The Signature of Selection Mediated by Expression on Human Genes (original) (raw)
Abstract
As the efficacy of natural selection is expected to be a function of population size, in humans it is usually presumed that selection is a weak force and hence that gene characteristics are mostly determined by stochastic forces. In contrast, in species with large population sizes, selection is expected to be a much more effective force. Evidence for this has come from examining how genic parameters vary with expression level, which appears to determine many of a gene's features, such as codon bias, amino acid composition, and size. However, not until now has it been possible to examine whether human genes show the signature of selection mediated by expression level. Here, then, to investigate this issue, we gathered expression data for >10,000 human genes from public data sets obtained by different technologies (SAGE and high-density oligonucleotide chip arrays) and compared them with gene parameters. We find that, even after controlling for regional effects, highly expressed genes code for smaller proteins, have less intronic DNA, and higher codon and amino acid biases. We conclude that, contrary to the usual supposition, human genes show signatures consistent with selection mediated by expression level.
It is usually assumed that in humans, gene characteristics such as gene length and amino acid composition are mostly determined by stochastic processes (Eyre-Walker 1991; Sharp et al. 1995; Smith and Hurst 1999). The only sources of significant selective pressure would be those related to protein function optimization. Because protein synthesis has an associated cost to the cell, selection should favor changes in gene sequences that make protein synthesis more efficient or reduce its costs. The strength of selection related to protein synthesis efficiency should be higher for those genes transcribed in large quantities. Observations from several unicellular and invertebrate species have shown that expression profiles of genes covary with a variety of sequence parameters (Akashi 2001) such as gene length (Coghlan and Wolfe 2000; Jansen and Gerstein 2000), codon usage bias (Gouy and Gautier 1982; Sharp et al. 1986; Duret and Mouchiroud 1999; Coghlan and Wolfe 2000), and amino acid composition (Jansen and Gerstein 2000; Akashi and Gojobori 2002). These patterns have been interpreted as evidence of selection acting to increase protein synthesis efficiency and to reduce associated costs.
In human and other mammalian species, it has been suggested that gene sequences should not show the effects of natural selection to increase protein synthesis efficiency because of their small population sizes. Therefore, no relationship between expression and gene characters is expected (Eyre-Walker 1991; Sharp et al. 1995; Smith and Hurst 1999). Some evidence of selection acting on codon usage in mammalian genes has been reported in the past, but these studies are based on samples of limited size and/or do not test directly whether codon usage is related to activity levels of genes (Eyre-Walker 1991; Debry and Marzluff 1994; Iida and Akashi 2000). Recently, it was shown (Castillo-Davis et al. 2002) that expression patterns are related to intron sizes in human genes. However, this study does not take into account the possible influence of regional mutational biases influencing the local level of insertions and deletions. In addition, some reservations should be taken when using data derived from EST libraries used in this study to estimate levels of activity. Here, using two independent data sets of gene expression and correcting for regional effects, we provide a systematic analysis to clarify whether human genes show signatures consistent with expression-mediated selection.
If selection is acting on gene sequences, then we expect them to be modified to maximize expression efficiency. This effect should be particularly pronounced for highly expressed genes. To address this issue, we compared estimates of expression against gene characteristics. For this purpose, we assembled expression data from publicly available SAGE libraries from NCBI collected at different laboratories and representing 22 different tissues (see Methods). Serial Analysis of Gene Expression technology (SAGE) allows the measurement of expression profiles for large numbers of genes in a relatively unbiased way by avoiding gene-specific mRNA screening (Velculescu et al. 1995). In addition, we also used the comprehensive analysis of gene expression data using high-density oligonucleotide array technology recently released and representing 29 different tissues (Su et al. 2002; see Methods). Because in this data set all tissues were tested for the same genes, there is no sampling bias caused by the screening for different sets of genes in different tissues.
RESULTS
Transcription-Translation Efficiency and Gene Expression
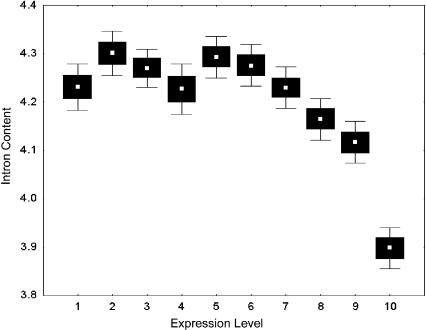
Does selection act on coding sequences of genes to maximize translation and/or transcription efficiency and gene position? If so, then we may expect highly expressed genes to produce shorter proteins to reduce translation costs. This is what we find: Genes of higher expression produce only short proteins, and we find a significant negative correlation between protein length and mean level of expression (R = 0.182, p < 0.0001; N = 8212; see Table 1). Similarly, if transcription is costly, we might expect selection to act on intron length (Hurst et al. 1996). We found that, indeed, highly expressed genes have reduced total intron content (R = 0.181, p < 0.0001; N = 7967; Fig. 1). We found that intron and protein length are correlated. To examine whether both exon and intron lengths are independently related to expression levels, we performed multiple regression analyses correcting for intron and protein length, respectively. Expression levels are significantly related to intron and protein lengths after correction (β = -0.221, p < 0.0001; β = -0.100, p < 0.0001).
Table 1.
Results From Multiple-Regression Analyses of Level of Gene Expression and Length With Gene Parameters When Including GC3 Content
| Dependent variable | Database | Pearson correlation with level of expression (p < 0.0001) | Effect of expression when controlling for regional effects (p < 0.0001)a |
|---|---|---|---|
| Protein length b | SAGE | R = 0.182 | β = −0.175 |
| Chip Array | R = 0.194 | β = −0.200 | |
| Intron length b | SAGE | R = 0.181 | β = −0.403 |
| Chip Array | R = 0.198 | β = −0.369 | |
| Codon bias (MCB) b | SAGE | R = 0.122 | β = 0.019 |
| Chip Array | R = 0.180 | β = 0.032 | |
| AA complexity b | SAGE | R = 0.062 | β = −0.006 |
| Chip Array | R = 0.045 | NS |
Figure 1.
Intron content and expression level in human genes. Genes were split into 10 groups of an equal number of cases according to expression level. White dots represent the mean expression value for each group. Black boxes and error bars show the standard error with 68% and 95% of confidence.
The compact nature of highly expressed genes is then consistent with the activity of selection. If selection has acted to maximize the efficiency of translation (as suggested by the correlation with protein size), we might also expect patterns of gene expression to be related to codon bias, as they are in several unicellular and invertebrate species (Gouy and Gautier 1982; Sharp et al. 1986; Duret and Mouchiroud 1999). In these species, certain tRNAs are more abundant than others, and selection favors, in highly expressed genes, the codons that match the most abundant isoacceptor (Sprinzl et al. 1996) or the most accurate one (Dix and Thompson 1989; Akashi 1994), thus resulting in a correlation between codon bias and expression level.
In mammals, evidence of codon usage bias and its possible relation to expression profiles has remained scarce. In these species, there is a great degree of heterogeneity in base composition along the genome (Bernardi 1995), and codon usage bias in mammalian genes has been interpreted as the result of regional base composition variations (Eyre-Walker 1991; Sharp et al. 1995; Smith and Hurst 1999). Nevertheless, some previous studies indicate that codon bias might be related to expression profiles. Two studies, one using histone genes, which are highly expressed (Debry and Marzluff 1994), and a second comparing codon preferences of alternatively spliced and constitutive exons (Iida and Akashi 2000), conclude that codon choice in highly expressed genes/constitutive exons deviates from the expected distribution, from flanking regions/alternatively spliced exons, respectively. Further support for a possible relationship between codon usage and expression levels of genes comes from studies in which the expression of nonmammalian genes in mammalian cells has been dramatically increased by the replacement of rare codons in the mammalian genome with common ones. This method of “codon optimization” has been used to increase expression of several genes (Levy et al. 1996; Wells et al. 1999; Zhou et al. 1999). All of these studies used a very limited sample size, and therefore their findings cannot be generalized to all mammalian genes.
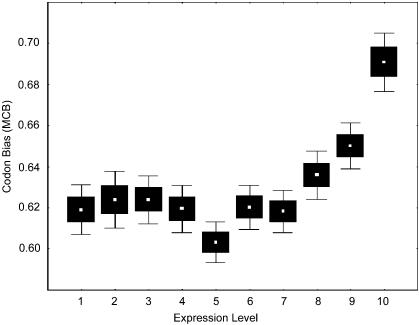
Are the above isolated cases or is there is a broad relationship between expression and codon choice? In a previous study using a sample size of >2000 human genes, we showed that for most of the genes, codon usage bias is significantly higher than expected from background nucleotide composition (Urrutia and Hurst 2001). We now examine whether this residual bias is related to expression levels. To test this, we compared expression levels with codon bias in our gene data set. We measured codon bias using the methods of KM (Karlin and Mrazek 1996) and MCB (Urrutia and Hurst 2001; see Methods). Unlike more conventional measures (e.g., ENC), these two methods attempt to correct for background nucleotide variation. MCB has the advantage over KM of being less biased by amino acid composition. When correcting for nucleotide bias, we found that codon bias is correlated with level of expression (for KM, R = 0.130, p < 0.0001; for MCB, R = 0.122, p < 0.0001; N = 6071; Fig. 2). In a previous study (Urrutia and Hurst 2001), we showed that the MCB method is biased by protein length. Therefore, we assessed the correlation of expression level and codon bias after correction by length of protein. The correlation of MCB index with expression level remains significant after correcting for length of protein (β = 0.048, p < 0.0001).
Figure 2.
Codon bias (MCB) and level of expression. Codon usage bias MCB after correcting for background nucleotide content. Genes were split into 10 groups of an equal number of cases according to expression level. White dots represent the mean expression value for each group. Black boxes and error bars show the standard error with 68% and 95% of confidence.
Protein Synthesis and Expression Rates
Because of differences in the costs and biochemical properties associated with amino acid biosynthesis and/or with acquisition through the diet, we might expect genes expressed in large quantities to have a biased amino acid usage from that expected by their base composition. Evidence for a relation between expression levels and amino acid biases has been reported for yeast and bacteria (Jansen and Gerstein 2000; Akashi and Gojobori 2002). We examined the amino acid composition of genes and its relation to expression patterns. We observed a significant relation between amino acid usage and expression level for 16 out of 20 amino acids (after Bonferroni correction; see Table 2). However, because amino acid composition is also affected by background GC content (Singer and Hickey 2000), we corrected for the effect of GC3 content. All relationships remained significant even after correcting for gene length and GC3 content (after Bonferroni correction; see Table 2). It may be expected that the bias in the use of amino acids that we found would correspond to the avoidance of expensive to produce or scarce amino acids. Dufton (1997) developed an index of amino acid size/complexity based on the molecular weight and the shape of amino acids. We used this index as an indirect estimate of amino acid cost and examined its relationship to expression level. We find that, indeed, there is a tendency to avoid the use of complex amino acids in highly expressed genes (R = 0.062, p < 0.0001; N = 6223; Table 1). More accurate estimates of the true cost of amino acid synthesis/acquisition for mammals would allow us to resolve the extent of the relationship between expression and amino acid choice.
Table 2.
Amino Acid Usage and Expression Level (SAGE), Multiple Regression Analysis
| Amino acid | One letter code | Pearson correlation with expression (p < 0.0001) | Effect of expression when controlling for GC3 content and gene length (p < 0.0001) |
|---|---|---|---|
| Alanine | A | 0.100 | β = 0.336 |
| Arginine | R | NS | — |
| Aspargine | N | NS | — |
| Aspartic acid | D | 0.100 | β = 0.562 |
| Cysteine | C | −0.077 | β = −0.458 |
| Glutamine | Q | −0.071 | β = −0.256 |
| Glutamic acid | E | 0.055 | β 0.561 |
| Glycine | G | 0.084 | β 0.418 |
| Histidine | H | −0.118 | β = −0.349 |
| Isoleucine | I | NS | — |
| Leucine | L | −0.105 | β = −0.888 |
| Lysine | K | 0.126 | β = 1.049 |
| Methionine | M | 0.071 | β = −0.142 |
| Phenylalanine | F | −0.032 | β = −0.155 |
| Proline | P | −0.045 | β = −0.401 |
| Serine | S | −0.145 | β = 0.785 |
| Threonine | T | 0.045 | β = −0.086 |
| Tryptophan | W | −0.071 | β = −0.211 |
| Tyrosine | Y | NS | — |
| Valine | V | 0.055 | β = 0.227 |
Gene Position and Density Are Related to Expression Level
It has been previously reported that highly expressed genes tend to cluster in the human genome (Caron et al. 2001; Lercher et al. 2002). We confirm this: When we compared expression patterns of pairs of adjacent genes, we found significant similarity in expression level (R = 0.09, p < 0.001; N = 4376; see Methods). Note that all pairs of duplicated genes were removed (see Methods). We found that intergene spacers tend to be shorter for highly expressed genes (R = 0.029, p < 0.0001; N = 8076). This may possibly reflect an adaptation for more efficient gene transcription, but might alternatively reflect some regional mutational bias that tends to compact sequences in these regions, or differences in recombination rates (Hey and Kliman 2002). In addition, intron and protein lengths are correlated to intergenic distances (data not shown). Therefore, it is necessary to ask whether, controlling for regional effects, there remains a significant relationship between both intronic and protein sizes and expression level. On a multiple regression test, expression level is a relevant predictor of both protein and intron lengths after correcting them for intergenic length (see Table 1).
We have recently reported that highly expressed genes tend to be in GC-rich regions of the genome (Lercher et al. 2002); consistent with this, we found that highly expressed genes tend to have a higher GC content in the adjacent intergenic regions (R = 0.065, p < 0.0001; N = 6430; see Methods). Expression data derived from SAGE technologies could overestimate expression measures for GC-rich genes (Margulies et al. 2001). However, we find a similar pattern with chip-array-technology-derived data, for which no systematic errors have been reported, indicating that the relationship between expression level and GC content is not an artifact. We assessed, nevertheless, the relationship between expression level and protein and intron lengths, controlling for GC effects. The results of multiple regression analysis, however, show that level of expression is a relevant parameter for the above-discussed gene characteristics after correcting for GC content (see Table 1). Similar results were obtained correcting for GC3s (data not shown).
The control for GC content, in addition, in part controls for ancestral recombination rates (Marais 2003). But we also corrected our results using present estimates of recombination rates (Kong et al. 2002). There is a weak tendency for highly expressed genes to be situated in regions of higher recombination (r = 0.013, p < 0.0001, N = 7987). Expression level remains a relevant predictor of gene parameters after incorporation of recombination rate in the multiple regression analysis (see Table 1). However, as noted (Marais 2003), the present measures are both noisy and may well have little correlation to ancestral recombination rates; hence, interpretation of the above results from the best direct estimates must be limited.
DISCUSSION
Here we have evaluated the interaction between expression level of human genes and gene sequence characteristics. In sum, we find that highly expressed genes code for small proteins, have little intronic content, high codon bias, and tend to encode cheaper amino acids. These signatures found in human genes are consistent with the action of selective pressures to maximize protein synthesis efficiency in highly expressed genes.
In addition, we confirmed previous results on gene sorting by expression patterns and the relationship between expression patterns and GC content. We performed multiple regression analysis to rule out the possibility that these regional characters could potentially explain the relationship between expression patterns and gene characteristics. The relationship between expression level and intron and protein size can only in part be accounted for by regional compaction effects. Biases in codon and amino acid usage are not accounted for by GC bias or gene size. The relationship between expression rates and amino acid composition could be partly due to functional properties of the proteins associated with different expression levels.
Although the effects presented here are weak, it should be noted that similar results were obtained with two independent databases of gene expression obtained with different methodologies. In addition, in doing this work we have assumed a conservative approach when correcting all results for intergene spacers and GC content corrections not done in previous analysis (Castillo-Davis et al. 2002). The compaction of intergenic regions of highly expressed genes, however, need not reflect a mutation bias, but selective forces directly or indirectly related to expression patterns. In addition, codon bias has been estimated taking out any compositional biases, but these themselves could be partly driven by selection. Moreover, the correspondence between libraries representing the same tissue obtained with the same method is usually high (r > 0.80), whereas the correspondence of data obtained with different methods is low (r < 0.60; data not shown). These discrepancies are likely to add noise to our analyses and possibly derive from errors in the correspondence between oligonucleotides and/or tags and the gene represented. This should not affect our conclusions.
The results presented on gene length and expression patterns are consistent with those obtained in other multicellular eukaryotes but differ from observations in unicellular eukaryotes and bacteria, in which intron (Vinogradov 2001) and protein sizes (Moriyama and Powell 1998) are positively correlated to expression estimates. The patterns in unicellular organisms might be caused by increased expression gained by the inclusion of functional elements important for transcription regulation or splicing efficiency. The reversal of this along the evolutionary scale could be explained by the increased gene and genome size where most intergenic and intron sequences do not possess a function in gene regulation.
Where do our results leave the usual supposition that human population sizes are too small for selection to affect the properties that we have analyzed? Our results are probably largely in agreement with this general position. It is most notable that many of the results that we describe are not strong effects and in many cases appear to affect only the most highly expressed set of genes. We can imagine two reasons for this. First, only in this subset of genes is selection strong enough to have an appreciable effect. Classical theory postulates that for deleterious mutations not to be deterministically eliminated by selection, the selective coefficient, s, must be less than 1/2_N_e. Should these mutations not be eliminated, they would lead to genes tending to move away from optimal structure and codon usage. In the human genome, there may well be more mutations that are effectively neutral than in flies (humans having a smaller _N_e), there nonetheless remains a respectable number of genes (the most highly expressed) in which s » 1/2_N_e for many mutations affecting level of expression. Second, and not mutually exclusively, we may be witnessing, in some part, the decay of selected features. As many of the features concerned may take time to reach equilibrium, we would expect that the most highly expressed genes would still retain many of the features of the prior action of selection. Analysis of the population genetics of insertion mutations in introns in highly expressed genes would then be interesting as the former model predicts that they may still be under counter selection, whereas the latter predicts that they may instead be effectively neutral.
METHODS
Sequence Information
Sequence information was obtained for genes from human genome annotations from build 30 of the NCBI site (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/). Although 26,297 genes were considered for the analysis, data for each parameter were not obtained for all of the genes; therefore, the actual number of genes used in comparisons varied as indicated in the text. Nucleotide sequences were retrieved from the Fa file from the NCBI site (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/). Nucleotide composition was determined, and KM (Karlin and Mrazek 1996) and MCB values of codon bias were obtained after nucleotide corrections were obtained according to methods stated elsewhere (Urrutia and Hurst 2001). Nucleotide expectations for codon usage were based on the coding sequence of each gene and obtained according to Urrutia and Hurst (2001). This is preferable to using noncoding regions as these typically contain repetitive elements, regulatory sequences, and even RNA genes that would bias base composition. In all cases in which more than one alternative transcript was available, the largest was analyzed. Incomplete sequences were removed from analysis.
Intron-Exon Boundaries
Intron-exon boundaries, intergenic length, and the identity of neighboring genes were established from contig annotations from the human genome sequence (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/). All overlapping genes were removed. Because contigs are not always continuous, adjacent genes were not determined for genes that were either the first or last genes within their contig.
Intergenic GC Content
The intergenic GC content was obtained from masked chromosome assembly in fasta format of build 30 at the NCBI site (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/). A section adjacent to each side of each gene of a minimum 500 bp and a maximum of 5000 bp was used to estimate intergenic GC content. The GC content was not calculated for all overlapping genes. The analyses presented here refer to global GC content percentage, but similar results were obtained with the GC content of nonrepetitive sequences only or GC3s from coding regions of genes.
Duplicated Genes
Duplicated genes were removed from the analysis of adjacent genes. All genes were blasted against the two adjacent genes using the BLASTN downloadable version from the NCBI site (http://www.ncbi.nlm.nih.gov/BLAST/). All pairs of adjacent genes with an expected value of sequence similarity <0.01 were removed from analysis. The correlation coefficient was obtained from the comparison of rates of expression of adjacent genes, in which the order of the genes of each pair was randomly assigned. The correlation coefficient shown for expression rates of adjacent genes refers to the mean value of 100 of such correlations.
Recombination Data
Recombination data were obtained from Kong et al. (2002). The recombination rate indexes for each gene were derived from composing the recombination rates of the nearest marker at each side. The relative weight for each adjacent marker was determined by the distance separating the gene from the marker at each side.
Expression Data
SAGE expression data were collected from the NCBI site (http://www.ncbi.nlm.nih.gov/SAGE/). Only tags that matched to a single gene were taken into account. In addition, because tags were matched against reported sequences in GenBank and only a small percentage of these sequences contain a poly(A), tags containing poly(A) tails would only be matched against a small subset of the sequences. Therefore, all tags that ended in a stop codon followed by more than five As were discarded. All genes for which only one tag was detected in all libraries were also eliminated from the analysis as they potentially represent a sequencing error. Only libraries from normal tissues (noncancerous) were used in the study (43). Transcript counts for libraries corresponding to the same tissue were joined, and tags per million were then calculated for each gene. The data on 8220 genes for 22 tissues were taken into account: brain, cerebellum, spinal cord, skin, vascular, T-cells, lymphocyte, muscle, retina, cornea, mammary glands, heart, lung, kidney, stomach, liver, pancreas, colon, peritoneum, uterus, ovary, and prostate. Those corresponding to the same tissue were averaged before obtaining a global measure of expression level.
High-density oligonucleotide array data was collected from the gene expression atlas site (http://expression.gnf.org). For any gene to be counted as expressed in a given tissue, a cutoff value on the expression index of 20 was defined. The data for 101 samples were available, corresponding to 28 noncancerous tissues: cerebellum, brain, cerebral cortex, caudate nucleus, amygdale, thalamus, corpus callosum, spinal cord, whole blood, testis, pancreas, placenta, pituitary gland, thyroid, prostate, ovary, uterus, dorsal root ganglia, salivary gland, trachea, lung, thymus, spleen, adrenal gland, kidney, liver, heart, umbilical vein, and endothelial cells.
From SAGE and chip array data, we could define two measures of level of expression: Peak expression, which is the highest value of expression of a gene in any tissue, and mean level of expression, the mean quantity of expression of a gene in all tissues in which it is expressed (if divided among all tissues, then this measure would be dependent on breadth of expression). As these two measures proved to be highly correlated (R = 0.99; data not shown), only mean expression is referred to here as level of expression. However, the results presented also apply to peak of expression of genes (data not shown).
The figures presented here refer to the analysis of SAGE data; similar results were also obtained when using chip-array data unless otherwise indicated in text and tables. Similar data are obtained when only genes not known to undergo alternative splicing are taken into account (data not shown). Indexes of expression level and lengths of coding and noncoding regions were log-transformed prior to analyses.
Acknowledgments
We thank Brian Charlesworth, Laurent Duret, Hiroshi Akashi, and four anonymous referees for their helpful comments. We thank the BBSRC (L.D.H.) and the CONACyT and ORS (A.O.U.) for funding.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.641103\. Article published online before print in September 2003.
References
- Akashi, H. 1994. Synonymous codon usage in _Drosophila melanogaster_—Natural selection and translational accuracy. Genetics 136**:** 927-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Gene expression and molecular evolution. Curr. Opin. Gene Dev. 11**:** 660-666. [DOI] [PubMed] [Google Scholar]
- Akashi, H. and Gojobori, T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. 99**:** 3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, G. 1995. The human genome: Organization and evolutionary history. Ann. Rev. Genet. 29**:** 445-476. [DOI] [PubMed] [Google Scholar]
- Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.C., van Asperen, R., Boon, K., Voute, P.A., et al. 2001. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science 291**:** 1289-1292. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis, C.I., Mekhedov, S.L., Hartl, D.L., Koonin, E.V., and Kondrashov, F.A. 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31**:** 415-418. [DOI] [PubMed] [Google Scholar]
- Coghlan, A. and Wolfe, K.H. 2000. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16**:** 1131-1145. [DOI] [PubMed] [Google Scholar]
- Debry, R.W. and Marzluff, W.F. 1994. Selection on silent sites in the rodent H3 histone gene family. Genetics 138**:** 191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, D.B. and Thompson, R.C. 1989. Codon choice and gene-expression—Synonymous codons differ in translational accuracy. Proc. Natl. Acad. Sci. 86**:** 6888-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufton, M.J. 1997. Genetic code synonym quotas and amino acid complexity: Cutting the cost of proteins? J. Theor. Biol. 187**:** 165-173. [DOI] [PubMed] [Google Scholar]
- Duret, L. and Mouchiroud, D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, Arabidopsis. Proc. Natl. Acad. Sci. 96**:** 4482-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A. 1991. An analysis of codon usage in mammals: Selection or mutation bias? J. Mol. Evol. 33**:** 442-449. [DOI] [PubMed] [Google Scholar]
- Gouy, M. and Gautier, C. 1982. Codon usage in bacteria—Correlation with gene expressivity. Nucleic Acids Res. 10**:** 7055-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J. and Kliman, R.M. 2002. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics 160**:** 595-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L.D., McVean, G.T., and Moore, T. 1996. Imprinted genes have few and small introns. Nat. Genet. 12**:** 234-237. [DOI] [PubMed] [Google Scholar]
- Iida, K. and Akashi, H. 2000. A test of translational selection at `silent' sites in the human genome: Base composition comparisons in alternatively spliced genes. Gene 261**:** 93-105. [DOI] [PubMed] [Google Scholar]
- Jansen, R. and Gerstein, M. 2000. Analysis of the yeast transcriptome with structural and functional categories: Characterizing highly expressed proteins. Nucleic Acids Res. 28**:** 1481-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin, S. and Mrazek, J. 1996. What drives codon choices in human genes? J. Mol. Biol. 262**:** 459-472. [DOI] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31**:** 241-247. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31**:** 180-183. [DOI] [PubMed] [Google Scholar]
- Levy, J.P., Muldoon, R.R., Zolotukhin, S., and Link, C.J. 1996. Retroviral transfer and expression of a humanized, red-shifted green fluorescent protein gene into human tumor cells. Nat. Biotech. 14**:** 610-614. [DOI] [PubMed] [Google Scholar]
- Marais, G. 2003. Biased gene conversion: Implications for genome and sex evolution. Trends Genet. 19**:** 330-338. [DOI] [PubMed] [Google Scholar]
- Margulies, E., Kardia, S., and Innis, J. 2001. Identification and prevention of a GC content bias in SAGE libraries. Nucleic Acids Res. 29**:** e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E.N. and Powell, J.R. 1998. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucleic Acids Res. 26**:** 3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M., Tuohy, T.M.F., and Mosurski, K.R. 1986. Codon usage in yeast—Cluster-analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14**:** 5125-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M., Averof, M., Lloyd, A.T., Matassi, G., and Peden, J.F. 1995. DNA-sequence evolution—The sounds of silence. Phil. Trans. R Soc. Lond. B 349**:** 241-247. [DOI] [PubMed] [Google Scholar]
- Singer, G.A.C. and Hickey, D.A. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17**:** 1581-1588. [DOI] [PubMed] [Google Scholar]
- Smith, N.G.C. and Hurst, L.D. 1999. The effect of tandem substitutions on the correlation between synonymous and nonsynonymous rates in rodents. Genetics 153**:** 1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Steegborn, C., Hubel, F., and Steinberg, S. 1996. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 24**:** 68-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99**:** 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia, A.O. and Hurst, L.D. 2001. Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics 159**:** 1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. 1995. Serial analysis of gene-expression. Science 270**:** 484-487. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A.E. 2001. Intron length and codon usage. J. Mol. Evol. 52**:** 2-5. [DOI] [PubMed] [Google Scholar]
- Wells, K.D., Foster, J.A., Moore, K., Pursel, V.G., and Wall, R.J. 1999. Codon optimization, genetic insulation, and an rtTA reporter improve performance of the tetracycline switch. Transg. Res. 8**:** 371-381. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Liu, W.J., Peng, S.W., Sun, X.Y., and Frazer, I. 1999. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 73**:** 4972-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/; Chromosome annotations, human genome, NCBI.
- ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/; RNA file, human genome, NCBI.
- http://expression.gnf.org; Expression Atlas.
- http://www.ncbi.nlm.nih.gov/BLAST/; BLAST tools, NCBI.
- http://www.ncbi.nlm.nih.gov/SAGE/; SAGE, NCBI.