Phylogenetically Older Introns Strongly Correlate With Module Boundaries in Ancient Proteins (original) (raw)
Abstract
The hypothesis that some (but not all) introns were used to construct ancient genes by exon shuffling of modules at the earliest stages of evolution is supported by the finding of an excess of phase-zero intron positions in the boundary regions of such modules in 276 ancient proteins (defined as common to eukaryotes and prokaryotes). Here we show further that as phase-zero intron positions are shared by distant taxa, and thus are truly phylogenetically ancient, their excess in the boundaries becomes greater, rising to an 80% excess if shared by four out of the five taxa: vertebrates, invertebrates, fungi, plants, and protists.
We recently studied the distribution of introns in homologs of 276 ancient unrelated proteins of known three-dimensional structure and found a significant, but small, excess of phase-zero intron positions in the boundary regions of modules 15–35 Å in diameter (Fedorov et al. 2001). This correlation holds only for phase-zero introns, which lie between codons, and not for phase-one or phase-two introns, lying after the first or second base. We interpreted these results in terms of a mixed model of intron origin: That some of the phase-zero introns were used in exon shuffling of modules to make domains at the beginning of evolution, whereas many of the phase-zero introns and all of the phase-one and phase-two introns had been added after the creation of these ancient genes. If the excess of introns in the boundary regions is due to the presence of a small number of ancient introns amid a larger number of more recently added introns, then if one could identify such ancient introns by their phylogenetic pattern, one expects them to show a greatly enhanced likelihood of lying near module boundaries. Here we show that this prediction is true.
Modules are compact subregions of the peptide chain, described as lying within a maximum diameter (Go 1981), in general much smaller than a “domain.” Modules can be defined for any arbitrary diameter, and introns have long been shown to correlate with boundaries of modules defined over a large range of diameters, from 15–30 Å. These geometrically defined Go-modules have been shown to code for quasi-independently folding units of a protein, as determined by energy landscape analysis (Panchenko et al. 1996) and as such give a useful definition of modular pieces of protein structure. Analysis of intron positions with respect to protein module boundaries was described by de Souza et al. (1996, 1998) and Fedorov et al. (2001).
The “ancient” proteins have homologs in both prokaryotes and eukaryotes. They have no introns in the prokaryotes, but have introns in the complex eukaryotes. In an introns-late model (Palmer and Logsdon 1991; Logsdon 1998), all of these introns must have been added to the original gene, so there can be no exon shuffling in their history, because the eukaryotic forms are colinear to the prokaryotic sequence. However, in an introns-early model (Gilbert 1987), some or all of these introns might be left over from the exon-shuffling events that created the gene before the separation of prokaryotes and eukaryotes. This picture of the modular substructure of domains being created by exon shuffling using phase-zero introns is not in contradiction to the use of phase-one introns later in evolution in the shuffling of domains (Patthy 1999; Kaessmann et al. 2002).
RESULTS AND DISCUSSION
Our large sample of 3328 phase-zero introns (Fedorov et al. 2001) from homologs of 276 ancient proteins is collected from diverse species, traditionally divided into five distant taxa: vertebrates, invertebrates, fungi, plants, and protists (following de Souza et al. 1996, 1998). We define four nested sets of these 3328 introns based on the coincidence of intron positions between these distant groups. Set 2 consists of 550 introns whose positions match in at least two of the five taxa. Set 3 consists of 118 introns whose positions match in at least three taxa. Lastly, set 4 consists of 29 positions at which introns are found in at least four taxa. Intron position coincidences among diverse taxa may be explained either by maintenance of an ancestral intron or by parallel insertion in multiple lineages (Palmer and Logsdon 1991). As more and more different taxa are found with a common intron position, the former possibility becomes more likely and the latter less so. Therefore, our sets should be more and more enriched in intron positions which existed at least at the early stages of eukaryotic evolution, and perhaps before.
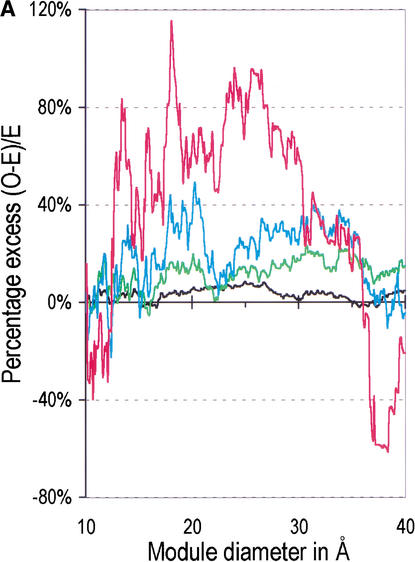
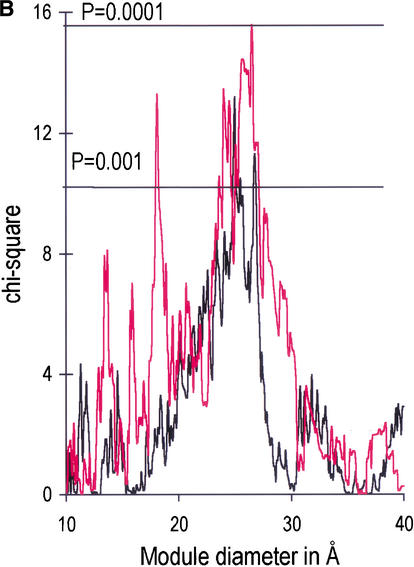
Figure 1A, 1B shows the correlations of these sets with the module boundary regions of the 276 ancient proteins. Introns have been shown to correlate with boundaries of modules defined for a large range of diameters, from 10–40 Å (de Souza et al. 1996, 1998; Fedorov et al. 2001), and we thus show the correlation for this entire range. As we saw before, for all of the phase-zero introns, there is an excess of introns within the boundary regions which reaches 10% above the random expectation. In addition, Figure 1A, 1B shows that each of the sets of matched introns shows a greater preference for the boundary regions. The set of 550 pairs shows an excess that reaches about 20%. The 118 triple matches show excesses up to 30%–40%. Finally, the most “ancient” set of 29 quadruple-matched phase-zero introns has an ∼80% excess in the module boundaries. Figure 1b shows the chi-square values against the random insertion model for these excesses. They are all statistically very significant in the 20–30 Å diameter range. (For clarity, the figure shows the χ2 curve for the set of 3328 and the set of 29. The other two curves are very similar.)
Figure 1A.
The correlation of intron positions with module boundaries for 276 ancient, nonrelated genes. Black line: Initial set of 3328 phase-zero introns. Green line: Set 2, comprising 550 phase-zero introns whose positions match in the genes of at least two of five taxa (vertebrates, invertebrates, fungi, plants, and protists). Blue line: Set 3, comprising 118 phase-zero introns whose positions match in at least three taxa. Red line: Set 4, comprising 29 phase-zero introns whose positions match in at least four taxa. (A) The percentage excess of intron positions over the random expectation in module boundaries ([O-E]/E * 100%) as a function of module diameter, where O is the observed number of intron positions in module boundaries, and E the expected number. The horizontal axis gives the module diameter. The range of module diameters from 10–40 Å corresponds approximately to polypeptide chains from 5–45 amino acid residues in length. (B) χ2 values for the excess of introns in module boundaries. Thresholds of P = 0.001 (for χ2 = 10.8) and P = 0.0001 (for χ2 = 15.1) are marked. Only the curves for sets 1 and 4 are given, for clarity; sets 2 and 3 are similar.
Are the differences between the patterns for the various subsets (sets 2–4) and the full set (set 1) significant? To determine this, we asked how often one would expect to draw a subset with an average excess equal to or larger than that for a given subset. For each subset, we generated 10,000 subsets (100,000 for set 4) of an equal number of intron positions as the real subset, and we calculated their average excess over the range 15–35 Å. As Table 1 shows, each subset exhibits a correlation with module boundaries significantly stronger than would be expected from such a random sample.
Table 1.
Statistical Significance of the Intron Excess in Module Boundaries
| Number of introns | Average excess (15-30 Å) | _p_-value | |
|---|---|---|---|
| Set 1 | 3328 | 4.3% | |
| Set 2 | 550 | 10.8% | 0.025 |
| Set 3 | 118 | 23.4% | 0.0044 |
| Set 4 | 29 | 68.0% | 0.00001 |
For the phase-one and phase-two introns, we did not find any significant correlation or excess of the corresponding matched sets with module boundary regions of ancient genes. Neither did we find any effects with the matched introns of different phases with module boundaries of eukaryote-specific genes (see our Web page, www.mcb.harvard.edu/gilbert/intron_subsets).
This greater and greater excess in module boundary regions is exactly what one expects if the excess is due to a population of ancient introns that define module boundaries, as would be the case if these genes had been assembled by exon shuffling between modules using phase-zero introns. These findings are not explicable within the framework of an introns-late theory, which claims that all introns are inserted into genes at the latest stages of evolution. If one were to argue that perhaps there is a weak selective advantage to introns, once inserted, being maintained at module boundaries, so that old introns would be more likely to lie at module boundaries, one must provide reasons for that not being the case for phase-one and phase-two introns. It is far more likely that phase-zero introns were used before the branching of the eukaryotes to create proteins by exon shuffling. Since the proteins we examine are shared by both eukaryotes and prokaryotes, their homology and colinearity suggest that they were created by exon shuffling at the early stages of evolution, predating the major divergences of eukaryotes and prokaryotes. This hypothesis was recently supported by Kaessmann et al. (2002), who showed that phase-zero introns occur preferentially at the borders of protein domains (Pham classification) of ancient proteins.
METHODS
We analyze intron patterns with a computer program, INTERMODULE, that divides each three-dimensional structure into modules of a given diameter and defines boundary regions between the module cores. The program then calculates the excess of introns in the boundary regions, compared to a random expectation, for the entire set of introns and proteins (de Souza et al. 1998). Our set of 276 ancient proteins and the sample of 3328 phase-zero introns mapped onto these proteins was described previously (Fedorov et al. 2001).
We used Monte Carlo methods to determine whether our sets 2–4 are significantly more correlated with module boundaries than is the phase-zero set as a whole. For each set of phylogenetically conserved positions, we generated 10,000 random subsets of the full set of 3328 phase-zero positions, each subset containing the same number of positions as the real set (550, 118, and 29 intron positions for sets 2, 3, and 4, respectively). Then we studied the correlation of the introns in these random subsets with module boundaries. For each real subset (2–4), the fraction of the corresponding 10,000 random subsets which have correlations as strong as the real subset is the probability of seeing the observed correlation at random.
Acknowledgments
All calculations were performed with computer programs written in PERL and C. A full set of our results is available on our Web page: www.mcb.harvard.edu/gilbert/intron_subsets.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1008203\. Article published online before print in May 2003.
References
- de Souza, S.J., Long, M., Schoenbach, L., Roy, S.W., and Gilbert, W. 1996. Intron positions correlate with module boundaries in ancient proteins. Proc. Natl. Acad. Sci. 93**:** 14632–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, S.J., Long, M., Klein, R.J., Roy, S., Lin, S., and Gilbert, W. 1998. Toward a resolution of the introns early/late debate: Only phase zero introns are correlated with the structure of ancient proteins. Proc. Natl. Acad. Sci. 95**:** 5094–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, A., Cao, X., Saxonov, S., de Souza, S., Roy, S.W., and Gilbert, W. 2001. Intron distribution difference for 276 ancient and 131 modern genes suggests the existence of ancient introns. Proc. Natl. Acad. Sci. 98**:** 13177–13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W. 1987. The exon theory of genes. Cold Spring Harbor Symp. Quant. Biol. 52**:** 901–905. [DOI] [PubMed] [Google Scholar]
- Go, M. 1981. Correlation of DNA exonic regions with protein structural units in haemoglobin. Nature 291**:** 90–92. [DOI] [PubMed] [Google Scholar]
- Kaessmann, H., Zollner, S., Nekrutenko, A., and Li, W.-H. 2002. Signatures of domain shuffling in the human genome. Genome Res. 12**:** 1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon, J.M. 1998. The recent origin of spliceosomal introns revised. Curr. Opin. Genet. Dev. 8**:** 637–648. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D. and Logsdon, J.M. 1991. The recent origin of introns. Curr. Opin. Genet. Dev. 1**:** 470–477. [DOI] [PubMed] [Google Scholar]
- Panchenko, A.R., Luthey-Schulten, Z., and Wolynes, P.G. 1996. Foldons, protein structural modules, and exons. Proc. Natl. Acad. Sci. 93**:** 2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthy, L. 1999. Genome evolution and the evolution of exon-shuffling—A review. Gene 238**:** 103–114. [DOI] [PubMed] [Google Scholar]