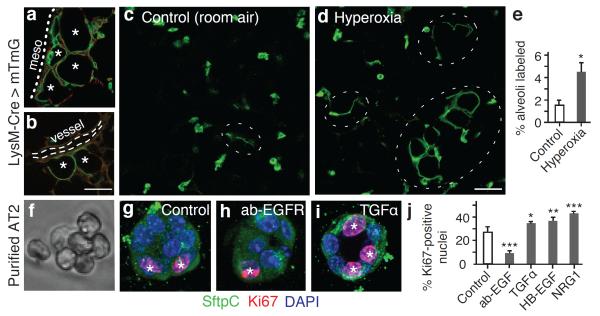
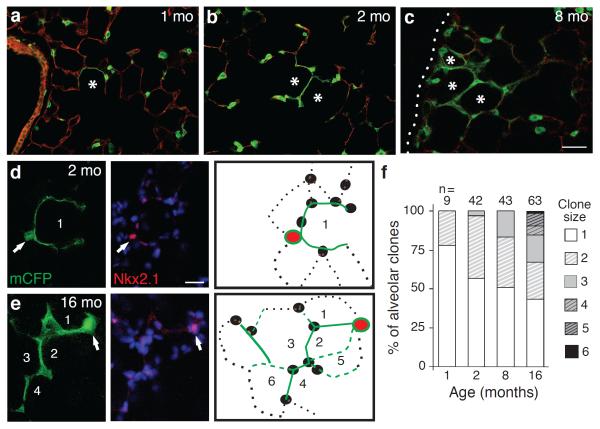
To investigate the frequency and spatial localization of AT1 renewal by AT2 cells, we labeled lungs as above using LysM-Cre and analyzed them at 1, 2, 4, 8, and 16 months of age. Less than 1% of AT1 cells expressed the AT2 lineage tag at 1 month, 3.9% at 4 months and 7.5% at 16 months. AT1 cell replacement occurred preferentially in alveoli abutting arterioles and in the lung periphery (Figure 3a,b). Wherever new AT1 cells arose, the alveolus was replaced essentially in half or its entirety, suggesting alveoli include just one or two AT1 cells. With aging, adjacent alveolar units were also renewed, evidenced by slowly enlarging clusters of AT1 cells marked with the AT2 lineage tag (Figure 2a–c). These patches of AT1 replacement were indistinguishable from surrounding areas, except for AT1 labeling with the AT2 lineage tag; similar results obtained with SftpC-Cre-ER (Extended Data Figure 5). We conclude that AT1 replacement by AT2 cells occurs intermittently and in a spatially patchy distribution in “renewal foci” that slowly enlarge over time, with perivascular and peripheral regions serving as relative “hot spots.”
Figure 3. Activation of AT2 stem cell function in vivo and proliferation in vitro.
(a,b) Renewal foci (AT2 lineage label, green; other cells, red) commonly involve alveoli (asterisks) in peripheral (a, mesothelium, dots) and perivascular (b, dashes) domains. Bar, 50um (a,b). (c–e) Alveolar regions under room air (c) or after hyperoxia (88% O2, 5 days) to injure AT1 cells (d). Note increased foci (ovals) after injury. Bar, 50um (c,d). Quantitation (e) shows increased alveolar surface (mean±SEM) from AT2-lineage labeled cells after hyperoxia. p <0.05 (Mann-Whitney U test). (f,g) Freshly isolated AT2 cells (f, phase contrast) cultured four days in Matrigel (g) proliferate, shown by Ki67 staining (red, asterisks), but maintain AT2 marker expression (SftpC, green). (h–j) Images (h,i) and quantification (500 cells per biological replicate, n=4,3,3,3,4) (j) of proliferation with EGFR blocking antibody (ab-EGFR, 2.5 μg/mL) and EGF ligands indicated (4 μM). Bar, 10um (f–i). Mean±SEM; *, p<0.05; **, p<0.01; ***, p<0.001 (Tukey's multiple comparisons test).
Figure 2. Mature AT2 cells renew AT1 cells in clonal foci.
(a–c) AT2 lineage-labeled AT1 cell foci (green) enlarge with aging, incorporating adjacent alveoli (asterisks). Dotted line, mesothelium. Bar, 150um (a–c). (d,e) AT2 cells were sparsely marked with Confetti reporter, and mCFP-labeled foci (green) are shown co-stained for AT2 marker Nkx2.1 (red) at age 2 (d) and 16 (e) months. Note “founder” AT2 cell (arrow) and its labeled AT1 progeny (green). Numbers, incorporated alveoli. Entire clone schematized in right panel. Red, founder AT2; green lines, AT1 daughters visible (solid) or outside (dashed) focal plane; black, unlabeled AT1 (dotted lines) and AT2 (ovals) cells. Bar, 20um (d,e). (f) Clone size increases with aging (p=0.03,Kruskal-Wallis test). n, clones scored; clone size, number of incorporated alveoli.
Renewal foci derive from single, differentiated AT2 cells with stem cell properties
Each renewal focus could derive from a single AT2 cell, in which case the cells would be clonally related, or from multiple AT2 cells. To distinguish these possibilities, we used a Confetti Cre-dependent reporter that stochastically expresses one of four fluorescent proteins in each cell that undergoes recombination. We analyzed the membrane CFP lineage marker and found that foci were similar in size and progressive enlargement to those observed using the single color reporter, indicating renewal foci derive from a single “founder” AT2 cell. Because of fortuitously inefficient recombination of the Confetti reporter using LysM-Cre (~2% of AT2 cells expressed mCFP at 16 months), we were able to distinguish marked AT2 cells within renewal foci. Most contained one or two AT2 cells and multiple AT1 cell progeny colonizing up to six contiguous alveoli (Figure 2d–f). We also observed small clonal foci comprised only of AT2 cells (Extended Data Figure 6b,c), implying they are capable of self-renewing without forming AT1 cells (self-duplication). Founder AT2 cells expressed mature AT2 markers including LAMP-1, a lysosome-associated protein, suggesting they continue to produce surfactant (Extended Data Figure 6a), and they didn't express Clara, ciliated, and neuroendocrine markers. Their only distinguishing feature was their nucleus, which was often slightly larger and stained more intensely for Nkx2.1 than nearby AT2 cells (see also Figure 2d,e). We conclude that each renewal focus derives from a single, self-renewing AT2 cell that can generate multiple AT1 and/or AT2 cells, is long-lived, and remains closely associated with its progeny, as evidenced by its persistence in large foci at advanced ages (Figure 2e). Because founder cells appear to be mature (LysM-Cre lineage positive), functional (SftpC-Cre-ER lineage and LAMP-1 positive) AT2 cells that share the essential properties of conventional stem cells (multipotent, self-renewing, persist for life), they are “bi-functional” stem cells, executing both differentiated and regenerative functions.
Rapid and extensive alveolar repair by AT2 cells induced by acute AT1 cell injury
Elevated oxygen tension is toxic to AT1 but not AT2 cells24. Exposure of two month old mice carrying the AT2 lineage tag to 88% oxygen for 120 hours tripled the number of renewed AT1 cells (Figure 3c–e). This finding shows that AT2 cells become activated following hyperoxic injury, and supports the idea that although normally only a rare subset of AT2 cells executes a stem cell function, others can be recruited to repair alveolar damage. Whether every AT2 cell can be activated this way could not be determined because more severe hyperoxia was lethal.
Oncogenic KrasG12D activates AT2 self-renewal and efficiently transforms AT2 cells
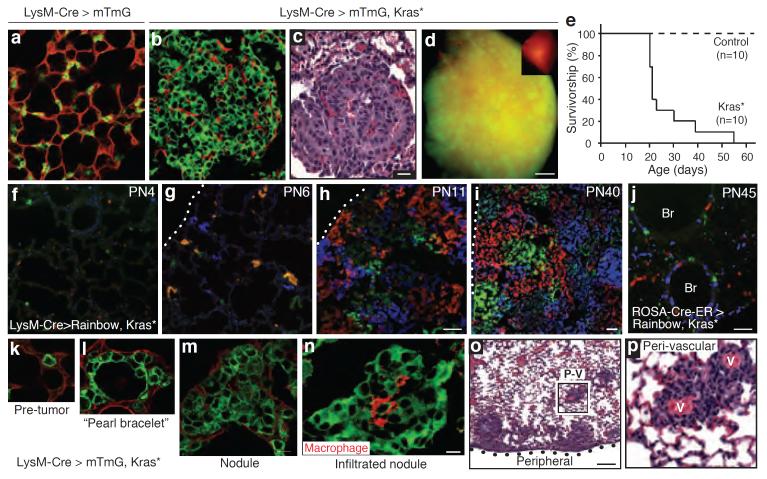
Adenocarcinoma, the major form of lung cancer, is associated with activating mutations in Kras or Egfr25. Typically located in peripheral lung regions, nearly all tumor cells express SftpC, leading to long-standing speculation that they originate from AT2 cells or their progenitors. However, the identity of the tumor initiating cell(s) remains controversial12,26–28. To test the effect of oncogenic Kras G12D on mature AT2 cells, a conditional Kras LSL-G12D allele was activated using LysM-Cre along with the mTmG Cre-dependent reporter. Tumor nodules grew rapidly throughout the lungs (Figure 4a–d), with dense replacement of virtually the entire alveolar region by one month after induction and death shortly thereafter (Figure 4e). When lungs were examined in the first few days following induction, we found with a Rainbow multi-color reporter that nearly every epithelial cell expressing the AT2 lineage tag proliferated, demonstrating highly efficient AT2 transformation by Kras G12D (Figure 4f–i). The biggest tumors were found in peripheral and perivascular regions, sites where physiological AT1 renewal by AT2 cells was commonly observed (compare Figures 3a,b and 4o,p). Similar results obtained using SftpC-Cre-ER to activate Kras G12D in adult mice (Extended Data Figure 7a,c). By contrast, when we used CCSP-Cre-ER, most Clara cells were unaffected or divided minimally, whereas some at bronchoalveolar junctions formed small clonal adenomas (Extended Data Figure 7b). We also used ubiquitously expressed ROSA-Cre-ER to activate the Kras G12D allele at random, resulting in many singlets and minimally affected cells throughout the lung, even 18 days after induction (Figure 4j). The transformed AT2 cells comprising the adenomas continued to express AT2 markers (Nkx2.1, SftpD) and did not turn on a Clara marker (CCSP) or, with rare exceptions, AT1 markers (Pdpn, LEL) (Extended Data Figure 8). Thus, oncogenic Kras G12D appears to selectively and permanently induce AT2 self-renewal, without deprogramming the cells to the bipotent progenitor or causing reprogramming to AT1 or Clara cell fates.
Figure 4. Transformation of mature AT2 cells by activated Kras.
LysM-Cre>mTmG control (a) and LysM-Cre>mTmG,KrasLSL-G12D/+ (abbreviated Kras*) (b,c) lungs, showing proliferation AT2 cells (b, green cells) compressing surrounding cells (red) and forming adenomas (c, H&E stain). Bar, 20um (a–c). (d) Lung lobe as in B showing widespread infiltration by tumor (green). Inset, control lung. Bar, 1mm. (e) Survival curves. (f–i) LysM-Cre>Rainbow,Kras* lungs at indicated ages (PN, postnatal day). Note rapid clonal (single color) expansion of labeled AT2 cells with minimal cell mixing. Bar, 50um (f–h), 100um (i). Dotted line, mesothelium. (j) PN45 ROSA-Cre-ERT2>Rainbow,Kras* lung 18 days (d) after induction. Note minimal expansion. Br, bronchi; bar, 50um. (k–n) A recombined (green) AT2 cell (k) generates progeny that spread laterally, giving “pearl bracelet” appearance (l). Cells later “heap up” into nodules (m), some infiltrated by macrophages (n; red, F4/80). Bar, 10um (k–n). (o,p) H&E stain shows robust tumor growth peripherally and peri-vascularly (PV). Boxed area (p) shows proliferated cells around blood vessels (v). Bar, 200um (o).
By examining lineage-tagged Kras G12D mutant lungs at progressive stages, we could infer the cellular mechanism of adenoma formation (Figure 4k–m). Proliferation of the activated AT2 cell generates daughter cells that spread laterally yet maintain a monolayer (“lepidic” expansion), the first histological sign of the tumor (Figure 4l). Later, cells heap up and form a nodule (“hilical” expansion) that obliterates the lumen and begins compressing and invading adjacent alveoli (Figure 4m). Infrequent clones developed into large, well-formed papillary structures, though most formed simple adenomas29. At advanced stages, a striking pattern of large, closely-packed, single color tumors was evident, implying minimal cellular exchange between neighboring tumor foci (Figure 4i). Some advanced tumors became infiltrated by macrophage “nests” (Figure 4n), a poor prognostic sign30.
EGF signaling controls self-renewal of AT2 cells in culture
We profiled the transcriptome of bipotent progenitors and AT2 cells. Both express Egfr, two other EGFR family members (Erbb2, Erbb3) (Supplementary Table 2), and receptors for many other signals (Supplementary Table 2,3). Purified AT2 cells (Figure 3f) retain a robust ability to both proliferate (>25% Ki67-positive, Figure 3g,j) and to differentiate into AT1-like cells (>95% thin, flat morphology and Aquaporin 5-positive, n=240 cells) (Extended Data Figure 6d). Addition of purified EGF ligands (TGFα, HB-EGF, NRG1) stimulated AT2 proliferation, whereas EGFR blocking antibody inhibited proliferation (Figure 3h–j). None of these treatments induced differentiation into AT1 cells. Thus, EGF signaling is a critical and selective regulator of AT2 proliferation, at least under these conditions.
Discussion
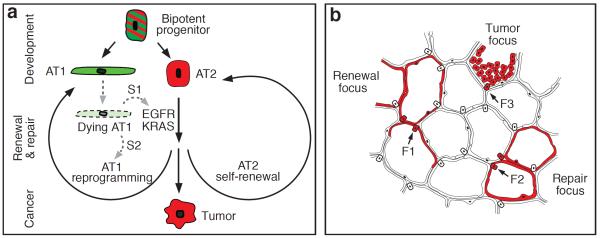
Our characterization of alveolar progenitor and stem cells throughout the lifespan supports a model in which AT1 and AT2 cells arise independently during development from a bipotent progenitor (Figure 5a). Several weeks after birth, when alveolar development is complete, there is a switch and mature AT2 cells become a renewable source of AT1 and AT2 cells. But these postnatal renewal events are rare and occur in monoclonal foci (“alveolar renewal focus”, Figure 5b) that slowly expand over months or years. Only a small fraction (~1%) of mature AT2 cells normally express this stem cell function and they divide intermittently (~40 day doubling time) and supply only local domains, giving an overall renewal rate of just 7% of alveoli per year. This stem cell function is more broadly induced by AT1 injury, indicating that other AT2 cells may have similar potential (“alveolar repair focus”, Figure 5b) that can be activated by dying AT1 cells. Classical pneumonectomy experiments support the idea that most AT2 cells possess latent regenerative capacity31, and AT2 ablation induces self-duplication of surviving AT2 cells9 (“AT2 replacement focus”). Our data do not exclude alternative sources of new alveolar cells, especially following severe injuries14 that regionally deplete AT2 cells.
Figure 5. Model of alveolar progenitors and stem cells in development, maintenance, and cancer.
(a) Bipotent progenitors expressing some AT1 (green) and AT2 (red) markers differentiate into AT1 or AT2 cells. Mature AT2 cells function as stem cells intermittently activated for alveolar renewal and repair. Dying AT1 cells are proposed to produce a signal (S1) transduced by EGFR-KRAS that activates division of nearby AT2 cell (self-renewal); another signal (S2) reprograms a daughter into an AT1 cell. Activating mutations of Egfr or Kras in AT2 cell drives constitutive self-duplication, forming tumor of AT2-like cells. (b) Rare AT2 cells function as stem cells, giving rise to clonal renewal foci (red, left) that slowly enlarge, with persistence of founder AT2 cell (F1). With injury, additional AT2 cells (F2) are recruited to generate repair foci (red, right). Activating Kras mutation in AT2 cell (F3) initiates tumor focus (red, top).
If many or all AT2 cells can serve as stem cells, why does only a minority execute this function for maintenance, producing large monoclonal foci? Perhaps alveolar turnover is coupled with a stem cell hierarchy whereby an initially activated AT2 cell suppresses nearby AT2 cells and becomes the dominant stem cell. It is also unclear why renewal foci slowly enlarge over time, since there is no obvious recurrent injury. Perhaps foci are programmed anatomical domains of alveolar renewal, with new cells moving out from a specialized niche akin to intestinal crypts albeit with much slower turnover. Whatever the explanation, there are clearly regional influences as both renewal and tumor foci are preferentially located in perivascular and peripheral lung domains, possible sources of stem cell signals32.
EGFR signaling selectively stimulates proliferation of AT2 cells in vitro, and oncogenic Kras G12D permanently and selectively activates proliferation in vivo, efficiently transforming AT2 cells into rapidly growing monoclonal tumors (“lung tumor focus”, Figure 5b). By virtue of their large numbers, class susceptibility, and robust adenomatous response to Kras G12D, AT2 cells may constitute the major cell type responsible for human lung adenocarcinoma, making them among the most dangerous cells in the body.
We propose that the oncogenic potential of AT2 cells is a direct consequence of their stem cell function: AT2 cells are poised to function as alveolar stem cells and EGFR/KRAS signaling regulates the self-renewal part of the stem cell program (and the related process of self-duplication); another signaling pathway (Supplementary Table 2) must control AT2 reprogramming to AT1 fate (Figure 5a). This model predicts that dying AT1 cells secrete an EGF that initiates self-renewal, plus another signal for fate reprogramming. Dying AT2 cells presumably produce only the former. It will be important to identify these signals and the events they control. This could suggest new strategies for early detection and treatment of tumors and for replenishing diseased alveoli.
Methods
Mouse strains
CD1 or B6 (Charles River Laboratories) were the wild type strains. The Cre recombinase system was used to manipulate gene and marker expression in vivo in specific mouse lung cell types and lineages in mixed strain backgrounds, using a knock-in allele that expresses Cre or a tamoxifen-inducible Cre-Estrogen Receptor (ER or ERT2) fusion crossed to a Cre-dependent target gene. For Cre-ER lines, tamoxifen (Sigma) was dissolved in corn oil by sonication, stored in 100 ul aliquots at −80°C, and 1 mg (Rosa26-Cre-ERT2, CCSP-Cre-ER), 2 or 4 mg (Shh-Cre-ERT2) or 1, 2, or 3 mg (SftpC-Cre-ERT2-rtTA) injected intraperitoneally at specified ages to induce recombination in cells throughout the transgene expression domain. The Cre expression strains used were: Shh-Cre35, which recombines throughout the endoderm-derived lung epithelium from the beginning of lung development; Shh-Cre-ERT235 expressed in early lung endoderm then restricts to distal tip epithelium during branching morphogenesis22; LysM-Cre36 expressed in mature AT2 cells, macrophages, and neutrophils; SftpC-Cre-ERT2-rtTA4 expressed in the bipotent alveolar progenitor and developing and mature AT2 cells, as well as in rare bronchiolar cells; CCSP-Cre-ER16 expressed in Clara cells and rare SftpC-expressing bronchiolar cells; and, and Rosa26-Cre-ERT237 expressed throughout the lung and rest of the body. The Cre-responsive target genes used were: R26-EYFP38, a ubiquitously expressed transgene that expresses EYFP following Cre recombination; R26-mTmG39, which expresses membrane-targeted tdTomato before recombination and membrane-targeted EGFP after recombination; R26-Confetti40, which expresses either RFP, EYFP, nuclear GFP, or membrane CFP after recombination; R26-tdTomato41, which expresses tdTomato after recombination; and R26-Rainbow42, which expresses GFP before recombination and either CFP, mOrange, or mCherry after recombination. Kras LSL-G12D is a knock-in at the Kras locus in which the wild type Kras coding sequence is replaced by a lox-STOP-lox-Kras G12D allele34, which following Cre recombination results in endogenously regulated expression of a constitutively-active KRAS protein. LysM-EGFP43 is an EGFP knock-in at the Lyz2 locus expressed by mature AT2 cells, macrophages, and neutrophils. Genotyping was performed by PCR of DNA extracted from ear clips using the Red-Xtract Kit (Sigma) and published primer sets. Mice were housed in filtered cages and all experiments were performed in accordance with approved IACUC protocols.
Hyperoxic lung injury
8 week old female mice were housed in a sealed Plexiglas chamber with oxygen delivered by continuous flow to maintain levels at 88% for five days, after which the animals were returned to room air to recover for three weeks before analyzing. Female siblings maintained under room air were used as controls. AT1 cell renewal was quantitated as described below by blinded scoring of the number of alveoli in which AT1 cells expressed the AT2 cell lineage label in three randomly selected lung volumes (100–300 um thick lung slices in randomly selected 20x fields) from two animals per group.
Lung collection, fixation, and processing
Embryos were staged by vaginal plugging of the mother, with noon on the day of appearance of the plug taken as embryonic day (E) 0.5. Individual embryos were also staged by fetal crown-rump length at time of sacrifice. Lungs were removed en bloc and a scalpel used to excise the tip of the accessory lobe, after which lungs and the tip were fixed in methanol:DMSO (4:1) with rocking overnight at 4°C, bleached in methanol:DMSO:30% H202 (4:1:1) for 5 hours at room temperature, then dehydrated and stored in methanol at −20°C until staining.
Postnatal mice were euthanized by carbon dioxide inhalation, the abdominal aorta severed, and sternotomy performed. Phosphate buffered saline (PBS; Ca2+- and Mg2+-free, pH 7.4) was gently perfused into the right ventricle by manual pressure using a syringe with a 21 gauge needle to clear the pulmonary vasculature. For mice up to 7 days old, lungs were removed en bloc, the accessory lobe tip excised, lobes separated and trimmed to create a flat base, glued to a tissue disk with cyanoacrylate, then submerged in ice cold PBS and sliced at a thickness of 50 to 250 um using a vibrating microtome (Leica). Lung slices were fixed in methanol:DMSO as above. For direct imaging without immunostaining, slices were fixed in PFA (2% in PBS) overnight at 4°C then stored in Vectashield (Vector) at 4°C.
For mice 8 days or older, lungs were collected and processed as above except that following clearance of the pulmonary vasculature the ventral trachea was incised and cannulated with a blunt needle that was secured in place by tying a suture around the trachea. Lungs were then gently inflated to full capacity with molten low melting point agarose (Sigma, 2% in PBS) after which the cannula was withdrawn and the suture tightened to prevent leakage. Ice-cold PBS was dripped into the thorax to solidify the agarose, then the inflated lungs were removed en bloc and processed as above.
Lungs from mice carrying the Rainbow transgene were cleared and inflated as above, immersion fixed in PFA (4% in PBS) overnight at 4°C, cryoprotected in 30% sucrose overnight at 4°C, then submerged in OCT (Tissue Tek) in an embedding mold, frozen on dry ice, and stored at −80°C. Sections of 10 and 30 μm obtained using a cryostat (Leica CM3050S) were collected on glass slides, mounted in Prolong Gold (Invitrogen) and stored at 4°C after curing at room temperature overnight. Lungs for tumor histology were collected without perfusion, formalin-fixed, paraffin-embedded, then sections obtained using a microtome were collected on glass slides, stained with hematoxylin and eosin (H&E) and mounted.
Lung fluorescent immunostaining and microscopy
Indirect immunohistochemistry was performed as described44 with the following modifications. For signal amplification of whole-mount lung tips and pieces, a horseradish peroxidase (HRP)-conjugated secondary antibody was used (Jackson ImmunoResearch) and detected by tyramide signal amplification (TSA, Plus Fluorescein System, PerkinElmer NEL741001KT). Primary antibodies raised in mouse were detected using the Mouse-on-Mouse Kit (Vector). For lung slices, secondary antibodies conjugated to an Alexa fluor dye (A488, A555, or A633; Invitrogen) were used. DAPI (5 ng/ml) was included during incubation with secondary antibodies. Staining with biotin-conjugated lectins was performed as described for primary antibodies then detected with streptavidin conjugated to an Alexa fluor dye (Invitrogen).
Primary antibodies against the following antigens (used at 1:200 dilution unless otherwise noted) were: pro-SftpC (rabbit, Chemicon AB3786), GFP (chicken, Abcam AB13970), F4/80 (rat, Serotec MCA497G; clone CI:A3-1), CGRP (guinea pig, Europroxima 2263BGP470-1), acetylated tubulin (mouse, Sigma T6793; clone 6-11B-1), Lysozyme (rabbit, Thermo Scientific RB372A), Rage (rat, R&D MAB1179), E-cadherin (rat, Life Technologies 131900; clone ECCD-2), CCSP (rabbit, EMD Millipore 07-623; goat, provided by B. Stripp), Pdpn (hamster, DSHB 8.1.1; 1:20), Aqp5 (rabbit, Calbiochem 178615), Muc1 (hamster, Thermo Scientific HM1630, clone MH1 (CT2); rabbit, Novus NBP1-02974), Cathepsin H (goat, R&D AF1013), Ki67 (rat, DAKO M7249, clone TEC-3), Nkx2.1 (rabbit, Millipore 07-601; mouse, Thermo Scientific MS-699-P, clone 8G7G3/1), SftpB (rabbit, Chemicon AB3780), SftpD (rabbit, Chemicon AB3434), Abca3 (mouse, Seven Hills 13-H2-57), Lamp-1 (rat, DSHB 1D4B) and Lamp-2 (rat, DSHB GL2A7). Biotin-conjugated lectins used were: Lycopersicon esculentum (Vector B-1175) and Ricinis communis I (Vector B-1085).
After staining, lung slices were immersed in Vectashield (Vector) and either placed in a glass depression slide with a coverslip gently lowered on top and its edges secured with scotch tape, or into a chambered coverslip (Falcon). Images were acquired using a Leica Sp2 upright or Sp5 inverted laser scanning confocal microscope with LCS software. Cryosectioned Rainbow lungs were imaged using a Zeiss Axiophot upright fluorescence microscope with Axiovision 4.2 software. Intact lungs were visualized with a Leica MZ16FA fluorescent stereomicroscope and images captured with a Retiga 2000R (Q Imaging) camera using Image-Pro Plus (Media Cybernetics) software. H&E stained sections were imaged using a compound light microscope equipped with a digital camera. Images were processed (pseudocolored, levels and contrast adjusted, overlaid) using Adobe Photoshop.
Electron microscopy and ultrastructural analysis
E19 accessory lobe tips were collected as described above then fixed in 3% glutaraldehyde/0.1M cacodylate (pH 7.4) overnight at 4°C, post-fixed in 1% OsO4 for 1 hour at 4°C, washed, stained in 1% uranyl acetate overnight at 4°C, dehydrated to 100% ethanol, infiltrated with Embed 812 resin (EMS), then oriented in casts and polymerized at 65°C for 24 hours. Blocks were sectioned using an ultramicrotome (Leica Ultracut) until fields containing partially sacculated airway tips were identified. Specimens were visualized with a transmission electron microscope (Jeol TEM1230) and images captured with a CCD camera then processed using Adobe Photoshop. Individual epithelial cells were scored for the presence or absence of glycogen vacuoles, lamellar bodies, and cuboidal versus attenuated morphology.
Assigning alveolar cell marker classes
Each of the alveolar antigens and lectins was identified by systematically searching the published literature using PubMed. We then purchased all antibodies and lectins from our list that were commercially available and tested their expression in the adult lung against the canonical markers of AT1 (T1α/Pdpn) and AT2 (pro-SftpC) cells. In cases where we could not perform double-labeling with a canonical marker because the antisera were raised in the same species, we compared expression against other markers we had already validated that were raised in a different species. For AT1 markers that co-localized with Pdpn, we further attempted to confirm these were not also expressed by endothelial or AT2 cells by directly comparing their expression against anti-GFP in lung slices from Shh-Cre> mTmG mice, which provides pan-epithelial membrane marking. In total, we tested 77 antibodies raised against 48 antigens and seven lectins for robustness of signal, then for AT1 or AT2 selective marking. We then analyzed the expression patterns of our panel of validated AT1 and AT2 cell markers in alveolar progenitors at the E17 accessory lobe tip, which we selected because its distinctive shape provides reliable landmarks for identifying the same airway position in different lung specimens. Only two expression profiles were observed, absent or ubiquitous. Of those expressed ubiquitously, each progenitor cell would subsequently selectively downregulate either the AT1 or the AT2 subset, distinguishing pre- or early AT2 or AT1 cells, respectively. We designated these marker classes P1E, P2E, P1L, and P2L, based on the specificity and relative timing of their restriction (initial expression in Progenitors (P) with restriction to AT1 (1) or AT2 (2) cells Early (E) or Late (L) in the differentiation process). Next, the AT1 and AT2 markers that were absent in progenitors at E17 initiated selective expression in nascent AT1 or AT2 cells at E18, so we designated these two classes A1L and A2L (initial expression in AT1 (1) or AT2 (2) cells, Late (L) onset of expression). Classes were assigned based on analysis of five terminal airways (100 cells) for each developmental stage indicated.
Quantitation of AT1 cell renewal
20x alveolar fields were randomly selected and scored for the presence and extent of AT1 cell labeling by GFP. An alveolar unit with at least half of the epithelial AT1 surface marked by the lineage tag was considered labeled, and contiguous extension of the tag into adjacent alveoli was considered part of the same focus. All scoring was performed with blinding to the experimental condition (for hyperoxia versus room air) and age (for clonal analysis). Percent of alveoli labeled was determined by counting the number of positive alveoli in a given volume of tissue then dividing by the total number of alveoli scored, with the latter estimated by assuming a cuboidal shape and average diameter of 75 um per alveolus.
Bipotent progenitor and AT2 cell isolation
To quantify alveolar progenitor populations and isolate bipotent progenitors for expression profiling, E18 lungs from a single litter (~6 embryos) were removed en bloc without perfusion and a scalpel used to collect peripheral lobe edges enriched for alveolar cells. Lung tissue was rinsed briefly in PBS, minced with a razor blade into 1 mm3 fragments, suspended in 5 ml of digestion buffer consisting of Elastase (3 U/mL; Worthington Biochemical Corporation) and DNase I (0.33 U/mL; Roche) in DMEM/F12, incubated with frequent agitation at 37 °C for 45 minutes, triturated briefly with a 5 ml pipette, then an equal volume of DMEM/F12 supplemented with 10% FBS and penicillin-streptomycin (1 U/mL, Thermo Scientific) was added and the cell suspension passed through a 100 μm mesh filter (Fisher) to remove residual tissue fragments, then centrifuged at 400 × g for 10 minutes. The pelleted cells were resuspended and incubated in red blood cell lysis buffer (BD Biosciences) for 2 minutes, passed through a 40 μm mesh filter (Fisher), centrifuged at 400 × g for 10 minutes, then resuspended in sorting buffer (PBS supplemented with 0.05% BSA and 2 mM EDTA). Antibodies against Muc1 (hamster, Thermo Scientific HM-1630-P1ABX; 1:100) conjugated using Alexa Fluor A488 Antibody Labeling Kit (Life Technologies A-20181), PE-conjugated Pdpn (hamster, eBiosciences 12-5381-80, clone eBio8.1.1; 1:100), and Sytox Blue cell viability stain (Invitrogen) were added and the cells incubated for 15 minutes, after which cell populations were quantified by flow cytometry (Aria II, BD Biosciences) and viable Muc1+/Pdpn+ progenitors sorted directly into cell lysis buffer for RNA isolation.
To isolate LysM-lineage AT2 cells for expression profiling, adult (PN70) LysM-Cre> tdTomato mice were euthanized and pulmonary vasculature perfused clear (described above). The trachea was cannulated with a 20 gauge needle and lungs gently inflated with digestion buffer, then removed en bloc and lobes individually rinsed briefly in PBS, minced, suspended in digestion buffer, and a cell suspension prepared (described above). A PE-conjugated antibody against EpCAM (rat, eBiosciences 12-5791-81, clone G8.8; 1:100) was added and after incubation, EpCAM+/Tomato+ AT2 cells were sorted by FACS directly into cell lysis buffer for RNA isolation.
To isolate AT2 cells for culturing, a cell suspension (generated as above) from adult (PN50) B6 mouse lung was depleted of CD45+ cells using a magnetic-activated cell sorter (MACS, Miltenyi Biotec) and CD45 microbeads (rat monoclonal, 30F11.1, Miltenyi Biotech 130-052-301) according to manufacturer instructions, incubated with a biotinylated antibody against EpCAM (rat, eBiosciences 13-5791-81; clone G8.8), then enriched for EpCAM+ cells using streptavidin-coated microbeads (Miltenyi Biotech 130-048-102) as described45,46. Immunostaining for SftpC (rabbit, Chemicon AB3786; 1:100) was used to determine purity, with 90% of cells positive for this AT2 cell type marker.
To determine if Shh+ embryonic progenitors were present in adult lung, we administered 4 mg tamoxifen to an adult (PN154) Shh-Cre-ER> mTmG mouse then collected and examined the lungs six days later for cells expressing the lineage mark, which were not observed. We also isolated distal epithelial cells from lungs of adult (PN70) B6 mice (n=3) as above, performed FACS to select EpCAM+ cells, then quantitated the fraction of Muc1+/Pdpn+ cells.
AT2 cell culture
Purified AT2 cells (described above) were seeded at a density of ~10,000 cells per well in #1 8-well coverglass chambers (Labtek) containing DMEM/F12 media supplemented with 10% fetal bovine serum. Cells were cultured at 37°C for 16 days (with media replacement every two days), during which they acquired an AT1-like cell phenotype47. Alternatively, AT2 cells were cultured on a layer of Matrigel (BD) in serum-free media48 (DMEM/F12 supplemented with L-glutamine, non-essential amino acids, and penicillin-streptomycin) at 37°C for 72 hours, after which these conditions were either maintained or one of the EGF ligands TGFα (4 uM, R&D Systems 239-A-100), HB-EGF (4 uM, R&D Systems 259-HE-050), NRG1 (4 uM, R&D Systems 5898-NR-050) or a function blocking anti-EGFR antibody (rat, Abcam AB231, clone ICR10; 2.5 ug/mL) was added and the cells cultured for an additional 24 hours, then fixed (Cytofix/Cytoperm, BD) and immunostained for SftpC (rabbit, Chemicon AB3786; 1:100) and Ki67 (rat, eBiosciences 41-5698-80, clone SolA15; 1:100) to identify proliferating AT2 cells. Stained samples were imaged using a Zeiss LSM 780 confocal microscope. Total and Ki67 positive nuclei were quantified (500 cells per well) for each condition in a blinded manner.
Alveolar progenitor and AT2 cell gene expression profiling
RNA was extracted from E18 bipotent progenitors or adult AT2 cells (isolated as described above, with yields of approximately 35,000 cells) using the RNeasy Micro Kit (Qiagen), cDNA synthesized using a 2 cycle cDNA synthesis kit (Affymetrix) and analyzed on a Mouse Genome 430 2.0 Array (Affymetrix). RNA quality and quantity assessment, cDNA synthesis, probe preparation, labeling, hybridization and image scan were performed by the Protein and Nucleic Acid (PAN) facility at Stanford School of Medicine. Intensity values acquired from the Expression Console (Affymetrix) were analyzed using Gene Expression Commons49 (gexc.stanford.edu).
Supplementary Material
Extended Data Figures
Extended Data Figure 1. Mature and developing structure of the lung and alveoli. (a) Mature lung showing close up (inset) of transition from bronchial tree to alveoli (modified from www.mayoclinic.com/IM02960>www.mayoclinic.com/IM02960). (b) Alveoli are surrounded by dense capillaries networks in which de-oxygenated blood (blue) from pulmonary arteries is oxygenated (red) and then returned to the heart through pulmonary veins (modified from hendrosmk.wordpress.com>hendrosmk.wordpress.com). (c) E16.5 Shh-Cre> mTmG mouse lung in which GFP is expressed throughout the epithelium. Lobes are labeled (RAcc, right accessory; RCr, right cranial; RMid, right middle; RCa, right caudal; L, left) and boxed region shows the tip of the accessory lobe that was used for developmental analyses. By E16.5, the bronchial tree has formed and one day later sacculation begins as squamous AT1 and cuboidal AT2 cells mature to generate a functional gas exchange interface. Saccules subsequently undergo subdivision (“secondary septation”) into mature alveoli that provides an extensive gas-exchange surface. Bar, 1 mm; E, embryonic day. (d) Schematic of cell morphogenesis during sacculation (sac) (from Burri, 1998). Progenitors form flat AT1 cells adjacent to capillaries and AT2 cells specialized to secrete surfactant. (e,f) Schematics and images (e',f') of E-cadherin-stained accessory lobe tips at E18.5 and postnatal day 2 (PN2) showing longitudinal (e) and en face (f) views of maturing AT1 (1) and AT2 (2) cells. P, proximal; D, distal; red, cell junctions (jxn); yellow, apical surfaces. Note lack of AT1 cells distally in sacculating airway. Bar, 10um (e',f'). (g) Quantitation of ultrastructural classification of cell types in sacculating airways in E18.3 lungs (see Figure 1r–u). Values shown are the numbers of each progenitor and cell type observed with the indicated ultrastructural features. No cells (n=36) had features of an AT2>AT1 intermediate (AT1/2) or mature AT2 cell.
Extended Data Figure 2. Clonal analysis of alveolar progenitor cells and lineage marking and tracing alveolar type 2 (AT2) cells with LysM-Cre. (a,b) Shh-Cre-ER> mTmG embryos were induced in utero with a limiting dose (2 mg) of tamoxifen (tamox) at E15 to pulse-label epithelial cells at the distal lung tips (alveolar progenitors) with GFP (0.2 labeled cells per embryonic lung lobe) shortly before the onset of differentiation then examined 34 days later at PN30. (a) An isolated clone (dashed circle) expressing the GFP lineage tag (green) in a PN30 lung. (b) Close up of boxed region showing several flat AT1 cells (open arrows) and a cuboidal AT2 cell (filled arrows) within the alveolar clone, indicating that the tagged progenitor was bipotent. Tagged cells are interspersed with unrecombined cells (tdTomato, red). E, embryonic day; PN, postnatal day; Bar, 50 um. (c–f) Close-ups of alveoli of one (c, e) and two month (mo) old (d, f) LysM-Cre> mTmG lungs stained for the AT2 lineage tag (GFP, green) and the AT2 (c, d) or AT1 (e, f) markers indicated. Note that at 1 month lineage marked cells (green) express the AT2 (c) but not the AT1 marker (e). At 2 months (d, f), the lineage mark is also observed in some flat AT1 cells. Filled arrows, AT2 cells; open arrows, AT1 cells. Bar, 20 um (c–f).
Extended Data Figure 3. Proliferation analysis of bipotent progenitors and alveolar epithelial cells. Late gestational (E17.5, a) and early postnatal (PN7, 14, 21; b–d) lungs stained for Nkx2.1 (green) for epithelial and Ki67 (red) for actively cycling cells. Note essentially exclusive labeling, indicating minimal proliferation of (a) bipotent progenitors or (b–d) AT1 and AT2 cells. Arrow, a rare proliferating AT2 cell. Dashes outline distal epithelial tips; dotted line indicates mesothelium. E, embryonic day; PN, postnatal day; Bar, 35 um.
Extended Data Figure 4. Quantitation of cell type labeling and long-term lineage contribution of LysM-Cre and SftpC-Cre-ER marked cells. (a) Alveolar region of a PN3m Shh-Cre> R26EYFP mouse lung co-stained for GFP (green, epithelial cytoplasm) and LysM (red). Inset shows close-up of boxed region. LysM is detected in cytoplasm of many AT2 cells (solid arrow) but not AT1 cells (open arrow). (b) Bronchoalveolar lung region of a PN2m LysM reporter mouse expressing GFP from the endogenous locus stained for E-cadherin (red) to mark airway epithelium and GFP (green) to mark LysM-expressing cells. Note AT2 (filled arrows) but not bronchiolar cells (arrowheads mark bronchoalveolar junction (Badj)) express the LysM reporter. (c) PN17m LysM-Cre> mTmG lung stained for E-cadherin (red) and the AT2 lineage marker (green). Note many marked AT2 cells (solid arrows) but absence of lineage-marked cells in the terminal bronchiole (arrowheads, Badj). (d,e) Lungs from LysM-GFP (d) and LysM-Cre> mTmG (e) mice of the indicated ages stained for ciliated (acetylated tubulin, acTub, red) and neuroendocrine (NE) cell (CGRP, blue) markers and GFP (green) show no co-expressing cells, indicating ciliated (*) and NE (arrowhead) cell types do not express LysM and are not maintained by AT2 cells. Br, bronchus. (f–h) Lungs from LysM-Cre> mTmG mice of the indicated ages stained for the AT2 lineage tag (green), the AT2 cell marker SftpC (red), and the Clara cell marker CCSP (blue) shows SftpC+/CCSP+ (double-positive) cells (*) at the Badj, some of which are tagged. Marked double-positive cells are solitary (g) or in doublets (h). (i) Lung from PN1m SftpC-Cre-ER> mTmG mouse (administered 1 mg tamoxifen at PN19) analyzed 13d after pulse-labeling and stained as in D. Note no co-expressing cells, indicating that ciliated (*) and NE (arrowhead) cell types are not tagged. (j–l) Lungs from mice labeled as in I analyzed 13d (j) and 192d (k, l) later stained as in F. Note double-positive SftpC+/CCSP+ cells (*) at the Badj, some of which are pulse-labeled (j). After 192d chase, marked double-positive cells are solitary (k) or in doublets (l). (m) Quantification of lung cell types marked under different labeling and lineage trace conditions. The number of marked and total cells of each type scored (and Badj analyzed for SftpC+/CCSP+ cells) are shown for each genotype and age analyzed. For the SftpC-Cre-ER line, the dosage of tamoxifen (tamox) and interval time until analysis is also indicated. Because bronchial maintenance involves proliferation without significant cell dispersion1 (as we find for alveolar maintenance), the presence of marked SftpC+/CCSP+ cells primarily in isolation (>90%) at advanced ages suggests they did not contribute significantly to physiologic bronchiolar renewal. d, days; PN, postnatal; m, months. Bar, 50 um (a–l), 10 um (insets in A).
1 Giangreco, A. et al. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA 106, 9286–9291 (2009).
Extended Data Figure 5. Lineage tracing alveolar type 2 (AT2) cells using SftpC-Cre-ER. SftpC-Cre-ER> mTmG mice were administered 1 mg tamoxifen (Tamox) at PN18 then analyzed later by staining for the lineage label (GFP, green) and AT2 cells (SftpC, red). (a) 13d later only AT2 cells are marked, whereas (b) after 212d flat AT1 cells (open arrows) also express the AT2 lineage tag. A peripheral renewal focus (mesothelium indicated by dotted line) involving multiple alveoli (asterisks) is shown, similar to results using LysM-Cre. Quantitation revealed that 94% of AT2 cells were marked 13d after tamoxifen induction and 97% after 192d (see Extended Data Figure 5m), suggesting that the SftpC+ population is maintained by self-duplication, and that new AT2 cells do not derive from another cell population during physiologic aging. PN, postnatal day; d, day; m, month; Bar, 100 um.
Extended Data Figure 6. Alveolar type 2 (AT2) founder cell functional marker expression and self-duplication in vivo, and reprogramming into AT1 cells in vitro. (a–c) Postnatal (PN) 16 month LysM-Cre> Confetti mouse lung stained for mCFP lineage tag (green), AT2 cell marker (Nkx2.1, red), and DAPI (blue) to identify clonal renewal foci. (a) AT2 founder cell (solid arrow) associated with a daughter AT1 cell (open arrow) shown to express the A2L marker LAMP-1 (white), a protein associated with surfactant-containing lysosomes in mature, secretory AT2 cells. (b,c) A clonal focus (b, boxed region) in which an AT2 cell has generated two additional AT2 cells (solid arrows) but no AT1 cells (c, close up of boxed region), demonstrating isolated self-duplication without AT1 cell reprogramming in vivo. m, months. Bars, 25 um (b,c). (d) Freshly isolated AT2 cells (Figure 3f) cultured four days on glass with 10% serum adopt flat AT1 morphology (E-cadherin, green) and initiate AT1 marker expression (Aqp5, not shown). Bar, 10 um.
Extended Data Figure 7. Clonogenic response to activated Kras using SftpC-Cre-ER and CCSP-Cre-ER. Lungs of SftpC-Cre-ER (a, c) and CCSP-Cre-ER (b) mice carrying oncogenic KrasLSL-G12D/+ (Kras*) and either Rainbow (Rbw) (a, b) or mTmG (c) alleles, injected with 3 mg (a), 1 mg (b), or 2 mg (c) of tamoxifen (tamox) at indicated ages to induce Kras* expression and clonal lineage marking of StfpC- and CCSP-positive cells, respectively. Lungs were analyzed after 17d at PN43 (a), 39d at PN81 (b), and 53d at PN169 (c). Note that multifocal tumors result when SftpC-Cre-ER mice are induced at 25 (a) and 116 (c) days of age, while induction of Kras* in CCSP-Cre-ER mice at 42 days (b) shows many unresponsive cells and doublets throughout the bronchi (Br) as well as small clonal tumors located at bronchoalveolar duct junctions (Badj). Dotted line indicates mesothelium; PN, postnatal day; Bar, 100 um.
Extended Data Figure 8. Marker expression analysis of alveolar type 2 (AT2) cell derived adenomas. LysM-Cre> mTmG, KrasLSL-G12D/+ (abbreviated Kras*) lungs stained for the AT2-lineage marker (GFP, green), the nuclear stain DAPI (blue), and the indicated AT2, AT1, and Clara cell markers (red). Note tumor cells (green) maintain expression of AT2 markers (a, b) and do not turn on Clara (e) or AT1 markers (c, d), except possibly rare cells (*). c' and d' show control (non-tumor) regions with normal AT1 staining (arrows). Bars, 10 um.
Supplementary_Tables
03
Acknowledgments
We thank Andres Andalon for technical assistance; Hal Chapman (SftpC-Cre-ER-rtTA), Brigid Hogan (CCSP-Cre-ER), Hiroo Ueno and Irv Weissman (Rainbow), Hans Clevers (Confetti), Liqun Luo (mTmG), and Julien Sage (KrasLSL-G12D) for strains; Barry Stripp for goat anti-CCSP antibody; Hernan Espinoza for annotated gene lists; and Ross Metzger, Hal Chapman, and members of the Krasnow Lab for discussions and comments on the manuscript; and Maria Petersen for help preparing figures and the manuscript.
funding This work was supported by a Parker B. Francis Foundation Fellowship and NIH 5KO8HL084095 Award (T.D.), NIH T32HD007249 (D.B.), and an NHLBI U01 Progenitor Cell Biology Consortium grant (M.K.). M.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions T.D. conducted the experiments except the gene expression profiling and AT2 cell cultures, which were done by D.B. T.D. and M.K. conceived the experiments, analyzed the data and wrote the manuscript.
References
- 1.Bertalanffy FD, Leblond CP. Structure of respiratory tissue. Lancet. 1955;269:1365–1368. doi: 10.1016/s0140-6736(55)93164-0. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Rock JR, Hogan BL. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- 4.Chapman HA, et al. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamson IY, Bowden DH. Derivation of type 1 epithelium from type 2 cells in the developing rat lung. Lab Invest. 1975;32:736–745. [PubMed] [Google Scholar]
- 6.Spencer H, Shorter RG. Cell turnover in pulmonary tissues. Nature. 1962;194:880. doi: 10.1038/194880a0. [DOI] [PubMed] [Google Scholar]
- 7.Evans MJ, Bils RF. Identification of cells labeled with tritiated thymidine in the pulmonary alveolar walls of the mouse. Am Rev Respir Dis. 1969;100:372–378. doi: 10.1164/arrd.1969.100.3.372. [DOI] [PubMed] [Google Scholar]
- 8.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:E1475–1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Renewal of alveolar epithelium in the rat following exposure to NO2. Am J Pathol. 1973;70:175–198. [PMC free article] [PubMed] [Google Scholar]
- 11.Adamson IY, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab Invest. 1974;30:35–42. [PubMed] [Google Scholar]
- 12.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 13.McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci U S A. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar PA, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajstura J, et al. Evidence for human lung stem cells. N Engl J Med. 2011;364:1795–1806. doi: 10.1056/NEJMoa1101324. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Rawlins EL, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 18.Burri PH, Moschopulos M. Structural analysis of fetal rat lung development. Anat Rec. 1992;234:399–418. doi: 10.1002/ar.1092340310. [DOI] [PubMed] [Google Scholar]
- 19.Buckingham S, McNary WF, Jr., Sommers SC. Pulmonary Alveolar Cell Inclusions: Their Development in the Rat. Science. 1964;145:1192–1193. doi: 10.1126/science.145.3637.1192. [DOI] [PubMed] [Google Scholar]
- 20.Williams MC, Dobbs LG. Expression of cell-specific markers for alveolar epithelium in fetal rat lung. Am J Respir Cell Mol Biol. 1990;2:533–542. doi: 10.1165/ajrcmb/2.6.533. [DOI] [PubMed] [Google Scholar]
- 21.Hughes GM. Ultrastructure of the lung of Neoceratodus and Lepidosiren in relation to the lung of other vertebrates. Folia Morphol (Praha) 1973;21:155–161. [PubMed] [Google Scholar]
- 22.Miller LA, Wert SE, Whitsett JA. Immunolocalization of sonic hedgehog (Shh) in developing mouse lung. J Histochem Cytochem. 2001;49:1593–1604. doi: 10.1177/002215540104901213. [DOI] [PubMed] [Google Scholar]
- 23.Messier B, Leblond CP. Cell proliferation and migration as revealed by radioautography after injection of thymidine-H3 into male rats and mice. Am J Anat. 1960;106:247–285. doi: 10.1002/aja.1001060305. [DOI] [PubMed] [Google Scholar]
- 24.Bowden DH, Adamson IY, Wyatt JP. Reaction of the lung cells to a high concentration of oxygen. Arch Path. 1968;86:671–675. [PubMed] [Google Scholar]
- 25.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland KD, Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:4910–4915. doi: 10.1073/pnas.1112499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin C, et al. Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One. 2012;7:e53817. doi: 10.1371/journal.pone.0053817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikitin AY, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 30.Takanami I, Takeuchi K, Kodaira S. Tumor-associated macrophage infiltration in pulmonary adenocarcinoma: association with angiogenesis and poor prognosis. Oncology. 1999;57:138–142. doi: 10.1159/000012021. [DOI] [PubMed] [Google Scholar]
- 31.Brody JS, Burki R, Kaplan N. Deoxyribonucleic acid synthesis in lung cells during compensatory lung growth after pneumonectomy. Am Rev Respir Dis. 1978;117:307–316. doi: 10.1164/arrd.1978.117.2.307. [DOI] [PubMed] [Google Scholar]
- 32.Ding BS, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 37.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 38.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 40.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726. [PubMed] [Google Scholar]
- 44.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Messier EM, Mason RJ, Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp Lung Res. 2012;38:363–373. doi: 10.3109/01902148.2012.713077. [DOI] [PubMed] [Google Scholar]
- 46.Fujino N, et al. A novel method for isolating individual cellular components from the adult human distal lung. Am J Respir Cell Mol Biol. 2012;46:422–430. doi: 10.1165/rcmb.2011-0172OC. [DOI] [PubMed] [Google Scholar]
- 47.Dobbs LG, Williams MC, Brandt AE. Changes in biochemical characteristics and pattern of lectin binding of alveolar type II cells with time in culture. Biochim Biophys Acta. 1985;846:155–166. doi: 10.1016/0167-4889(85)90121-1. [DOI] [PubMed] [Google Scholar]
- 48.Sugahara K, Mason RJ, Shannon JM. Effects of soluble factors and extracellular matrix on DNA synthesis and surfactant gene expression in primary cultures of rat alveolar type II cells. Cell Tissue Res. 1998;291:295–303. doi: 10.1007/s004410050999. [DOI] [PubMed] [Google Scholar]
- 49.Seita J, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One. 2012;7:e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figures
Extended Data Figure 1. Mature and developing structure of the lung and alveoli. (a) Mature lung showing close up (inset) of transition from bronchial tree to alveoli (modified from www.mayoclinic.com/IM02960>www.mayoclinic.com/IM02960). (b) Alveoli are surrounded by dense capillaries networks in which de-oxygenated blood (blue) from pulmonary arteries is oxygenated (red) and then returned to the heart through pulmonary veins (modified from hendrosmk.wordpress.com>hendrosmk.wordpress.com). (c) E16.5 Shh-Cre> mTmG mouse lung in which GFP is expressed throughout the epithelium. Lobes are labeled (RAcc, right accessory; RCr, right cranial; RMid, right middle; RCa, right caudal; L, left) and boxed region shows the tip of the accessory lobe that was used for developmental analyses. By E16.5, the bronchial tree has formed and one day later sacculation begins as squamous AT1 and cuboidal AT2 cells mature to generate a functional gas exchange interface. Saccules subsequently undergo subdivision (“secondary septation”) into mature alveoli that provides an extensive gas-exchange surface. Bar, 1 mm; E, embryonic day. (d) Schematic of cell morphogenesis during sacculation (sac) (from Burri, 1998). Progenitors form flat AT1 cells adjacent to capillaries and AT2 cells specialized to secrete surfactant. (e,f) Schematics and images (e',f') of E-cadherin-stained accessory lobe tips at E18.5 and postnatal day 2 (PN2) showing longitudinal (e) and en face (f) views of maturing AT1 (1) and AT2 (2) cells. P, proximal; D, distal; red, cell junctions (jxn); yellow, apical surfaces. Note lack of AT1 cells distally in sacculating airway. Bar, 10um (e',f'). (g) Quantitation of ultrastructural classification of cell types in sacculating airways in E18.3 lungs (see Figure 1r–u). Values shown are the numbers of each progenitor and cell type observed with the indicated ultrastructural features. No cells (n=36) had features of an AT2>AT1 intermediate (AT1/2) or mature AT2 cell.
Extended Data Figure 2. Clonal analysis of alveolar progenitor cells and lineage marking and tracing alveolar type 2 (AT2) cells with LysM-Cre. (a,b) Shh-Cre-ER> mTmG embryos were induced in utero with a limiting dose (2 mg) of tamoxifen (tamox) at E15 to pulse-label epithelial cells at the distal lung tips (alveolar progenitors) with GFP (0.2 labeled cells per embryonic lung lobe) shortly before the onset of differentiation then examined 34 days later at PN30. (a) An isolated clone (dashed circle) expressing the GFP lineage tag (green) in a PN30 lung. (b) Close up of boxed region showing several flat AT1 cells (open arrows) and a cuboidal AT2 cell (filled arrows) within the alveolar clone, indicating that the tagged progenitor was bipotent. Tagged cells are interspersed with unrecombined cells (tdTomato, red). E, embryonic day; PN, postnatal day; Bar, 50 um. (c–f) Close-ups of alveoli of one (c, e) and two month (mo) old (d, f) LysM-Cre> mTmG lungs stained for the AT2 lineage tag (GFP, green) and the AT2 (c, d) or AT1 (e, f) markers indicated. Note that at 1 month lineage marked cells (green) express the AT2 (c) but not the AT1 marker (e). At 2 months (d, f), the lineage mark is also observed in some flat AT1 cells. Filled arrows, AT2 cells; open arrows, AT1 cells. Bar, 20 um (c–f).
Extended Data Figure 3. Proliferation analysis of bipotent progenitors and alveolar epithelial cells. Late gestational (E17.5, a) and early postnatal (PN7, 14, 21; b–d) lungs stained for Nkx2.1 (green) for epithelial and Ki67 (red) for actively cycling cells. Note essentially exclusive labeling, indicating minimal proliferation of (a) bipotent progenitors or (b–d) AT1 and AT2 cells. Arrow, a rare proliferating AT2 cell. Dashes outline distal epithelial tips; dotted line indicates mesothelium. E, embryonic day; PN, postnatal day; Bar, 35 um.
Extended Data Figure 4. Quantitation of cell type labeling and long-term lineage contribution of LysM-Cre and SftpC-Cre-ER marked cells. (a) Alveolar region of a PN3m Shh-Cre> R26EYFP mouse lung co-stained for GFP (green, epithelial cytoplasm) and LysM (red). Inset shows close-up of boxed region. LysM is detected in cytoplasm of many AT2 cells (solid arrow) but not AT1 cells (open arrow). (b) Bronchoalveolar lung region of a PN2m LysM reporter mouse expressing GFP from the endogenous locus stained for E-cadherin (red) to mark airway epithelium and GFP (green) to mark LysM-expressing cells. Note AT2 (filled arrows) but not bronchiolar cells (arrowheads mark bronchoalveolar junction (Badj)) express the LysM reporter. (c) PN17m LysM-Cre> mTmG lung stained for E-cadherin (red) and the AT2 lineage marker (green). Note many marked AT2 cells (solid arrows) but absence of lineage-marked cells in the terminal bronchiole (arrowheads, Badj). (d,e) Lungs from LysM-GFP (d) and LysM-Cre> mTmG (e) mice of the indicated ages stained for ciliated (acetylated tubulin, acTub, red) and neuroendocrine (NE) cell (CGRP, blue) markers and GFP (green) show no co-expressing cells, indicating ciliated (*) and NE (arrowhead) cell types do not express LysM and are not maintained by AT2 cells. Br, bronchus. (f–h) Lungs from LysM-Cre> mTmG mice of the indicated ages stained for the AT2 lineage tag (green), the AT2 cell marker SftpC (red), and the Clara cell marker CCSP (blue) shows SftpC+/CCSP+ (double-positive) cells (*) at the Badj, some of which are tagged. Marked double-positive cells are solitary (g) or in doublets (h). (i) Lung from PN1m SftpC-Cre-ER> mTmG mouse (administered 1 mg tamoxifen at PN19) analyzed 13d after pulse-labeling and stained as in D. Note no co-expressing cells, indicating that ciliated (*) and NE (arrowhead) cell types are not tagged. (j–l) Lungs from mice labeled as in I analyzed 13d (j) and 192d (k, l) later stained as in F. Note double-positive SftpC+/CCSP+ cells (*) at the Badj, some of which are pulse-labeled (j). After 192d chase, marked double-positive cells are solitary (k) or in doublets (l). (m) Quantification of lung cell types marked under different labeling and lineage trace conditions. The number of marked and total cells of each type scored (and Badj analyzed for SftpC+/CCSP+ cells) are shown for each genotype and age analyzed. For the SftpC-Cre-ER line, the dosage of tamoxifen (tamox) and interval time until analysis is also indicated. Because bronchial maintenance involves proliferation without significant cell dispersion1 (as we find for alveolar maintenance), the presence of marked SftpC+/CCSP+ cells primarily in isolation (>90%) at advanced ages suggests they did not contribute significantly to physiologic bronchiolar renewal. d, days; PN, postnatal; m, months. Bar, 50 um (a–l), 10 um (insets in A).
1 Giangreco, A. et al. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA 106, 9286–9291 (2009).
Extended Data Figure 5. Lineage tracing alveolar type 2 (AT2) cells using SftpC-Cre-ER. SftpC-Cre-ER> mTmG mice were administered 1 mg tamoxifen (Tamox) at PN18 then analyzed later by staining for the lineage label (GFP, green) and AT2 cells (SftpC, red). (a) 13d later only AT2 cells are marked, whereas (b) after 212d flat AT1 cells (open arrows) also express the AT2 lineage tag. A peripheral renewal focus (mesothelium indicated by dotted line) involving multiple alveoli (asterisks) is shown, similar to results using LysM-Cre. Quantitation revealed that 94% of AT2 cells were marked 13d after tamoxifen induction and 97% after 192d (see Extended Data Figure 5m), suggesting that the SftpC+ population is maintained by self-duplication, and that new AT2 cells do not derive from another cell population during physiologic aging. PN, postnatal day; d, day; m, month; Bar, 100 um.
Extended Data Figure 6. Alveolar type 2 (AT2) founder cell functional marker expression and self-duplication in vivo, and reprogramming into AT1 cells in vitro. (a–c) Postnatal (PN) 16 month LysM-Cre> Confetti mouse lung stained for mCFP lineage tag (green), AT2 cell marker (Nkx2.1, red), and DAPI (blue) to identify clonal renewal foci. (a) AT2 founder cell (solid arrow) associated with a daughter AT1 cell (open arrow) shown to express the A2L marker LAMP-1 (white), a protein associated with surfactant-containing lysosomes in mature, secretory AT2 cells. (b,c) A clonal focus (b, boxed region) in which an AT2 cell has generated two additional AT2 cells (solid arrows) but no AT1 cells (c, close up of boxed region), demonstrating isolated self-duplication without AT1 cell reprogramming in vivo. m, months. Bars, 25 um (b,c). (d) Freshly isolated AT2 cells (Figure 3f) cultured four days on glass with 10% serum adopt flat AT1 morphology (E-cadherin, green) and initiate AT1 marker expression (Aqp5, not shown). Bar, 10 um.
Extended Data Figure 7. Clonogenic response to activated Kras using SftpC-Cre-ER and CCSP-Cre-ER. Lungs of SftpC-Cre-ER (a, c) and CCSP-Cre-ER (b) mice carrying oncogenic KrasLSL-G12D/+ (Kras*) and either Rainbow (Rbw) (a, b) or mTmG (c) alleles, injected with 3 mg (a), 1 mg (b), or 2 mg (c) of tamoxifen (tamox) at indicated ages to induce Kras* expression and clonal lineage marking of StfpC- and CCSP-positive cells, respectively. Lungs were analyzed after 17d at PN43 (a), 39d at PN81 (b), and 53d at PN169 (c). Note that multifocal tumors result when SftpC-Cre-ER mice are induced at 25 (a) and 116 (c) days of age, while induction of Kras* in CCSP-Cre-ER mice at 42 days (b) shows many unresponsive cells and doublets throughout the bronchi (Br) as well as small clonal tumors located at bronchoalveolar duct junctions (Badj). Dotted line indicates mesothelium; PN, postnatal day; Bar, 100 um.
Extended Data Figure 8. Marker expression analysis of alveolar type 2 (AT2) cell derived adenomas. LysM-Cre> mTmG, KrasLSL-G12D/+ (abbreviated Kras*) lungs stained for the AT2-lineage marker (GFP, green), the nuclear stain DAPI (blue), and the indicated AT2, AT1, and Clara cell markers (red). Note tumor cells (green) maintain expression of AT2 markers (a, b) and do not turn on Clara (e) or AT1 markers (c, d), except possibly rare cells (*). c' and d' show control (non-tumor) regions with normal AT1 staining (arrows). Bars, 10 um.
Supplementary_Tables
03