Sensitive capture of circulating tumour cells by functionalised graphene oxide nanosheets (original) (raw)
. Author manuscript; available in PMC: 2014 May 12.
Published in final edited form as: Nat Nanotechnol. 2013 Sep 29;8(10):735–741. doi: 10.1038/nnano.2013.194
Abstract
The spread of cancer throughout the body is driven by circulating tumour cells (CTCs)1. These cells detach from the primary tumour and move from the blood stream to a new site of subsequent tumour growth. They also carry information about the primary tumour and have the potential to be valuable biomarkers for disease diagnosis and progression, and for the molecular characterization of certain biological properties of the tumour. However, the limited sensitivity and specificity of current methods to measure and study these cells in patient blood samples prevent the realization of their full clinical potential. The use of microfluidic devices is a promising method for isolating CTCs2, 3; however, the devices are reliant on three-dimensional structures, which limit further characterization and expansion of cells on the chip. Here we demonstrate an effective approach to isolate CTCs from blood samples of pancreatic, breast and lung cancer patients, by using functionalised graphene oxide nanosheets on a patterned gold surface. CTCs were captured with high sensitivity at low concentration of target cells (73% ± 32.4 at 3–5 cells/mL blood).
In early stage cancers, CTCs are present in the blood stream at a low concentration and epithelial cell adhesion molecule (EpCAM) expression, along with other cancer related biomarkers, may be heterogeneous4, 5. Hence, methods for detecting CTC in early stage cancer patients need to be more sensitive, reliable and specific6. Existing microfluidic-based technologies have improved the recovery of CTC from cancer patients2, 3; however, these technologies rely on micro-structures, trapping arrays2 or microfilters7, limiting downstream analysis and further culture. The development of a novel platform that enhances CTC isolation, is sensitive, allows imaging of captured CTCs and enables cell culture would dramatically increase the use of CTCs in diagnostics and prognostics.
Nanomaterials offer excellent opportunities to improve the sensitivity of biomolecule detection due to their high surface area to volume ratio and similar size to biomolecules8, 9. Recently, silicon-nanopillars10, quartz-nanowires11 and TiO2 nanofibers12 have been used to trap CTCs, with enhanced capture efficiency for nanomaterials with higher aspect ratios; however, capture yields were found to be lower than those observed for microfluidic-based CTC chips. Graphene oxide (GO) is a promising nanomaterial in application such as drug delivery13, biosensing14 and nanocomposites15. Its ease of surface modification using PEG-based chemistry16, control over the size of its sheets using sonication and filtration17 and its unique optical properties18 make GO an attractive material for use in biomolecule detection.
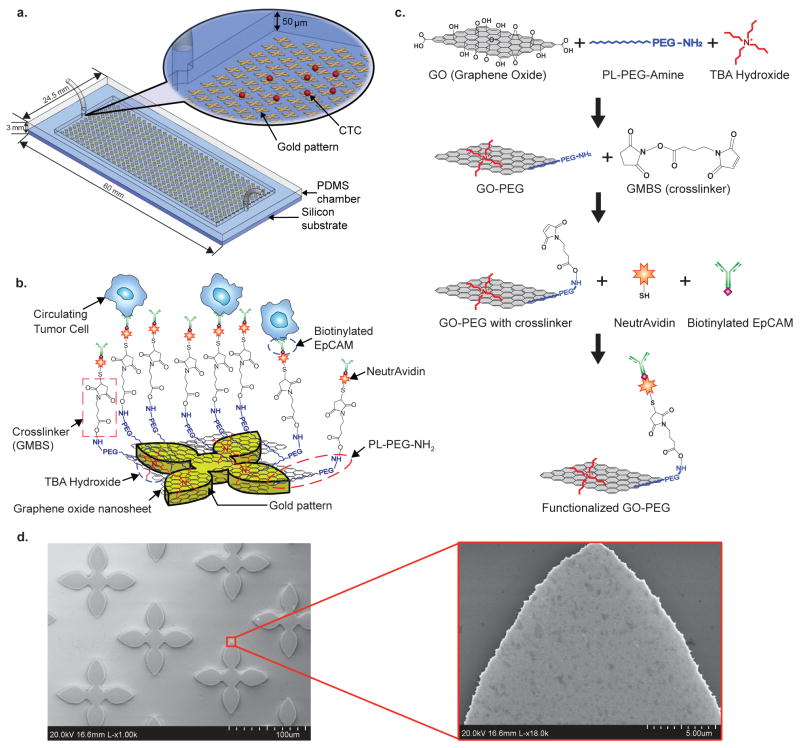
The GO chip we present here takes advantage of this novel nanomaterial for sensitive capture of CTCs using functionalised GO nanosheets on a flat substrate (Fig. 1a–b). The silicon substrate has 58,957 flower-shaped gold patterns with a dimension of 100 μm × 100 μm. The distance between each structure in a column is 150 μm and the overall size of the microfluidic device is 24.5 mm × 60 mm × 3 mm. The PDMS (polydimethylsiloxane) layer forms a microfluidic chamber with a 50 μm height and a total volume of 45 μL. Unlike other CTC capture devices with microposts (50~100 μm-thick)2, the effective functionalised surface created here enables the device to be a simple, flat and chamber-like structure (Supplementary Fig. S1). GO nanosheets are adsorbed onto the patterned gold surface, and then chemically functionalised with EpCAM antibodies. The GO functionalisation in this study uses a method described by Li et al. with some modifications (Fig. 1c)19. GO nanosheets are non-covalently functionalised by phospholipid-polyethylene-glyco-amine (PL-PEG-NH2) and the hydrophobic lipid chains of PL-PEG-NH2 are strongly immobilized onto the GO surface. Tetrabutylammonium (TBA) hydroxide is added for intercalation and complete exfoliation of GO. TBA cations and the amino group of PL-PEG-NH2 interact with the patterned gold surface by electrostatic attraction20. N-γ-maleimidobutyryloxy succinimide ester (GMBS) is introduced, which has _N_-hydroxysuccinimide (NHS) esters that react with amine groups of GO-PEG to form amide bonds. The CTCs are then captured using the following NeutrAvidin and biotinylated EpCAM antibody interactions. SEM images reveal that gold patterns were covered with functionalised GO nanosheets (Fig. 1d). This shows the high selectivity of GO adsorbed onto the gold patterns rather than silicon dioxide substrate as well as the uniform assembly and saturation density of GO on the gold pattern.
Figure 1. GO chip and functionalisation/characterisation of GO.
a, Schematic diagram of the GO chip. b, Schematic showing the conjugation chemistry between functionalised GO nanosheets and EpCAM antibodies. GO nanosheets are adsorbed onto the gold pattern. The GMBS crosslinker binds to PL-PEG-NH2 onto the GO nanosheets. The NeutrAvidin is connected to the GMBS and biotinylated EpCAM. c, Preparation procedures of the functionalized GO. d, SEM image of gold patterns. Inset: magnified SEM image of adsorbed GO nanosheets on gold patterns.
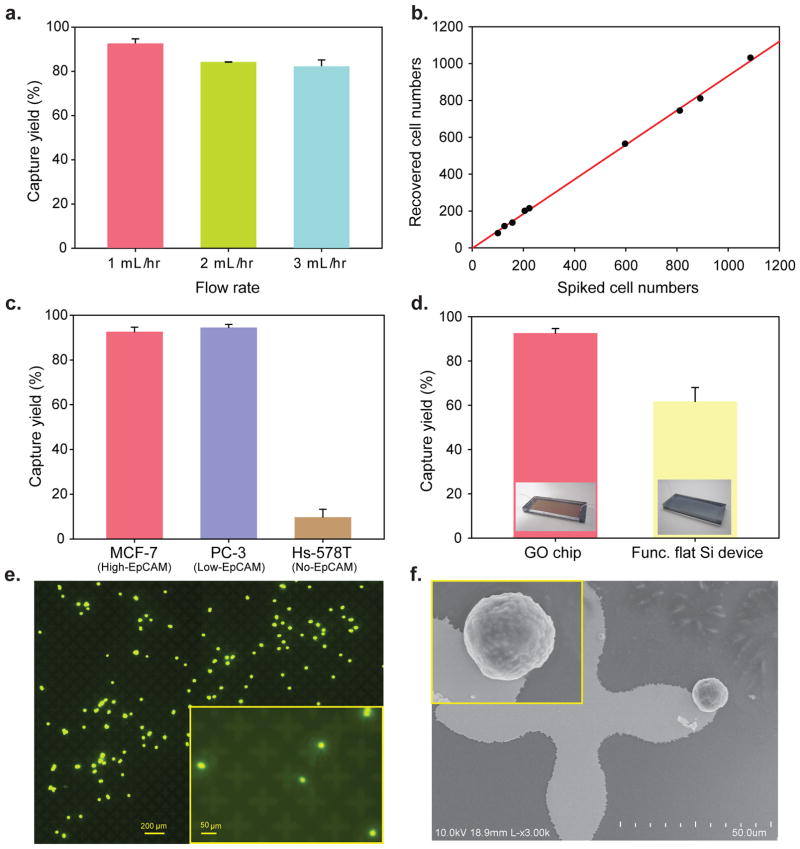
Human breast cancer cell lines (MCF-7, Hs-578T, originally from ATCC) and a human prostate cancer cell line (PC-3, originally from ATCC) were labeled with a fluorescent cell tracker dye, spiked into buffer at varying concentrations, and flowed through the GO chip. The captured cells in the GO chip and the non-captured cells collected in the waste were then counted. Different flow rates were applied to find the optimal flow rate (Fig. 2a), and at the 1–3 mL/hr rate range, the capture yield was over 82.3% (n=3). 100–1000 MCF-7 cells per milliliter were then spiked into buffer solution and captured at a flow rate of 1 mL/hr (Fig. 2b). There was little difference in capture yield between MCF-7 cells (high EpCAM expression cells) and PC3 cells (relatively low EpCAM expression cells), whereas the capture yield was less than 10% for a non-EpCAM expressing cell line Hs-578T21 (Fig. 2c). Next, the functionalised GO chip and a functionalised flat silicon device without microposts were compared to examine how GO might increase the capture yield (Fig. 2d). The results from spiked cells in buffer solution clearly showed that the GO chip increased capture efficiency. Furthermore, it can be seen that cells were specifically captured on the flower-shaped gold surfaces functionalised with GO. (Fig. 2e–f, Supplementary Fig. S5bc).
Figure 2. Characterization of the GO-chip with cells in buffer solution.
a, Flow rate dependency on capture efficiency of MCF-7 cells. Error bars represent the standard deviation of three replicates. b, Capture efficiency of MCF-7 cells at the 1 mL/hr. The red solid line is a fit to the result expected. c, Comparison of different cell lines; MCF-7 cells (high EpCAM expression), PC-3 cells (low EpCAM expression), and Hs-578T cells (no EpCAM expression). Error bars represent the standard deviation of three replicates. d, Cell recovery of MCF-7 cells compared to functionalized flat silicon device. Error bars represent the standard deviation of three replicates. Inset: photograph of GO chip (left) and photograph of functionalized flat silicon device (right). e, Fluorescence image of the captured MCF-7 cells. Inset: magnified (10x) fluorescence image of the captured MCF-7 cells. f, SEM image of the captured MCF-7 cell on the gold pattern. Inset: magnified SEM image of the captured MCF-7 cell.
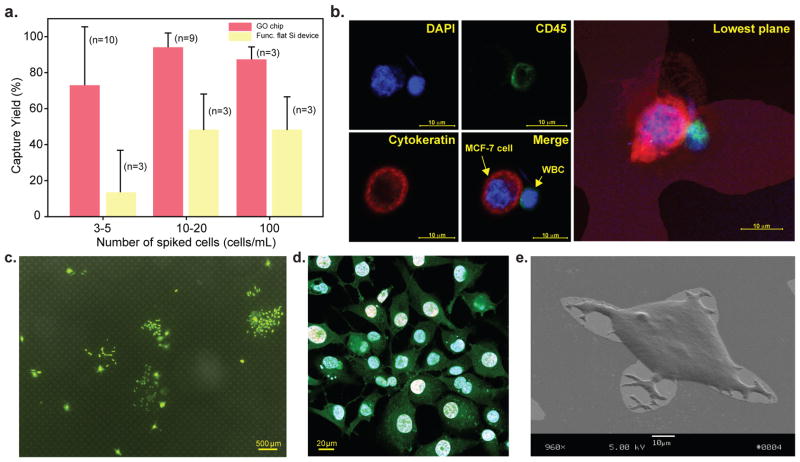
To further investigate capture efficiency in human blood samples, varying numbers of MCF-7 cells (3–5 cells, 10–20 cells, 100 cells) were spiked into 1 mL of whole blood and run through the GO chip and the functionalised flat silicon device (Fig. 3a, Supplementary Fig. S7). To differentiate captured CTCs and white blood cells, immunostaining was performed by using anti-cytokeratin and anti-CD45, where white blood cells were identified as positive for 4′, 6-diamidino-2-phenylindole (DAPI) and cluster of differentiation 45 (CD45, a common leucocyte antigen), while CTCs were identified as positive for DAPI and cytokeratin (CK), but negative for CD45 (Fig. 3b). The average recovery rates of 10–20 and 100 spiked cells per 1 mL were 94.2% (n=9) and 87.3% (n=3), respectively (Fig. 3a). In the case of 3–5 spiked cells per 1 mL, the average recovery rate was 73% (n=10) and five of the samples had 100% recovery rate (Supplementary Table S1). The higher standard deviation in the 3–5 spiked cell group may be explained by the inherent limitations in capture with cell numbers at low levels. The control functionalised flat silicon device had around 48% capture for both 10–20 and 100 spiked cells and only 13.3% capture for 3–5 spiked cells (Fig. 3a). Similar cell spike experiments were performed with the low EpCAM expressing PC-3 cell line and the recoveries were greater than 65% for both 3–5 and 10–20 spiked cells (Supplementary Fig. S8a). Furthermore, we compared the recovery of MCF-7 and PC-3 cells across the GO chip, the GO chip without gold patterns and the functionalised flat silicon device and found that the GO chip with gold patterns has higher sensitivity for low frequency (3–5 cells) cell recovery (Supplementary Fig. S8b). After MCF-7 cells were captured on the GO chip, they were cultured in the device for 6 days (Fig. 3c, Supplementary Fig. S9a–f). The captured cells spread on the surface of the substrate and showed evidence of active proliferation, measured by 5-Ethynyl-2′-deoxyuridine (EdU) staining shown in red fluorescence (Alexa Fluor 647)22 (Fig. 3d, Supplementary Fig. S9g–i). Scanning electron microscopy (SEM) images showed the cultured MCF-7 cells spread nicely onto the surface of the GO chip (Fig. 3e). The cultured cells were released from the chip by trypsinisation and further re-cultured in a 96 well plate (Supplementary Fig. S10).
Figure 3. Characterization of the GO-chip with MCF-7 cells spiked into whole blood.
a, Cell recovery MCF-7 cells spiked into 1 mL of whole blood at varying spike concentration from 3 to 100 cells/mL. Error bars represent the standard deviation of replicates. b, Fluorescence microscope image of MCF-7 and white blood cells stained with DAPI (blue), cytokeratin (red), and CD 45 (green). c, Fluorescence microscope image of 6-day-cultured MCF-7 cells. d, Fluorescence microscope image depicting proliferation of cells. e, SEM image of a captured and 6-day-cultured MCF-7 cell.
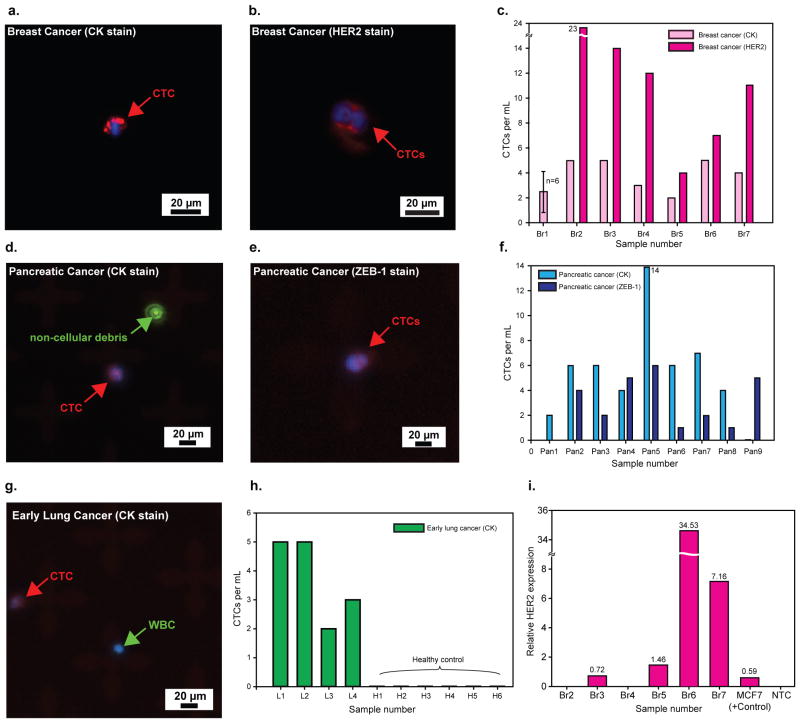
Fresh blood samples obtained from patients with metastatic breast cancer (n=7), early stage lung cancer (n=4) and metastatic pancreatic cancer (n=9) along with 6 age matched healthy individuals (Fig. 4, Supplementary Fig. S11) were processed on GO chips. Clinical data on these patients are available in Supplementary Table 2. CTCs in these samples were identified as DAPI positive nucleated cells staining positive for CK 7, 8 and negative for CD45 (Fig. 4a, d, g). Up to 23 CTCs/mL were captured from patient samples (n=39), with all patients analyzed (n=20) having ≥ 2 CTCs/mL (see Supplementary Table 2). We were unable to measure any CTCs in healthy controls (n=6, Fig. 4h, Supplementary Table S2). A small number of double positive cells (both CD45+ and CK+) were found in some patient samples. Given, very little has been known about the origin and significance of these cells23, 24, we excluded these from CTC enumeration. Two capture experiments were run in parallel for blood samples from breast cancer patients (n=6): one to collect the number of CK+ CTCs and another to stain cells with Human Epidermal Growth Factor Receptor 2 (HER2) along with CD45 to exclude the white blood cells (Fig. 4b). Almost all patients had a greater number of EpCAM+, HER2+ and CD45− cells (11 ± 6 CTCs/mL) than EpCAM+, CK+ and CD45− cells (3 ± 1 CTCs/mL) (Fig. 4c). Although both HER2-positive and HER2-negative primary breast tumour tissues were analyzed, CTCs from the respective blood samples showed a higher percentage of HER2-positive cells. It has been previously reported that the Veridex CellSearch platform is unable to capture CTCs in 30 to 35% of metastatic breast cancer patients1, 25, 26 and detected less than 5 CTCs per 7.5 mL in 70% of the samples with greater than 1 CTC per 7.5 mL. In a study comparing microfluidic approaches to CellSearch, the reported number of CTCs in metastatic breast cancer patients was ≤ 5 per 7.5 mL from 9 analyzed breast cancer patients27. For a similar cohort size, we detected 22 CTCs per 7.5 mL in metastatic breast cancer patients. Nine total pancreatic cancer patients were also analyzed, among which 7 were stage IV metastatic cancer patients, and 2 were patients with stage III disease. All patients had ≥ 2 CTCs/mL, with a mean of 5 ± 4 CTCs/mL. In addition to staining for CK, we analyzed samples in parallel for the epithelial to mesenchymal transition (EMT) marker, Zinc finger E-box-binding homeobox 1 (ZEB-1) along with CD45 to distinguish the white blood cells. Figure 4e presents a cluster of two CTCs expressing ZEB-1 which co-localized with the DAPI-stained nucleus. The average number of ZEB-1+ CTCs isolated was 2 ± 2 CTCs/mL. Among four early stage lung cancer patients analyzed, all of had ≥ 2 CTCs/mL, with a mean of 4 ± 1 CTCs/mL. To demonstrate downstream molecular assay feasibility, we extracted RNA from CTCs recovered from breast cancer patients. RNA extracted from four out of six breast cancer samples showed HER2 gene expression (Fig. 4i), whereas healthy control samples were negative for HER2. The bar chart shows the HER2 gene expression normalized with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in pure MCF-7 cells (100 cells), CTCs extracted from breast cancer patients. The negative controls (healthy blood run through the device) did not show the HER2 expression (up to 35 cycles).
Figure 4. Fluorescence microscope images and quantification of CTCs captured from cancer patient samples.
a, CTC captured from breast cancer patient #6 (Br6). b, CTCs captured from breast cancer patient #2 (Br2). c, Quantification of CTCs captured from breast cancer patients. d, CTC captured from pancreatic cancer patient #2 (Pan2); Fluorescence particle from Alexa Fluor 488 dye shows that CTC has no FITC (green). e, Two CTCs captured from pancreatic cancer patient #9 (Pan9). f, Quantification of CTCs captured from pancreatic cancer patients. g, CTC captured from early lung cancer patient #3 (L3). h, Quantification of CTCs captured from lung cancer patients and 6 healthy donors. i, HER2 gene expression (normalized to GAPDH) of captured CTCs by qRT-PCR. Four out of six patients showed expression higher than the gene expression of MCF-7 cells (100 cells).
In summary, we present a novel strategy using GO for sensitive planar CTC capture. Capture yields and detection sensitivities for single digit CTCs spiked into blood were much higher than the reported yields in the literature10. With functionalised GO nano-assemblies on the patterned gold surface as an effective tool, we were able to isolate, capture, identify and characterize rare CTCs in the blood of cancer patients for early cancer detection. The GO chip successfully isolates CTCs from early stage lung cancer patients along with advanced metastatic cancer patients. Recently two approaches were reported that are antibody independent and rely strictly on isolation of CTCs based on cell size27, 28. However their sensitivity in isolating CTCs in clinical samples was suboptimal compared to the capture efficiencies noted in this study. Furthermore, with filter-based techniques, downstream molecular assays still remain a significant challenge. Although larger clinical studies are warranted to validate our approach, the results demonstrate our ability to isolate CTCs from early stage cancer patients. Of interest, analysis of 6 breast cancer patient samples revealed both HER2-positive and HER2-negative cells primary tumours, but a relatively increased presence of HER2+ cells in the circulation. This warrants further mechanistic investigation, however studies have shown that metastatic lesions vary from the primary tumour in 5–30%29, 30 and that CTCs can be HER2-positive when the primary tumour is HER2-negative31–33, highlighting the importance of understanding the functional heterogeneity of cancer cells. Although further work is needed to refine the conditions, the extension of the platform to conventional cell culture techniques is a useful advantage. In summary, we demonstrate the efficacy of graphene oxide in a sensitive planar CTC capture device GO chip that provides a platform for functional studies of captured CTCs from blood.
Methods
Surface modification
GMBS solution was flowed through the PEG functionalised GO chip at a 20 μL/min flow rate using a syringe pump (Harvard Apparatus). After 30 minutes of incubation, the device was washed with ethanol at 100 μL/min. 50 μg/mL NeutrAvidin was prepared and flowed through the device at 20 μL/min. After 1 hour incubation, the device was flushed with phosphate buffered saline (PBS) at 100 μL/min to remove the excessive NeutrAvidin. Finally, biotinylated EpCAM antibody at a concentration of 20 μg/mL in PBS with 1% (w/v) BSA was flowed through the device for 10 minutes at 20 μL/min. After 1 hour incubation, PBS was flowed to wash then, 1% or 3% BSA solution in PBS was flowed at 100 μL/min for 5 minutes. After flowing BSA solution, the device was allowed to incubate for 30 minutes.
Cell culture and labeling
Tissue culture reagents were purchased from GIBCO Invitrogen Corporation/Life Technologies Life Sciences unless otherwise specified. MCF-7/Hs-578T and PC-3 cells were cultured in DMEM and DMEM/F12 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin solution. When cells reached more than 70–80% confluence, they were harvested and labeled with a green cell tracking dye (Invitrogen, CellTracker Green CMFDA, C7025). Subsequently, these fluorescence tracked cells were used to perform the capture efficiency experiments. For a low number cell spiking (3–20), cells were diluted in serum-free medium starting at an initial concentration of 1 × 105 cells/mL. 1 μL of the concentrated cell suspension was transferred to a low-attachment 96-well plate. The transferred cells were counted under the microscope, then immediately pipetted into a 1 mL of whole blood. After removing the cells from the 96-well plate, we counted the remaining cells at the same position. By subtracting these cells left behind from the original spot, the total number of cells spiked into blood was estimated.
Cytokeratin and CD45 staining
After flowing blood samples with low number of non-labeled cells, the captured cells were washed with PBS, fixed with 4% paraformaldehyde (PFA), permeabilized with 0.2% Triton-X and incubated for 30 minutes followed by PBS wash. The device was incubated for 30 minutes with 1 mL of blocking buffer containing 2% normal goat serum and 3% BSA. Anti-cytokeratin 7/8 (BD Biosciences) and anti-CD45 (BD Biosciences) were diluted to 5 μg/mL in 1% BSA. These antibodies were flowed through the GO chip for 20 minutes at 50 μL/min and incubated for 1 hour. After absorption of the primary antibody, the GO chip was washed with PBS. Anti-cytokeratin was probed with Alexa Fluor 488 IgG2a FITC (Invitrogen) and the anti-CD45 was probed with Alexa Fluor 546 IgG1 (Invitrogen). The secondary antibodies were diluted in 1% BSA at a 1:200 ratio, flowed through the GO chip for 20 minutes at 50 μL/min, incubated for 1 hour and followed by washing with PBS. To stain nuclei of the captured cells, DAPI (1:1000 dilution in PBS) was flowed for 20 minutes at 50 μL/min and the device was incubated for 15 minutes and washed with PBS.
Blood specimen collection
Blood samples were drawn from patients with tumours and healthy donors after obtaining informed consent under an IRB-approved protocol. All specimens were collected into EDTA tubes and were processed within 3 hours.
Cell Treatment with EdU
To measure cells’ ability to proliferate, Click-iT EdU Imaging Kit (Invitrogen, C10340) was used. After capturing cells, the GO chip was washed with PBS, and 10 μM EdU solution was added to the chip. The chip was incubated overnight, washed with PBS and followed by cell fixation with 4% PFA. After 15 minutes of incubation, the chip was washed with 3% BSA twice, followed by cell permeabilization with 0.5% Triton X-100 in PBS and incubated for 20 minutes. The chip was washed with 3% BSA twice and 0.5 mL of Click-iT reaction cocktail was added, followed by 30 minutes incubation and washing once with 3% BSA. For nucleus staining, 1 mL of 1X Hoechst 33342 solution was added and cells in the chip were incubated for 30 minutes and washed with 1 mL of PBS.
qRT-PCR analysis of captured CTCs
The RNA from captured CTCs either from spike samples or from patient’s blood samples was extracted from the GO chip using Arcturus Picopure RNA isolation kit according to the manufacturer’s instruction (ABI, Life Technologies). Then concentrated preparation of total RNA for each sample was used in RT reaction followed by pre-amplification of cDNAs using the pooled TaqMan Gene Expression assays of target genes and Cell-to-CT Kit according to the manufacturer’s instruction (Ambion, Life Technologies) on the Eppendorf mastercycler pro S instrument. Finally, gene expression experiments for each pre-amplified sample were performed using TaqMan Gene Expression Assays for GAPDH and HER2 (Life Technologies) in a multiplex qPCR setting on the ABI 7900HT instrument. Data were presented as mean Ct for mRNA expression level of studied genes.
Supplementary Material
1
2
Acknowledgments
We thank Kirk Herman and Stephanie Laurinec for procurement of patient blood samples. We also thank Shamileh Fouladdel for technical help of qRT-PCR. This work was supported by NIH Director’s New Innovator Award (1DP2OD006672-01), Career Development Program of the Gastrointestinal Specialized Program of Research Excellence (GI SPORE) award (CA130810), and 3M non-tenured Faculty Award. It was performed in part at the Lurie Nanofabrication Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation.
Footnotes
Author contributions
H.Y. and S.N. conceived and designed the study. H.Y. and T.K. fabricated devices and performed the experiments. T.K., Z.Z. and T.P. performed the cell culture. E.A. analyzed the RNA data. C.P., J.L., N.R., M.W., D.H., and D.S. prepared the clinical samples. H.Y. and S.N. analyzed the data and co-wrote the paper. All authors discussed the results and commented on the manuscript.
References
- 1.Cristofanilli M, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. New England Journal of Medicine. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 2.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maheswaran S, et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. New England Journal of Medicine. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willipinski-Stapelfeldt B, et al. Changes in Cytoskeletal Protein Composition Indicative of an Epithelial-Mesenchymal Transition in Human Micrometastatic and Primary Breast Carcinoma Cells. Clinical Cancer Research. 2005;11:8006–8014. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 5.Bednarz-Knoll N, Alix-Panabières C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer and Metastasis Reviews. 2012;31:673–687. doi: 10.1007/s10555-012-9370-z. [DOI] [PubMed] [Google Scholar]
- 6.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 7.Lin HK, et al. Portable Filter-Based Microdevice for Detection and Characterization of Circulating Tumor Cells. Clinical Cancer Research. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nano. 2007;2:469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 9.Xia XR, Monteiro-Riviere NA, Riviere JE. An index for characterization of nanomaterials in biological systems. Nat Nano. 2010;5:671–675. doi: 10.1038/nnano.2010.164. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, et al. Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angewandte Chemie. 2009;121:9132–9135. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SK, et al. Nanowire Substrate-Based Laser Scanning Cytometry for Quantitation of Circulating Tumor Cells. Nano Letters. 2012;12:2697–2704. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, et al. Electrospun TiO2 Nanofiber-Based Cell Capture Assay for Detecting Circulating Tumor Cells from Colorectal and Gastric Cancer Patients. Advanced Materials. 2012;24:2756–2760. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Robinson JT, Sun X, Dai H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J Am Chem Soc. 2008;130:10876–10877. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohanty N, Berry V. Graphene-Based Single-Bacterium Resolution Biodevice and DNA Transistor: Interfacing Graphene Derivatives with Nanoscale and Microscale Biocomponents. Nano Letters. 2008;8:4469–4476. doi: 10.1021/nl802412n. [DOI] [PubMed] [Google Scholar]
- 15.Ramanathan T, et al. Functionalized graphene sheets for polymer nanocomposites. Nat Nanotechnol. 2008;3:327–331. doi: 10.1038/nnano.2008.96. [DOI] [PubMed] [Google Scholar]
- 16.Dreyer DR, Park S, Bielawski CW, Ruoff RS. The chemistry of graphene oxide. Chemical Society Reviews. 2010;39:228–240. doi: 10.1039/b917103g. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, et al. Nano-Graphene Oxide for Cellular Imaging and Drug Delivery. Nano Res. 2008;1:203–212. doi: 10.1007/s12274-008-8021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eda G, Fanchini G, Chhowalla M. Large-area ultrathin films of reduced graphene oxide as a transparent and flexible electronic material. Nat Nano. 2008;3:270–274. doi: 10.1038/nnano.2008.83. [DOI] [PubMed] [Google Scholar]
- 19.Li X, et al. Highly conducting graphene sheets and Langmuir-Blodgett films. Nat Nano. 2008;3:538–542. doi: 10.1038/nnano.2008.210. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Wang X, Li X, Dai H. Chemical self-assembly of graphene sheets. Nano Research. 2009;2:336. [Google Scholar]
- 21.Sieuwerts AM, et al. Anti-Epithelial Cell Adhesion Molecule Antibodies and the Detection of Circulating Normal-Like Breast Tumor Cells. Journal of the National Cancer Institute. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lustberg M, Jatana KR, Zborowski M, Chalmers JJ. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 2012;195:97–110. doi: 10.1007/978-3-642-28160-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mego M, et al. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. International Journal of Cancer. 2011;129:417–423. doi: 10.1002/ijc.25690. [DOI] [PubMed] [Google Scholar]
- 26.Alix-Panabières C, Schwarzenbach H, Pantel K. Circulating Tumor Cells and Circulating Tumor DNA. Annual Review of Medicine. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 27.Ozkumur E, et al. Inertial Focusing for Tumor Antigen–Dependent and –Independent Sorting of Rare Circulating Tumor Cells. Science Translational Medicine. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs MG, et al. Analysis of Circulating Tumor Cells in Patients with Non-small Cell Lung Cancer Using Epithelial Marker-Dependent and -Independent Approaches. Journal of Thoracic Oncology. 2012;7:306–315. doi: 10.1097/JTO.0b013e31823c5c16. 310.1097/JTO.1090b1013e31823c31825c31816. [DOI] [PubMed] [Google Scholar]
- 29.Curigliano G, et al. Should liver metastases of breast cancer be biopsied to improve treatment choice? Annals of Oncology. 2011;22:2227–2233. doi: 10.1093/annonc/mdq751. [DOI] [PubMed] [Google Scholar]
- 30.Amir E, et al. Tissue confirmation of disease recurrence in breast cancer patients: Pooled analysis of multi-centre, multi-disciplinary prospective studies. Cancer Treatment Reviews. 2012;38:708–714. doi: 10.1016/j.ctrv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Fehm T, et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat. 2010;124:403–412. doi: 10.1007/s10549-010-1163-x. [DOI] [PubMed] [Google Scholar]
- 32.Riethdorf S, et al. Detection and HER2 Expression of Circulating Tumor Cells: Prospective Monitoring in Breast Cancer Patients Treated in the Neoadjuvant GeparQuattro Trial. Clinical Cancer Research. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 33.Pestrin M, et al. Final results of a multicenter phase II clinical trial evaluating the activity of single-agent lapatinib in patients with HER2-negative metastatic breast cancer and HER2-positive circulating tumor cells. A proof-of-concept study. Breast Cancer Res Treat. 2012;134:283–289. doi: 10.1007/s10549-012-2045-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2